Abstract
Many aspects of Epstein-Barr virus (EBV) and tumor biology have been studied in Burkitt's lymphoma (BL)-derived cell lines. However, in tissue culture, patterns of gene expression and C promoter-G (CpG) methylation often change and viral strain selection may occur. In this report, 10 cases of snap-frozen endemic BL tumors are characterized in terms of viral gene expression, promoter usage, methylation, and viral strain. EBNA1 and BamHI-A rightward transcripts (BART) were detected in 7 of 7 and LMP2A transcripts in 5 of 7 tumors with well-preserved RNA. Transcripts for the other EBNAs and for LMP1 were not detected in any tumor. These tumors differ from BL cell lines in that they lack a variety of lytic cycle transcripts. This pattern of viral gene expression in endemic BL is similar to that reported in peripheral blood mononuclear cells (PBMCs) from healthy EBV–seropositive individuals. EBNA1 transcripts originated from the Q promoter (Qp) but not C, W, or F promoters that drive transcription of EBNA1 in other circumstances. Whereas Cp has been previously shown to be entirely CpG methylated in BL, bisulfite genomic sequencing showed virtually no methylation in Qp. Type-A EBV was detected in 6 of 10 and type B in 4 of 10 cases. A previously reported 30bp deletion variant in the carboxyl terminal of LMP1 gene was detected in 5 of 10 cases. The association with both A and B strains contrasts with EBV–associated Hodgkin's disease, nasopharyngeal carcinoma, and post-transplant lymphoproliferative disease, which are much more consistently associated with A strain virus.
DENNIS BURKITT CALLED attention to the rapidly growing tumor in African children, that now bears his name, in 1958.1 The endemic form of Burkitt's lymphoma (eBL) that he described affects children in malarial areas of equatorial Africa and is consistently associated with Epstein-Barr virus (EBV).2-4 Elsewhere in the world the association with EBV is less consistent ranging from 20% to 80%.4-9
Studies of Burkitt's-derived cell lines have led to a variety of important advances, among them the identification of EBV itself, the initial characterization of the viral requirements for B-cell immortalization, and mapping of the c-myc locus to chromosome 8 (reviewed by I. Magrath1). EBV transcription and replication have been studied extensively in these cell lines. In fact, the three different types of EBV latent gene expression were also recognized first in Burkitt's cell lines.10-12 However, these studies have also underscored the inherent limitations of cell line research. Introduction of tumor cells into culture initiates a series of phenotypic and genetic changes. In early passage BL cell lines EBNA1 is the only nuclear antigen transcribed, whereas in late passage BL cell lines, the whole family of EBNAs is transcribed. The promoter driving EBNA1 expression is also different in some early and late passage cell lines. In early passage cell lines the Q promoter (Qp) is used, whereas in late passage cell lines the C promoter (Cp) can be used.13 In some circumstances the F promoter (Fp) or W promoter (Wp) may be used. Promoter switching can happen quite rapidly in culture. EBNA expression in newly infected B cells is initially driven by the Wp but within hours changes to Cp.14 Similarly, patterns of methylation change in culture. Thus, the study of cell lines, even early passage cell lines, cannot provide definitive information with regard to promoter usage or methylation status in vivo.
Whereas patterns of viral transcription have been studied directly in Hodgkin's disease (HD), nasopharyngeal carcinoma (NPC), peripheral T-cell lymphoma, nasal natural killer-(NK) and T-cell lymphoma, and other tumors,15-21 patterns of expression in Burkitt's tumors have only been studied in cell lines or by immunohistochemistry.3,6,22 Similarly, whereas studies of viral strain in NPC, HD, and other tumors have been carried out directly on tumor tissue, surveys of strain association with BL have been mainly carried out on cultured cell lines. The hazards of inference about strain on the basis of cultured cell lines is illustrated by studies of lymphocyte immortalization that show that A-strain virus is much more efficient at immortalization than B-strain virus, and thus will be more readily detected in lymphocyte immortalization assays.23 To characterize various aspects of EBV infection in eBL, we have studied DNA and RNA extracted directly from a collection of snap-frozen BL specimens from Ghana, Africa.
MATERIALS AND METHODS
Tumor samples and cell lines.
Ten BL tumor specimens were from the National Cancer Institute's BL tumor project at the University of Ghana at Accra, Ghana. BL1-5 have been described in an earlier report.24 NPC biopsy specimens from Taiwanese patients and post-transplant lymphoproliferative disease (PTLD) from patients at Johns Hopkins (Baltimore, MD) served as controls for EBV gene expression. The EBV–negative B-cell line, BJAB, was used as negative control for reverse-transcription polymerase chain reaction (RT-PCR). All cell lines were cultivated at 37°C in RPMI 1640 supplemented with 10% fetal bovine serum, 1 mmol/L glutamine, 100 U/mL penicillin and streptomycin. 5-azacytidine treatment for the Rael cell line (Rael-AzaC) was performed as previously described.25
RT-PCR for EBV transcripts.
The sequences of RT-PCR primers and internal probes are listed in Table 1. Total RNA was extracted from frozen BL tumors and cell line pellets by using TriZol (GIBCO BRL, Gaithersburg, MD). RT-PCR was performed by using random hexamers and the GeneAmp RNA PCR kit (Perkin Elmer-Cetus, Norwalk, CT). The PCR reaction involved an initial denaturation at 95°C for 3 minutes, followed by 40 cycles consisting of 94°C for 30 seconds, optimal annealing temperature for 1 minute, 72°C for 1 minute, and a final extension at 72°C for 10 minutes. The RT-PCR product was electrophoresed on a 1.8% agarose gel and alkali transferred with 0.4 mol/L NaOH onto HyBond N(+) membrane. The membrane was hybridized using 32P-labeled internal oligonucleotide probe and the Rapid-Hyb buffer system (Amersham, Arlington Heights, IL) at 52°C for 2 hours. The membrane was then washed and autoradiographed. The strength of the RT-PCR signal after Southern hybridization was graded as: ++ (strong signal after 1 hour exposure), + (strong signal after overnight exposure), and +/−(weak signal after overnight exposure).
Oligonucleotides Used in RT-PCR for EBV Transcripts
| Transcripts . | Oligos . | Oligonucleotide Sequence . | Coordinate in B95-8 . | Annealing Temperature . | Reference . |
|---|---|---|---|---|---|
| BART | A3 | 5′-AGAGACCAGGCTGCTAAACA | 157154-157173 | 55°C | 16 |
| A4 | 5-AACCAGCTTTCCTTTCCGAG | 159194-159175 | |||
| Probe | 5′-AAGACGTTGGAGGCACGCTG | 157359-157378 | |||
| EBNA1 | F | 5'-GGATCCGGAGGGGACCACTA | 62249-62268 | 55°C | 15 |
| Q′ | 5'-GCGGGATAGCGTGCGCTA | 62430-62447 | 5 | ||
| Q | 5′-GTGCGCTACCGGATGGCG | 62440-62457 | |||
| Y3 | 5′-TGGCGTGTGACGTGGTGTAA | 48397-48416 | |||
| K | 5′-CATTTCCAGGTCCTGTACCT | 107986-107967 | |||
| Probe | 5′-ATGCCCTGAGACTACTCTCT | 67563-67544 | |||
| EBNA2 | Y2 | 5′-ATTAGAGACCACTTTGAGCC | 47902-47921 | 58°C | |
| Y/P | 5′-CCCCATGTAACGCAAGATAG | 48534-48515 | |||
| Probe (Y3) | 5′-TGGCGTGTGACGTGGTGTAA | 48397-48416 | |||
| LMP1 | Primer | 5′-GTGACTGGACTGGAGGAGCC | 169341-169322 | 60°C | |
| Primer | 5′-GAGGGAGTCATCGTGGTGGTG | 168718-168738 | |||
| Probe | 5′-AGCCCTCCTTGTCCTCTA | 169325-169308 | |||
| LMP2A | T1 | 5′-GCAACACGACGGGAATGACG | 166824-166843 | 58°C | 33 |
| T1′ | 5′-TGGGAAGCGGCAGTGTAATC | 169809-169828 | |||
| T2 | 5′-AAACACGAGGCGGCAATAGC | 131-112 | |||
| Probe | 5′-ATCCAGTATGCCTGCCTGTA | 62-81 | |||
| BZLF1 | Primer | 5′-GGGAGAAGCACCTCAACCTG | 102826-102807 | 58°C | |
| Primer | 5′-TTGCTTAAACTTGGCCCGGC | 102447-102466 | |||
| Probe | 5′-AGCCAGAATC/CTGGAGGAAT | 102665-102656/ | |||
| 102530-102521 | |||||
| BHRF1 | Y2 (latent) | 5′-ATTAGAGACCACTTTGAGCC | 47902-47921 | 55°C | 50 |
| H2 (lytic) | 5′-GTCAAGGTTTCGTCTGTGTG | 53830-53849 | |||
| H3 | 5′-TTCTCTTGCTGCTAGCTCCA | 54461-54480 | |||
| Probe | 5′-CTGTCCCGTATACACAGGGC | 54425-54406 | |||
| BLLF1 | Primer | 5′-GTGGATGTGGAACTGTTTCCAG | 89934-89955 | 63°C | |
| (gp220) | Primer | 5′-CTGTATCCACCGCGGATGTCAC | 90753-90732 | ||
| Probe | 5′-AGTCCATCTCCATGGGACAA | 90682-90663 |
| Transcripts . | Oligos . | Oligonucleotide Sequence . | Coordinate in B95-8 . | Annealing Temperature . | Reference . |
|---|---|---|---|---|---|
| BART | A3 | 5′-AGAGACCAGGCTGCTAAACA | 157154-157173 | 55°C | 16 |
| A4 | 5-AACCAGCTTTCCTTTCCGAG | 159194-159175 | |||
| Probe | 5′-AAGACGTTGGAGGCACGCTG | 157359-157378 | |||
| EBNA1 | F | 5'-GGATCCGGAGGGGACCACTA | 62249-62268 | 55°C | 15 |
| Q′ | 5'-GCGGGATAGCGTGCGCTA | 62430-62447 | 5 | ||
| Q | 5′-GTGCGCTACCGGATGGCG | 62440-62457 | |||
| Y3 | 5′-TGGCGTGTGACGTGGTGTAA | 48397-48416 | |||
| K | 5′-CATTTCCAGGTCCTGTACCT | 107986-107967 | |||
| Probe | 5′-ATGCCCTGAGACTACTCTCT | 67563-67544 | |||
| EBNA2 | Y2 | 5′-ATTAGAGACCACTTTGAGCC | 47902-47921 | 58°C | |
| Y/P | 5′-CCCCATGTAACGCAAGATAG | 48534-48515 | |||
| Probe (Y3) | 5′-TGGCGTGTGACGTGGTGTAA | 48397-48416 | |||
| LMP1 | Primer | 5′-GTGACTGGACTGGAGGAGCC | 169341-169322 | 60°C | |
| Primer | 5′-GAGGGAGTCATCGTGGTGGTG | 168718-168738 | |||
| Probe | 5′-AGCCCTCCTTGTCCTCTA | 169325-169308 | |||
| LMP2A | T1 | 5′-GCAACACGACGGGAATGACG | 166824-166843 | 58°C | 33 |
| T1′ | 5′-TGGGAAGCGGCAGTGTAATC | 169809-169828 | |||
| T2 | 5′-AAACACGAGGCGGCAATAGC | 131-112 | |||
| Probe | 5′-ATCCAGTATGCCTGCCTGTA | 62-81 | |||
| BZLF1 | Primer | 5′-GGGAGAAGCACCTCAACCTG | 102826-102807 | 58°C | |
| Primer | 5′-TTGCTTAAACTTGGCCCGGC | 102447-102466 | |||
| Probe | 5′-AGCCAGAATC/CTGGAGGAAT | 102665-102656/ | |||
| 102530-102521 | |||||
| BHRF1 | Y2 (latent) | 5′-ATTAGAGACCACTTTGAGCC | 47902-47921 | 55°C | 50 |
| H2 (lytic) | 5′-GTCAAGGTTTCGTCTGTGTG | 53830-53849 | |||
| H3 | 5′-TTCTCTTGCTGCTAGCTCCA | 54461-54480 | |||
| Probe | 5′-CTGTCCCGTATACACAGGGC | 54425-54406 | |||
| BLLF1 | Primer | 5′-GTGGATGTGGAACTGTTTCCAG | 89934-89955 | 63°C | |
| (gp220) | Primer | 5′-CTGTATCCACCGCGGATGTCAC | 90753-90732 | ||
| Probe | 5′-AGTCCATCTCCATGGGACAA | 90682-90663 |
Bisulfite genomic sequencing.
DNA was treated with bisulfite as previously described.25 26 Briefly, the DNA was digested with EcoRI, denatured with NaOH, precipitated, and incubated with 3.1 mol/L sodium bisulfite (Sigma Chemical Co, St. Louis, MO) at 50°C for 16 hours in darkness. After the reaction, DNA was desalted and purified. The DNA was then alkaline treated with 0.3 mol/L NaOH and recovered. The bisulfite-treated DNA was PCR amplified with strand-specific primers (for the bottom strand): 5′-AACTAACCTAACTA AAAATAAAAC (corresponding to EBV coordinate 62179-62202), 5′-AATGTAAGGATAGTATGTATTATT (corresponding to EBV coordinate 62481-62458). The PCR products were electrophoresed, excised, purified, and then cloned into the pCR2.1-TA cloning vector (Invitrogen, Carlsbad, CA). Four to six colonies were analyzed for each DNA sample. Plasmid DNA was then extracted and sequenced.
Genotyping for EBNA3C and LMP1 gene deletion.
EBV subtypes (A, B) were determined with PCR by using primers spanning the EBNA3C region.27 Analysis of the previously reported 30bp deletion at the carboxyl terminus of LMP1 gene was performed by PCR.28 29 Primers were as follows: 5′-CCGCTGCCTCATAGCCC (168408-168392), 5′-TTAGCTGAACTGGGCCG (168174-168190). B95-8 was used as the control for type-A strain and wild-type LMP1 gene, whereas AG876 was the control for type-B EBV and the deleted LMP1 gene.
RESULTS
EBV gene expression in eBL.
Tissue RNA was well preserved in seven BL cases. RT-PCR for EBV transcripts showed EBNA1 transcript originating in Qp in all seven BL tumors (Fig 1). In one tumor, weak Cp/Wp-initiated EBNA1 and EBNA2 transcripts were also detected. No Fp-initiated EBNA1 transcript and no LMP1 transcripts were detected in any tumor. LMP2A but not LMP2B transcript was detected in five of seven cases. BamHI-A rightward transcripts (BART) were expressed in all seven cases. Weak lytic and latent BHRF1 transcripts, whose encoded protein is a Bcl-2 homolog, were detected in four cases. Lytic BLLF1 transcript that encodes a viral envelope glycoprotein gp220 was detected in three cases. These results are summarized in Table 2.
Autoradiography showing representative Southern blot hybridization of RT-PCR products for eBL tumors and BL cell lines.
Autoradiography showing representative Southern blot hybridization of RT-PCR products for eBL tumors and BL cell lines.
RT-PCR for EBV Transcripts in BL Tumors and Cell Lines
| Samples . | BART . | EBNA1 . | EBNA2 . | LMP1 . | LMP2 . | BZLF1 . | BHRF1 . | BLLF1 . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FUK . | QUK . | YUK . | 2A . | 2B . | Latent . | Lytic . | |||||||
| BL cell lines | Rael | ++ | + | ++ | − | − | − | ++ | − | +/− | + | ++ | ++ |
| Akata | ++ | ++ | ++ | − | − | − | ++ | − | + | − | ++ | ++ | |
| Chep | ++ | ++ | ++ | − | − | − | ++ | − | ++ | − | ++ | ++ | |
| Wan | ++ | ++ | ++ | ++ | + | ++ | ++ | − | ++ | +/− | ++ | ++ | |
| Wewak | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| Namalwa | ++ | ++ | ++* | ++ | ++ | ++ | − | − | − | ++ | +/− | − | |
| Rael-AzaC | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| Raji | ++ | ++ | ++* | ++ | ++ | ++ | +/− | ++ | +/− | ++ | ++ | + | |
| AG876 | ++ | ++ | ++ | ++ | ++ | ++ | ND | ++ | ++ | ND | ++ | ||
| LCL | IB4 | ++ | ++ | ++* | ++ | ++ | ++ | ++ | ++ | +/− | ++ | ++ | ++ |
| B95-8 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ND | ++ | ++ | |
| eBL | BL1 | ++ | − | ++ | − | − | − | + | − | − | +/− | +/− | +/− |
| BL2 | ++ | − | + | − | − | − | − | − | − | − | − | − | |
| BL6 | ++ | − | ++ | − | − | − | + | − | − | − | +/− | +/− | |
| BL7 | ++ | − | ++ | − | − | − | + | − | − | − | +/− | − | |
| BL8 | ++ | − | ++ | − | − | − | +/− | − | − | − | − | − | |
| BL9 | ++ | − | ++* | − | − | − | − | − | − | − | − | − | |
| BL10 | ++ | − | ++ | +/− | +/− | − | + | − | − | +/− | +/− | +/− | |
| NPC | NPC1 | ++ | +/− | ++ | − | − | − | + | − | − | − | − | − |
| NPC2 | ++ | +/− | ++ | − | − | − | ++ | − | − | − | +/− | − | |
| PTLD | PTLD1 | ++ | +/− | ++ | − | − | + | ++ | − | ++ | + | ++ | ++ |
| PTLD2 | ++ | +/− | ++ | − | − | − | − | − | ++ | +/− | ++ | + | |
| Samples . | BART . | EBNA1 . | EBNA2 . | LMP1 . | LMP2 . | BZLF1 . | BHRF1 . | BLLF1 . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FUK . | QUK . | YUK . | 2A . | 2B . | Latent . | Lytic . | |||||||
| BL cell lines | Rael | ++ | + | ++ | − | − | − | ++ | − | +/− | + | ++ | ++ |
| Akata | ++ | ++ | ++ | − | − | − | ++ | − | + | − | ++ | ++ | |
| Chep | ++ | ++ | ++ | − | − | − | ++ | − | ++ | − | ++ | ++ | |
| Wan | ++ | ++ | ++ | ++ | + | ++ | ++ | − | ++ | +/− | ++ | ++ | |
| Wewak | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| Namalwa | ++ | ++ | ++* | ++ | ++ | ++ | − | − | − | ++ | +/− | − | |
| Rael-AzaC | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| Raji | ++ | ++ | ++* | ++ | ++ | ++ | +/− | ++ | +/− | ++ | ++ | + | |
| AG876 | ++ | ++ | ++ | ++ | ++ | ++ | ND | ++ | ++ | ND | ++ | ||
| LCL | IB4 | ++ | ++ | ++* | ++ | ++ | ++ | ++ | ++ | +/− | ++ | ++ | ++ |
| B95-8 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ND | ++ | ++ | |
| eBL | BL1 | ++ | − | ++ | − | − | − | + | − | − | +/− | +/− | +/− |
| BL2 | ++ | − | + | − | − | − | − | − | − | − | − | − | |
| BL6 | ++ | − | ++ | − | − | − | + | − | − | − | +/− | +/− | |
| BL7 | ++ | − | ++ | − | − | − | + | − | − | − | +/− | − | |
| BL8 | ++ | − | ++ | − | − | − | +/− | − | − | − | − | − | |
| BL9 | ++ | − | ++* | − | − | − | − | − | − | − | − | − | |
| BL10 | ++ | − | ++ | +/− | +/− | − | + | − | − | +/− | +/− | +/− | |
| NPC | NPC1 | ++ | +/− | ++ | − | − | − | + | − | − | − | − | − |
| NPC2 | ++ | +/− | ++ | − | − | − | ++ | − | − | − | +/− | − | |
| PTLD | PTLD1 | ++ | +/− | ++ | − | − | + | ++ | − | ++ | + | ++ | ++ |
| PTLD2 | ++ | +/− | ++ | − | − | − | − | − | ++ | +/− | ++ | + | |
RT-PCR signal judged by autoradiography was indicated as ++, +, and +/−, as described in Materials and Methods.
Abbreviation: ND, not done.
Weakly positive or negative for QUK, but positive for Q'UK.
EBV gene expression in BL cell lines.
The patterns of gene expression in BL cell lines differed from those observed in BL tumors. Although similar to eBL, type-I cell lines (Rael, Akata, Chep) differed in that lytic transcripts (BZLF1, BHRF1, BLLF1) generally not detected in eBL tumor tissue were expressed. Cell lines (Wan, Wewak, and Namalwa) differed from eBL in expressing the LMP1 transcript and also differed from latency II tumors (NPC, HD, and nasal lymphoma) in that the Cp/Wp-initiated YUK transcript for EBNA1 was detected. Expression of EBNA2 and EBNA3C transcripts was also detected in these cell lines. The Wan cell line has been previously classified as type I/II.12 As expected, all viral transcripts studied were expressed in type-III BL cell lines (Rael-AzaC, Raji and AG876) and lymphoblastoid cell lines (LCL).
Methylation status of Qp.
Because Qp is the only promoter used for EBNA1 in eBL, the methylation status of its CpG sites was studied in eight cases. Twenty CpG sites in the minimal Qp and adjacent sequences were studied.30-32Among these CpG sites, all 16 CpG sites downstream of the Fp initiation site that include the whole Qp region were unmethylated in virtually all eBL samples studied (Fig 2). These sites were also unmethylated in the Rael cell line. However, the 4 CpG sites upstream of the initiation site of Fp were variably methylated in eBL and Rael cell line (Fig 3). Treatment with 5-azacytidine led to the disappearance of methylation at these 4 sites in Rael and an increase in Fp activity.
Summary of the methylation status of CpG sites in Qp region in eBL tumors. The top panel shows the structure of Qp and Fp. The region from CpG sites #8 to #15 corresponds to the minimal region required for Qp function.30-32 The CAAT and TATA box for Fp and an inverted CCAAT box for Qp are labeled. The transcription initiation sites of Qp (EBV coordinate 62422) and Fp (EBV coordinate 62230) are indicated. The binding sites for EBNA1 are shown by boxes. (m) indicates methylated CpG sites, (-) indicates unmethylated CpG sites.
Summary of the methylation status of CpG sites in Qp region in eBL tumors. The top panel shows the structure of Qp and Fp. The region from CpG sites #8 to #15 corresponds to the minimal region required for Qp function.30-32 The CAAT and TATA box for Fp and an inverted CCAAT box for Qp are labeled. The transcription initiation sites of Qp (EBV coordinate 62422) and Fp (EBV coordinate 62230) are indicated. The binding sites for EBNA1 are shown by boxes. (m) indicates methylated CpG sites, (-) indicates unmethylated CpG sites.
Genomic sequencing for bisulfite-treated DNA from eBL tumors for Qp. For bisulfite sequencing, the unmethylated C residue (or G at the opposite strand) within a CpG site will be converted to T (or A) in sequencing gel, whereas the methylated C residue is not changed. CpG sites 1 to 8 were sequenced from one direction, whereas CpG sites 9 to 20 were sequenced from another direction. All 16 CpG sites immediately downstream of the initiation site of Fp were unmethylated in BL1 and BL2. However, the 4 CpG sites upstream of the initiation site of Fp were methylated in BL2. Dark arrows indicate unmethylated CpG dinucleotides and open arrows indicate methylated CpG dinucleotides.
Genomic sequencing for bisulfite-treated DNA from eBL tumors for Qp. For bisulfite sequencing, the unmethylated C residue (or G at the opposite strand) within a CpG site will be converted to T (or A) in sequencing gel, whereas the methylated C residue is not changed. CpG sites 1 to 8 were sequenced from one direction, whereas CpG sites 9 to 20 were sequenced from another direction. All 16 CpG sites immediately downstream of the initiation site of Fp were unmethylated in BL1 and BL2. However, the 4 CpG sites upstream of the initiation site of Fp were methylated in BL2. Dark arrows indicate unmethylated CpG dinucleotides and open arrows indicate methylated CpG dinucleotides.
EBV genotyping.
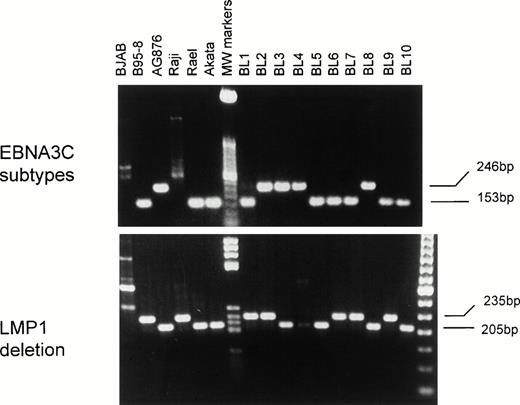
PCR amplification of the EBNA3C region yields a 153-bp band for type-A and a 246-bp band for type-B EBV. Six of 10 eBL cases harbored type-A virus, whereas 4 of 10 cases harbored type-B EBV (Fig 4). In 1 case (BL1), a strong type-A band and a very weak type-B band were both detected consistent with dual infection. PCR amplification of the carboxyl terminal region of LMP1 gene yields a 235-bp band for the wild-type LMP1 gene but a 205-bp band for the deleted type. The presence of the 30bp deletion was shown in 5 of 10 cases (Fig 4). There was no apparent relationship between the EBV strain and the presence or absence of the LMP1 carboxyl terminal deletion (Table 3).
Genotyping for EBNA3C subtypes and LMP1 gene deletion by PCR in eBL tumors and BL cell lines. A 153-bp band for type-A EBV and a 246-bp band for type B virus are shown. A 30-bp deletion at the carboxyl terminus of LMP1 gene is shown by the shorter PCR product of 205-bp, rather than a 235-bp band for the wild-type LMP1 gene.
Genotyping for EBNA3C subtypes and LMP1 gene deletion by PCR in eBL tumors and BL cell lines. A 153-bp band for type-A EBV and a 246-bp band for type B virus are shown. A 30-bp deletion at the carboxyl terminus of LMP1 gene is shown by the shorter PCR product of 205-bp, rather than a 235-bp band for the wild-type LMP1 gene.
EBV Genotyping for eBL Tumors
| Samples . | EBNA3C . | LMP1 gene . |
|---|---|---|
| B95-8 | A | W |
| AG876 | B | D |
| Raji | * | W |
| Rael | A | D |
| Akata | A | D |
| BL1 | A+(B) | W |
| BL2 | B | W |
| BL3 | B | D |
| BL4 | B | D |
| BL5 | A | D |
| BL6 | A | W |
| BL7 | A | W |
| BL8 | B | D |
| BL9 | A | W |
| BL10 | A | D |
| Total | 6A+4B | 5W+5D |
| Samples . | EBNA3C . | LMP1 gene . |
|---|---|---|
| B95-8 | A | W |
| AG876 | B | D |
| Raji | * | W |
| Rael | A | D |
| Akata | A | D |
| BL1 | A+(B) | W |
| BL2 | B | W |
| BL3 | B | D |
| BL4 | B | D |
| BL5 | A | D |
| BL6 | A | W |
| BL7 | A | W |
| BL8 | B | D |
| BL9 | A | W |
| BL10 | A | D |
| Total | 6A+4B | 5W+5D |
Abbreviations: A, type A EBV; B, type B EBV; W, wild-type LMP1 gene; D, deleted type LMP1 gene with a 30bp deletion at the carboxyl terminus; *, most of the EBNA3C gene is deleted in Raji EBV genome.
DISCUSSION
By RT-PCR, we have characterized the pattern of EBV gene expression in eBL tumors. In many regards the pattern of transcription corresponds to the latency I pattern, ie, EBNA1 is expressed from Qp whereas most of the rest of the genome is silent.10,11 However, both the BART and LMP2A transcripts were also detected in eBL tumors. This viral expression pattern is similar to that reported in peripheral blood mononuclear cells (PBMCs) isolated from healthy EBV–seropositive individuals. EBNA1 and LMP2A transcripts have been detected in PBMCs.33-35 Recently, BART transcripts have also been detected in PBMCs from a healthy individual (H.L. Chen, personal communication, June 1997). Although BL tumors are actively proliferating with a very high mitotic index while PBMCs, which are the locus of latent EBV infection in healthy seropositive individuals, are resting,36 37 these findings highlight the parallels in patterns of viral gene expression between the two situations.
The expression level of LMP2A RNA as detected by RT-PCR in eBL is similar to what we have observed in NPC and as reported by others,15,17 and is also similar to that reported in nasal lymphoma,20 HD,16 and T-cell lymphoma.18 Because immunologic reagents are not yet generally available to reliably detect the LMP2 antigen in tumor specimens, we do not know whether LMP2A protein is expressed in all tumor cells or just in a small percentage of them and what the protein expression level is. Although LMP2A RNA thus appears to be almost ubiquitous in EBV–infected cells, its role in the EBV life cycle remains ill defined. It appears not to be required for lymphocyte immortalization, because a mutant virus lacking functional LMP2A gene is able to infect and immortalize B cells.38 However, a recent study showed that the transforming ability of a mini–EBV without the LMP2A gene is greatly impaired and suggests a role for this protein in enhancing the transforming efficiency of EBV.39A role for LMP2A in protecting against lytic activation has also been suggested.40
Rightward transcripts (BART) originating in the BamHI-A region of the EBV genome were first recognized in 1989 in a nude mice-passaged NPC tumor, C15,41 and have since been detected in all EBV–associated tumors and cell lines.16-18,20,42,43Burkitt's tumor tissue has not been previously studied for this transcript, but in this tumor as in others that have been studied, BART transcripts are abundant. These transcripts are heavily spliced and the functional open-reading frames have yet to be fully defined.44,45 Recently, a new membrane protein encoded by one open-reading frame of BART, BARF0, has been identified, and its expression in one BL tumor biopsy has been shown by immunoblot.46
We also detected weak expression of other latent transcripts (Cp/Wp-initiated EBNA1 and EBNA2 transcripts) in one of the cases, and lytic transcripts (BHRF1 and BLLF1) in four cases. These weakly expressed transcripts may derive from a drift in viral gene expression in rare tumor cells, or the presence of EBV–infected tumor infiltrating lymphocytes.16,20,47 One of the limitations of RT-PCR as opposed to in situ detection techniques is that tissues with potentially heterogeneous patterns of gene expression are homogenized before characterization, sometimes allowing detection of transcripts expressed in rare cells. Unfortunately, the tumors studied here were snap-frozen in such a way as to preclude in situ analysis. Similarly, other investigators have reported heterogeneity in rare tumor cells in Burkitt's tumor specimens by immunohistochemical techniques.3 6
The major difference between patterns of EBV gene expression in type-I BL cell lines and eBL tumors was the higher level of expression of lytic cycle transcripts (BZLF1, BHRF1, BLLF1, and Fp transcripts) in BL cell lines. Tissue culture may permit drift toward lytic cycle expression that is prevented in patients by cytotoxic T-cell–mediated immune surveillance. Fp-initiated EBNA1 transcripts, first identified as latency transcripts in a type-I BL cell line (Rael) are now believed to be more appropriately characterized as lytic transcripts.48-50 In eBL tumors, Fp-initiated EBNA1 transcripts are absent and there is little evidence of ongoing lytic cycle gene expression as assessed by the absence or weak expression of BZLF1 and BLLF1 transcripts, respectively.
Transcription in EBV tumor cells appears to be regulated through CpG methylation. An inverse association between the activity of Cp, Wp, LMP1, EBER, and BHRF1 promoters, and CpG methylation has been noted previously and we have presented evidence that Cp methylation directly inhibits its transcription.25,51-56 The Cp, which drives EBNA expression, is active and hypomethylated in type-III BL cell lines and LCL,13,51 whereas it is silent and hypermethylated in BL tumors,24,56 type-I and -II BL cell lines,13,25 and other EBV–associated tumors with type-I and -II latency.24,57 Qp is the only active promoter for EBNA1 in eBL. Our bisulfite sequencing results showed that Qp, in contrast to Cp, is hypomethylated in virtually all eBL samples and in the Rael cell line. Recently, the methylation status of a 5 Kb region, including Fp and Qp, has also been reported in three type-I BL cell lines by genomic Southern hybridization and shown that Qp and Fp are located within a hypomethylated region.58 Our analysis of the methylation of the Qp region showed that although Qp is hypomethylated, Fp, which is only 200bp away from Qp, is actually variably methylated. This provides further evidence that Qp and Fp are distinct promoters30,32 and probably are regulated by different mechanisms. Qp has many features of a housekeeping promoter and the absence of methylation is consistent with this function.30 59
With regard to EBV strain, the nearly equal mix of type-A and type-B virus contrasts with studies of HD, NPC, and PTLD in America, Europe, and Asia, where type-A virus predominates and type-B is less frequently detected.60-62 However, this strain mix is in agreement with a previous report on eBL tumors,63 and similar to that detected in BL-derived cell lines from patients in equatorial Africa.60,64,65 It is also similar to the strain association of AIDS-associated EBV lymphomas, the latter having led to the suggestion that type-B EBV is more commonly seen in immunocompromised patients.63,66 Whether the virus type (A or B) detected in tumor tissue simply reflects the frequency of infection in the affected population or there is a specific virus-type tumor association in some instances remains to be determined. In one tumor we detected dual infection with type A and B viruses. Dual infections have been reported occasionally in other settings as well.61 67
LMP1 is a transforming gene product which interacts with tumor necrosis factor receptor (TNFR)-associated factors (TRAFs), and is involved in NF-κB activation.68 A 30-bp deletion at the carboxyl terminus (amino acids 346-355) of LMP1 gene was first reported in a Chinese NPC and has since been reported in a variety of EBV(+) tumors.28,29,69,70 This 30-bp deletion has been shown to be important to the transforming ability of LMP1 protein in Balb/3T3 cells. Its wild-type counterpart transformed Balb/3T3 cells only when driven by a strong promoter. 71-73 In addition, the deleted type LMP1 protein is reported to be non-immunogenic in a murine system.74 However, the TRAF interacting domains are still well conserved in the deleted LMP1,75 and the deleted LMP1 is as efficient as the wild type in activating NF-κB .76Although it has been suggested that this 30-bp deletion in the LMP1 gene is related to tumor pathogenesis or disease progress,29,70 these conclusions are still controversial. We detected this deletion in half of our eBL cases, but we have no information about the frequency of this deleted variant in healthy Ghanaians. Moreover, the LMP1 gene itself is not transcribed in BL tumor tissue, thus the pathogenetic importance, if any, remains uncertain. Overall, the deleted variant tends to be more frequently detected in Chinese and Japanese, but is also present in approximately half of Caucasians.60
In summary, the pattern of viral gene expression detected in primary eBL tumor [EBNA1 (Qp)(+), LMP2A(+), BART(+)] differs from that of BL cell lines in that the cell lines showed more lytic activation and more promiscuous use of EBNA1 promoters. The viral expression pattern in eBL tumor tissue is similar to what has been described in PBMCs isolated from healthy individuals and reinforces the notion that aspects of viral gene expression in BL may mirror viral gene regulation in latently infected normal B cells in vivo. In contrast to hypermethylation elsewhere in the EBV genome, the active Qp is hypomethylated in eBL tumors and BL cell lines. Type-A and type-B EBV are detected in eBL tumor tissue with approximately equal frequency and a 30bp carboxyl terminal deletion in the LMP1 gene is also common.
ACKNOWLEDGMENT
We thank the clinical and laboratory staff of the Burkitt's Tumor Project at the University of Ghana (Accra, Ghana) for collecting eBL specimens, and Dr. I-H Chen at the MacKay Memorial Hospital (Taipei, Taiwan) for providing the two NPC tumor specimens.
Support was provided by National Institutes of Health Grant No. R01 CA63532 (to R.F.A.). R.F.A. is a Leukemia Society Scholar.
Address correspondence to Richard F. Ambinder, MD, PhD, Johns Hopkins Oncology Center, 418 N. Bond St, Baltimore, MD 21231.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal