Abstract
We have analyzed 83 unrelated Hong Kong Chinese for the presence of genetic variants of factor V gene. Forty-three of them had a history of deep vein thrombosis. The DNA sequence variations of exons 7, 10, and 13, where the codons for Arg306, Arg506, and Arg679 are located, respectively, were studied by denaturing gradient gel electrophoresis. The G1691→A (Arg 506→Gln) mutation in exon 10 was not detectable in any of the 83 subjects. However, a high allelic frequency for the G1628→A (Arg 485→Lys) substitution was detectable in the same exon. We have also identified a novel DNA sequence mutation (A1090→G) in exon 7 that resulted in Arg 306→Gly substitution in 2 thrombotic patients and 1 nonthrombotic subject. Fresh blood samples were available from one of them for analysis of activated protein C resistance and the result was negative. Variation of DNA sequence was not found in exon 13 in any of our 83 subjects. The results of this study showed that, although the Arg 506→Gln mutation was rarely found in the Hong Kong Chinese population, a different mutation site such as A 1090→G in exon 7 of the factor V gene (Arg 306) may be of clinical importance.
HUMAN COAGULATION factor V acts as a cofactor in the conversion of prothrombin to thrombin in the coagulation process. In plasma, this high molecular weight glycoprotein is converted first to its active form, factor Va, by factor Xa and/or α-thrombin. Factor Va is then quickly inactivated by activated protein C (APC) for maintenance of hemostasis. This mechanism of factor Va inactivation is an ordered event.1 Factor Va is first cleaved at Arg 506 and then at Arg 306 and Arg 679. The peptide bond cleavage at Arg506 is essential for the subsequent optimal exposure of cleavage sites at Arg 306 and Arg 679. Peptide bond cleavage at Arg 306 appears to occur in a membrane-bound fashion and only accounts for the initial 70% loss of activity. The subsequent peptide bond cleavage at Arg 679 is responsible for the loss of remaining activity. Therefore, any defect on one or more of these three cleavage sites can potentially affect the APC inactivation process even though the factor V procoagulation activity may remain normal.
It has been reported that the most common defect among patients with deep vein thrombosis (DVT) is related to APC resistance.2-4A molecular defect resulting in APC resistance was identified as a single point mutation (G1691→A) in the factor V gene. It causes a single amino acid substitution (Arg 506→Gln) in the factor V molecule.5-7 This results in the loss of APC cleavage site at Arg 506 and subsequent ineffective peptide bond cleavage processes at Arg 306 and Arg 679. The rate of inactivation of mutant factor Va is diminished resulting in a higher level of thrombin. Patients with this genetic defect may develop a hypercoagulable state. It is well aware that the heterozygous state for this factor V gene mutation is associated with a fivefold to 10-fold increase in the risk of thrombosis and a 50- to 100-fold for the homozygous state.5,8,9 This genetic defect is found predominantly in Caucasian populations.5,8,10,11 This mutation and APC resistance is extremely rare in Asian populations.12
The effects of protein S and factor Xa on APC-catalyzed factor Va inactivation have also been studied. Protein S accelerates factor Va inactivation by selectively promoting the slow cleavage at Arg 306. Factor Xa protects factor Va from inactivation by APC by selectively blocking peptide bond cleavage at Arg 506. Inactivation of factor Va R506Q, which was isolated from the plasma of homozygous APC-resistant patient lacking the Arg 506 cleavage site, was stimulated by protein S but not affected by factor Xa. This confirms that the target sites of protein S and factor Xa involve Arg 306 and Arg 506, respectively. It has been observed that the large difference between the rates of APC-catalyzed inactivation of normal factor Va and factor Va R506Q were almost completely abolished in the presence of factor Xa and protein S. This may explain why, in the absence of other risk factors, APC resistance only results in a weak prothrombotic state.13There is further evidence to suggest that factor V gene abnormality responsible for the APC resistance may be not be a risk factor in the majority of some populations, but that it is a genetic risk factor in a significant minority and that it favors the clinical expression of protein C deficiency.14
So far, G1691→A mutation is the only genetic abnormality reported for the factor V gene resulting in APC resistance. In this study, we examined the whole exon 10 of factor V gene, in which the coding sequence for Arg 506 was located, of Hong Kong Chinese patients with or without DVT by denaturing gradient gel electrophoresis (DGGE) for detection of novel gene mutation. Also, the analysis was extended to exon 7 and 13, where Arg 306 and Arg 679 were respectively located.
MATERIALS AND METHODS
Forty-three unrelated Hong Kong Chinese patients with a definite history of DVT and 40 nonthrombotic patients were recruited for this study. High molecular weight DNA was extracted from peripheral blood of these 83 subjects.
Polymerase chain reaction (PCR) and Mnl/I restrictive enzyme digest.
The entire exon 10 containing the coding sequence of Arg 506 was amplified from genomic DNA by PCR. The two primers used were identical to those described by Ridker et al15 (5′ primer, acccacagaaatgatgccag; 3′ primer, tgcccattattagccaggag). The PCR product was a 233-bp fragment. In a total volume of 100 μL, the PCR reagents contained 1 μg genomic DNA, 50 pmol of each primer, 200 μmol/L of each deoxynucleoide triphosphate (Pharmacia), 1× Tag buffer (10 mmol/L Tris-HCl, pH 8.3, 50 mmol/L KCl, 1.5 mmol/L MgCl2, and 0.01% [wt/vol] gelatin), and 2.5 U of Tag polymerase (Perkin Elmer, Cetus, Norwalk, CT). The reaction was performed in a 0.6-mL microcentrifuge tube and a thermal cycler (Perkin Elmer, Cetus). The thermal cycle profile was 94°C for 5 minutes; 30 cycles of 94°C for 60 seconds, 50°C for 30 seconds, and 72°C for 90 seconds; followed by a final extension of 10 minutes at 72°C.
Ten microliters of PCR product was digested with 2 U of Mnl/I (New England Biolabs) at 37°C for 16 hours. The digestion was terminated by incubation at 65°C and the fragments were separated on a 6% polyacrylamide gel and visualized with ethidium bromide.
DGGE.
Sequence variations were detected by PCR and DGGE. For each segment of the coding exons 7, 10, and 13, a set of oligonucleotides composed of an upstream primer with an additional 40-bp G+C-rich sequence (GC-clamp) and a 20-mer downstream primer was used for PCR amplification. This G-C clamp allowed better resolution of the amplified fragments and more discriminating screening for sequence variations by DGGE.
The thermal profile consisted of 5 minutes of denaturation at 94°C and 30 cycles of 1 minute of denaturation at 94°C, 1 minute of annealing at 58°C for exon 7 (58°C for exon 10 and 56°C for exon 13), and 1 minute of extension at 72°C, followed by a final extension at 72°C for 5 minutes. The specificity and the size of the amplified fragments were checked on a 6% polyacrylamide gel.
Amplified PCR products were denatured at 94°C for 10 minutes and were allowed to reanneal at 37°C slowly over 30 minutes to form homoduplex and heteroduplex. The reannealed PCR products were then subjected to electrophoresis for 3 hours at 160 V, 60°C on a 6.5% polyacrylamide gel containing a 20% to 70% denaturing gradient (100% denaturant = 7 mol/L urea and 40% formamide in TEA buffer [40 mmol/L Tris, 20 mmol/L sodium acetate, 1 mmol/L EDTA, pH 7.6]).
DNA sequencing.
Direct PCR DNA sequencing was performed using the same primer sets. The amplified PCR fragment was purified from the polyacrylamide gel and used as template for sequencing reaction using a standard DNA sequencing kit (US Biochemical, Cleveland, OH).
The functional coagulation assay for APC resistance.
Resistance to APC was tested on platelet-poor plasma using commercial test kit from Chromogenix (Molnsal, Sweden), following the manufacturer's instructions. Activated partial thromboplastin time (APTT) was performed on the plasma samples with or without added APC and the sensitivity ration was calculated as APTT with APC/APTT without APC (normal, >2.1).12
RESULTS
DNA samples from 43 thrombotic and 40 nonthrombotic patients were studied for the presence of the G1691→A mutation of factor V gene by Mnl/I restriction enzyme digestion of the amplified gene. The mutation was not detectable in any of the samples. The same samples were also analyzed by DGGE. Abnormal patterns suggesting the presence of a genetic abnormality at exon 10 of the factor V gene was found in 41 of the 83 (49.3%) samples being studied. The results are shown in Fig 1. DNA sequencing of the normal fragments showed normal DNA sequences of exon 10 that was identical to those previously reported in the gene bank. The abnormal DNA fragments identified were found to have a G→A mutation at nucleotide 1628, which resulted in a change of the peptide sequence from Arg 485 to Lys. Among the 83 subjects, 8 (9.6%) were homozygous for the 1628 G → A mutation and 33 (39.8%) were heterozygous. The frequency distribution of this mutation was similar between the 43 thrombotic (4 homozygous and 17 heterozygous) and the 40 nonthrombotic (4 homozygous and 16 heterozygous) patients. Identical mutation has been previously reported by Gandrille et al14 in European populations but at a much lower frequency.
DGGE of the amplified exon 10 of factor V gene. Lanes 1 and 3, contol with a homozygosity for G1691→A mutation. Lane 2, control with a heterozygosity for G1691→A mutation. Lane 8, normal control. Lanes 9 and 11, homozygous for G1628→A mutation. Lanes 4 through 7, 10, and 12, heterozygous for G1628→A mutation. (Upstream primer, GC40-TCAGG CAGGA ACAAC ACCAT; downstream primer, GGTTA CTTCA AGGAC AAAAT.).
DGGE of the amplified exon 10 of factor V gene. Lanes 1 and 3, contol with a homozygosity for G1691→A mutation. Lane 2, control with a heterozygosity for G1691→A mutation. Lane 8, normal control. Lanes 9 and 11, homozygous for G1628→A mutation. Lanes 4 through 7, 10, and 12, heterozygous for G1628→A mutation. (Upstream primer, GC40-TCAGG CAGGA ACAAC ACCAT; downstream primer, GGTTA CTTCA AGGAC AAAAT.).
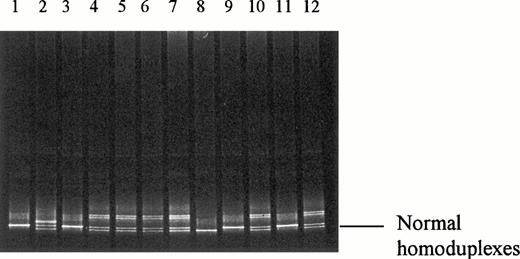
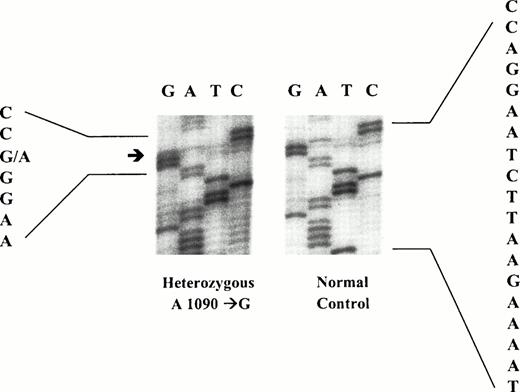
The PCR and DGGE technique were also applied to study the exons 7 of factor V gene, where the coding sequence of Arg306 was located. Abnormal patterns suggesting the presence of a genetic abnormality at exon 7 of the factor V gene were found in 3 of the 83 (3.6%) samples being studied. There were 2 positive cases in the thrombotic group and 1 in the nonthrombotic group. The results are shown in Fig 2. DNA sequencing of the normal fragments showed normal DNA sequences of exon 7 of factor V gene that were identical to those previously reported in the gene bank. DNA sequence analysis of the abnormal amplified fragment of exon 7 from these 3 patients showed a heterozygous A1090→G mutation (Fig 3). To confirm the mutation, a restriction enzyme digest of the amplified exon 7 of factor V gene withBstNI (New England Biolabs), which has a restriction site of CC∇AGG, were performed. The DNA fragments were studied by 6% polyacrylamide gel electrophoresis. The undigested DNA fragment was a 240-bp PCR product. A complete digestion of this 240-bp DNA fragment with BstNI was expected to yield two shorter fragments of 100 and 140 bp in length (Fig 4). The presence of an A1090→G mutation resulted in the loss of the cleavage site for BstNI. A BstNI-digested sample from a normal control on lane 1 produced the two fragments of 100 and 140 bp in length as expected. A BstNI-digested sample from a thrombotic patient on lane 2 yielded three DNA fragments (100, 140, and 240 bp), confirming heterozygosity for the A1090→G mutation. The additional 240-bp fragment represented the complete undigested PCR product, as the result of the loss of the enzyme cleavage site ofBstNI. Similar to the normal control one lane 1, aBstNI-digested sample from another thrombotic patient on lane 3 produced two DNA fragments of 100 and 140 bp in length, confirming the absence of A1090→G mutation. Fresh blood sample was available only from 1 of the 3 subjects with the mutation and the patient had history of DVT. The sample was taken while the patient was not on any anticoagulant. Functional coagulation assay for APC resistance was performed on the sample available and the APC sensitivity ratio was 3.2, which was normal. Other tests, including antithrombin III, protein C, protein S, and lupus anticoagulant, were also determined on the same blood sample and were all normal.
DGGE of the amplified exon 7 of factor V gene. Lane 1, normal control. Lanes 2 through 7, thrombotic patients. Lane 2 shows the heteroduplexes as the result of a heterozygous A1090→G mutation. (Upstream primer, [GC-40]-TGTCC TAACT CAGCT GGGAT; downstream primer, gtatg aaccc caaca actca.)
DGGE of the amplified exon 7 of factor V gene. Lane 1, normal control. Lanes 2 through 7, thrombotic patients. Lane 2 shows the heteroduplexes as the result of a heterozygous A1090→G mutation. (Upstream primer, [GC-40]-TGTCC TAACT CAGCT GGGAT; downstream primer, gtatg aaccc caaca actca.)
DNA sequence analysis of an abnormal amplified fragment of exon 7 of factor V gene. A heterozygous A1090→G mutation was identified.
DNA sequence analysis of an abnormal amplified fragment of exon 7 of factor V gene. A heterozygous A1090→G mutation was identified.
Polyacrylamide gel electrophoresis following theBstNI restriction enzyme digest of a 240-bp DNA fragment of exon 7 of factor V gene. Lane M, molecular weight marker. Lane 1,BstNI-digested sample from a normal control showing the two DNA fragments, 100 and 140 bp. Lane 2, BstNI-digested sample from a thrombotic patient heterozygous for A1090→G mutation. It shows three DNA fragments: 100, 140, and 240 bp. Lane 3,BstNI-digested sample from a thrombotic patient without the A1090→G mutation. Similar to lane 1, it shows only 2 DNA fragments, 100 and 140 bp. (Upstream primer, [GC-40]-TGTCC TAACT CAGCT GGGAT; downstream primer, gtatg aaccc caaca actca.)
Polyacrylamide gel electrophoresis following theBstNI restriction enzyme digest of a 240-bp DNA fragment of exon 7 of factor V gene. Lane M, molecular weight marker. Lane 1,BstNI-digested sample from a normal control showing the two DNA fragments, 100 and 140 bp. Lane 2, BstNI-digested sample from a thrombotic patient heterozygous for A1090→G mutation. It shows three DNA fragments: 100, 140, and 240 bp. Lane 3,BstNI-digested sample from a thrombotic patient without the A1090→G mutation. Similar to lane 1, it shows only 2 DNA fragments, 100 and 140 bp. (Upstream primer, [GC-40]-TGTCC TAACT CAGCT GGGAT; downstream primer, gtatg aaccc caaca actca.)
The PCR and DGGE analysis was finally used to study exon 13 of the factor V gene. It is the location of the codon of Arg 679. No abnormality was found in the samples from all 83 subjects being studied.
DISCUSSION
Factor V Leiden (Arg 506→Gln mutation) is known to be an important risk factor for venous thrombo-embolism. Its prevalence is known to vary widely among different populations. The mutation in heterozygous or homozygous state is relatively more common in some European populations. The allele frequency in European is estimated to be 4.4%, with the highest prevalence among Greeks (7%). However, it was only 0.6% in Asia Minor and the mutation was not found in Africa and Southeast Asia.11,12,16 This may partly explain the rarity of thrombo-embolic diseases in these populations, although other factors may also contribute. A previous study has demonstrated the lack of APC resistance and Arg 506→Gln mutation in healthy Hong Kong Chinese blood donors.12 None of our 43 thrombotic and 40 nonthrombotic Hong Kong patients in this study had the factor V Leiden mutation. Our result supports the findings of previous studies.12
An Arg 485→Lys mutation at exon 10 of the factor V gene was first described by Gandrille et al.14 They have found this sequence variation in 3 of 104 (2.9%) normal subjects and 7 of 113 (6.4%) patients with protein C deficiency. This mutation was not associated APC resistance in their healthy subjects and its allelic frequency in their protein C-deficient patients was not significantly different form that observed in healthy subjects. The Arg 485 to Lys substitution is apparently a neutral polymorphism in terms of procoagulant activity or susceptibility to APC. In this study, nearly half of our 83 subjects, including both thrombotic and nonthrombotic patients, were found to have this sequence variation, suggesting a much higher allelic frequency of this mutation in Hong Kong Chinese. Like many other polymorphism, this Arg 485 to Lys substitution appears to have a highly variable allelic frequency in different ethnic populations. This mutation was seen at similar frequency in our thrombotic and nonthrombotic subjects, supporting the hypothesis of a neutral polymorphism in terms of procoagulant activity as originally proposed by Gandille et al.14
We have detected in this study a previously unreported mutation (A1090→G) of the factor V gene at exon 7. This mutation results in a Arg 306 to Gly substitution. Among our 84 subjects, 2 thrombotic patients and 1 nonthrombotic patient were found to possess this mutation in heterozygous state. This A1090→G mutation in the codon AGG of Arg306 is a missense point mutation predicting a replacement of arginine by a glycine residue.17 The substitution may potentially affect APC cleavage at the Arg 306 site of the factor V molecule. As the inactivation of factor Va is a sequential event, initial peptide bond cleavage of residue Arg 506 results in the exposure of other cleavage sites. The subsequent cleavage of at residues Arg 306 and Arg 679 completes the factor Va inactivation process. With a normal Arg 506 in these subjects, the effect of a defect at Arg 306 is uncertain. Nevertheless, cleavage of Arg306 is required for complete inactivation of the factor Va molecule. An Arg 306 defect may result in a protein that is cleaved at Arg506 and Arg679 only but still possesses about 60% cofactor activity.14-18 Fresh plasma samples are only available from 1 of the 3 subjects with the mutation. The patient had history of DVT and the APC resistance assay on the sample was within normal limits. At this stage, the clinical significance of the substitution at Arg306 is not certain as the number of positive cases in our series is small and the defect is found in both thrombotic and nonthrombotic subjects. It is not yet possible to assess the thrombotic risk of subjects with this defect. Large-scale epidemiologic studies on this Arg306 mutation are warranted. The true allelic frequency distribution of this defect in Chinese and other populations remains to be determined. Further functional and clinical correlation is also required.
Supported by a Conference and Research Grant (CRCG) of the University Research Committee of the University of Hong Kong.
Address reprint requests to Raymond Liang, MD, Department of Medicine, Queen Mary Hospital, Hong Kong.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

![Fig. 2. DGGE of the amplified exon 7 of factor V gene. Lane 1, normal control. Lanes 2 through 7, thrombotic patients. Lane 2 shows the heteroduplexes as the result of a heterozygous A1090→G mutation. (Upstream primer, [GC-40]-TGTCC TAACT CAGCT GGGAT; downstream primer, gtatg aaccc caaca actca.)](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/4/10.1182_blood.v91.4.1135/4/m_blod4043702.jpeg?Expires=1767748712&Signature=UwWG7X-dZt7zNb9LDIynEBl7v-LZT8E016eWFblOmVNvmEM1-64geGVRDtfyG1v09BOBC0klN6biQbk0LxGh9InBkAl0VL2vHxqT3qQGAcMnsvo5oGzqcp9attrmT7SANWnQpY1iuhbE4ptnyB~GB8Y7shUch8MPXTg~NtEqs14iABEvQvAE~a8GzfbgvXyTTlU4CPVb4U4bAN16BrEy1cdy9czTJx0PX~PsWcq5FSDx9cmfV3YGgCoyzCkNjl41T6Q-CZZ~uUXxGIDTwueVNXpCBk6VUlz1f4j6L7hHdrc15j08G3e1IIkx-i-nA6LYgyh0BA50p31gwZsjTb6slw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Polyacrylamide gel electrophoresis following theBstNI restriction enzyme digest of a 240-bp DNA fragment of exon 7 of factor V gene. Lane M, molecular weight marker. Lane 1,BstNI-digested sample from a normal control showing the two DNA fragments, 100 and 140 bp. Lane 2, BstNI-digested sample from a thrombotic patient heterozygous for A1090→G mutation. It shows three DNA fragments: 100, 140, and 240 bp. Lane 3,BstNI-digested sample from a thrombotic patient without the A1090→G mutation. Similar to lane 1, it shows only 2 DNA fragments, 100 and 140 bp. (Upstream primer, [GC-40]-TGTCC TAACT CAGCT GGGAT; downstream primer, gtatg aaccc caaca actca.)](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/4/10.1182_blood.v91.4.1135/4/m_blod4043704.jpeg?Expires=1767748712&Signature=OvfVZ7~Tnco15~eYlRq97fWXK0zCyYQ~Er74zs0B1-uWQQ61-QrS3j~Rv8rqVgJ20pNGisso5Kw~qpQ7dMy6egEqb2WbuAoa7tXiPrQfy3jJ4FBfILVc6zck8c4mIssah2h3E6~uQ7zcfvY0T9K27FKF9KfmOwxICfl3Il75BS0ooBLKBmTjpHA803Ez44PP9FMh3mKlChNK4zWbPfAqzXTzLW5bldYqHJg5JVKPGDjIdzQ2DwgJ52Ub2xBRtiFmlz84P2TqcNvPEF1cSKk1cAGYZczNFZwxqmMf8-ZHCFNWaThAI--I280uQo4H-I054svoPxVCYtIFKo5sM6W~EQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal