Abstract
Cytokine-treated macrophages represent a useful model to unravel the molecular basis of reticuloendothelial (RE) iron retention in inflammatory conditions. In the present study, we showed that stimulation of murine macrophage J774 cells with interferon (IFN)-γ/lipopolysaccharide (LPS) resulted in a nitric oxide-dependent modulation of the activity of iron regulatory proteins (IRP)-1 and 2, cytoplasmic proteins which, binding to RNA motifs called iron responsive elements (IRE), control ferritin translation. Stimulation with cytokines caused a small increase of IRP-1 activity and a strong reduction of IRP-2 activity accompanied by increased ferritin synthesis and accumulation. Cytokines induced only a minor increase of H chain ferritin mRNA, thus indicating that IRP-2–mediated posttranscriptional regulation plays a major role in the control of ferritin expression. This was confirmed by direct demonstration that the translational repression function of IRP was impaired in stimulated cells. In fact, translation in cell-free extracts of a reporter transcript under the control of an IRE sequence was repressed less efficiently by IRP-containing lysates from cytokine-treated cells than by lysates from control cells. Our findings throw light on the role of IRP-2 showing that: (1) this protein responds to a stimulus in opposite fashion to IRP-1; (2) when abundantly expressed, as in J774 cells, IRP-2 is sufficient to regulate intracellular iron metabolism in living cells; and (3) by allowing increased ferritin synthesis, IRP-2 may play a role in the regulation of iron homeostasis in RE cells during inflammation.
IRON METABOLISM and physiology of cells of the mononuclear phagocyte system are closely connected. Macrophages, being both storage and recycling compartments of the metal, play a central role in iron metabolism. Indeed, alterations of iron handling by reticuloendothelial (RE) cells may also be involved in the pathogenesis of disorders of iron metabolism, such as genetic hemochromatosis.1 In turn, modifications of iron homeostasis are central to the function of RE cells in the inflammatory response. In fact, diversion of iron from the circulation to RE stores contributes to the production of reactive oxygen species essential for macrophage cytotoxic activity and, at the same time, limits iron availability to invading microorganisms.2
Intracellular iron homeostasis is controlled by iron regulatory proteins (IRP)-1 and 2, cytosolic proteins that bind iron-responsive elements (IRE) in the untranslated regions of ferritin, the major iron storage protein, and transferrin receptor (TfR) mRNA, which mediates iron uptake.3-5 Under conditions of limited iron supply, IRP binding to IRE blocks ferritin mRNA translation and at the same time stabilizes TfR mRNA. This results in increased cellular iron availability. Conversely, when cellular iron levels are high, IRP lose their binding activity, permitting ferritin mRNA translation and degradation of TfR mRNA, thus lowering iron levels in the free pool. In this condition, IRP-1 contains a 4Fe-4S cluster and functions as a cytoplasmic aconitase.6 When iron is scarce, the cluster is disassembled, aconitase activity is lost, and apo-IRP acquires RNA-binding activity. IRP-2 lacks the amino acids present at the active site of mitochondrial aconitase and consequently does not show enzymatic activity. IRP-1 and IRP-2 bind IRE with similar affinities and repress ferritin synthesis to the same extent, but their sensitivity to in vitro reducing agents and their mode of regulation by iron are different. In addition, they seem to bind distinct sets of RNA sequences. Furthermore, the varying abundance of the two forms in different tissues suggests that differential expression of the two IRP can play an important role in cellular iron metabolism (reviewed in Hentze and Kuhn,5 Henderson,7 and Harrison and Arosio8).
In addition to iron levels, other signals such as oxidative stress and nitric oxide (NO) can modulate the activity of both IRP and thus influence cellular iron metabolism (reviewed in Hentze and Kuhn,5 Harrison and Arosio,8 Pantopoulos et al,9 Drapier and Bouton,10 Rouault and Klausner,11 Hentze,12 Richardson and Ponka,13 and Domachowske14). In particular, NO, which targets metalloproteins, interacts with the 4Fe-4S cluster disrupting aconitase function and, accordingly, enhanced IRE-binding activity after induction of the NO pathway has been observed in several cellular systems (see Weiss,2 Pantopoulos et al,9 Drapier and Bouton,10 Richardson and Ponka,13 and Domachowske14 for review). A connection between NO, an important mediator of the inflammatory response, and IRP, the key regulator of iron homeostasis, may have important pathophysiologic ramifications in RE cells during the inflammatory response, as also demonstrated by the finding of an autoregulatory loop between iron metabolism and the NO pathway in activated macrophages.15 However, the NO-mediated activation of IRP that has been detected in interferon (IFN)-γ/lipopolysaccharide (LPS)-treated macrophages16-18is difficult to reconcile with the modifications of RE iron metabolism that occur in inflammatory states. In fact, increased IRP activity reduces ferritin synthesis17,18; it is expected that this, in turn, impairs the iron storage capacity of RE cells, whereas increased iron retention in the RE system is a well-known feature of inflammation.2 19
In the present study, we found that treatment of the murine J774 macrophages with cytokines triggers a selective, NO-mediated, downregulation of IRP-2 activity that results in increased synthesis and accumulation of ferritin.
MATERIALS AND METHODS
Reagents.
Minimum Essential Medium (MEM), fetal calf serum, (α-32P) uridine triphosphate (UTP), 32P deoxy cytidine triphosphate (CTP), 35S methionine and TRAN35S-LABEL were purchased from ICN Biomedicals (Opera MI, Italy), desferrioxamine (DFO), NG-monomethyl-L-arginine monoacetate (L-NMMA), S-nitroso-N-acetyl-D,L-penicillamine (SNAP), N-acetyl-D,L-penicillamine (NAP), LPS from Escherichia coliserotype 0111:B4 and ferric ammonium citrate were purchased from Sigma Chemical Co (Milano, Italy), and antiserum to mouse macrophage inducible nitric oxide synthase (iNOS) from Alexis Corp (Inalco S.p.A., Milano, Italy). Mouse recombinant IFN-γ and mouse recombinant tumor necrosis factor α (TNF-α) were supplied by Genzyme srl. (Cinisello, Italy), Hybond membranes and ECL Plus were supplied by Amersham Co (Milano, Italy). The kits for in vitro transcription and wheat germ extract were purchased from Promega Corp (Firenze, Italy).
Cell cultures and treatments.
The mouse macrophage cell line J774A.1 was grown in MEM supplemented with 10% heat-inactivated fetal calf serum, 2 mmol/L glutamine, 100 U/mL penicillin and 0.1 ng/mL streptomycin at 37°C in 5% CO2. Medium and all reagents were endotoxin-free. Near confluent cells (1.5 × 106) were stimulated for different time periods (4 to 24 hours) initially with various concentrations of LPS (0.1 to 10 μg/mL) and IFN-γ (10 to 200 U/mL) and then routinely with 1 μg/mL LPS plus 100 U/mL IFN-γ in the presence or absence of 250 μmol/L NMMA or 50 μmol/L DFO. Iron overload was obtained by culturing cells in the presence of 50 μg/mL ferric ammonium citrate for 24 hours. Cells were also exposed to 0.5 mmol/L SNAP or NAP for various time periods as described above. At the end of the treatments cells were harvested, pelletted, and stored at −80°C. TNF-α and nitrite levels in the supernatant were measured as previously described20 using the Wehi 164 Clone 13 cell line in an MTT tetrazolium cytotoxicity assay and the Griess reaction, respectively.
Western blot analysis.
Aliquots of the cytosolic extracts used for the determination of IRP activity containing equal amounts of proteins were electrophoresed in 10% acrylamide-sodium dodecyl sulfate (SDS) gels, electroblotted to Hybond polyvinylidene difluoride (PVDF) membranes (Amersham Co) and incubated with a 1:1,000 dilution of antiserum to mouse iNOS (Alexis Corp). iNOS was detected by chemiluminescence using an immunodetection kit (ECL Plus, Amersham Co) according to the manufacturer's instructions.
Generation of RNA transcripts in vitro.
Probes for bandshift assays were transcribed from linearized pSPT-fer containing the IRE of human ferritin H chain21 or CG28422 plasmids using T7 RNA polymerase in the presence of α-32P UTP using a commercially available kit (Promega Corp) as previously described.23 Unlabeled I-12.chloramphenicol acetyl transferase (CAT) and I-19.CAT mRNA,24 containing, respectively, the wild-type or mutated ferritin IRE sequence upstream of the chloramphenicol-acetyltransferase (CAT) open reading frame, were transcribed from linearized plasmids in the presence of m7GpppG using the same kit.
RNA-protein gel retardation assay.
Cells were lysed in the buffer described by Leibold and Munro,25 the lysate was centrifuged at 16,000g for 5 minutes and the supernatant was used for RNA-protein bandshift assays. Equal amounts of protein (2 μg as determined using the Bio Rad protein assay kit) were incubated, in the absence or presence of 2% 2-mercaptoethanol, with a molar excess of IRE probe and treated sequentially with RNase T1 and heparin as described previously.23 To detect IRP-2, 10 μg proteins were incubated with up to 2 × 105 cpm of32P labeled mutated CG284 probe in the presence of 300 ng tRNA and 1 mg/mL heparin.22 After separation on 6% nondenaturing polyacrylamide gels, RNA-protein complexes were visualized by autoradiography. For quantitation of IRP activity, radioactivity of bands excised from dried gels was determined by liquid scintillation counting.
Ferritin synthesis.
At the end of the treatments with cytokines or SNAP, 5 × 106 cells were incubated in methionine and cysteine-free medium for 2 hours at 37°C in the presence of 50 μCi/mL of35S methionine plus cysteine (TRAN35S-LABEL, ICN). Cells were then homogenized in 20 mmol/L Tris-HCl pH 7.4, 150 mmol/L NaCl, 1% Triton X-100, and 1 mmol/L phenylmethylsulfonylfluoride, and equal amounts of labeled proteins were immunoprecipitated using antibodies against recombinant mouse H and L ferritin subunits (see below). Immunoprecipitation products were run on 15% polyacrylamide gels and radioactivity in ferritin was revealed by fluorography and quantitated by densitometric scanning.
Analysis of ferritin mRNA levels.
Total cellular RNA was isolated as described elsewhere26and equal amounts of RNA were electrophoresed under denaturing conditions. To confirm that each lane contained equal amounts of total RNA, the ribosomal RNA content in each lane was estimated in ethidium bromide-stained gels. RNA was transferred to Hybond-N filters (Amersham Co), which were sequentially hybridized with the following32P-labeled DNA probes: rat ferritin L subunit pRLFL3 cDNA,27 rat ferritin H subunit H 1110 cDNA,28and mouse transferrin receptor cDNA clone pTfR-2.29 For quantitative determinations, autoradiographic bands in the linear range were scanned with a densitometer, and the values were calculated after normalization to the amount of ribosomal RNA.
In vitro translation.
Capped in vitro transcribed RNA (10 ng) was added to wheat germ extract (Promega Corp) in the presence or absence of different amounts of cytoplasmic extracts (10 or 20 μg protein) and incubated for 1 hour at 25°C in the presence of 15 μCi of 35S methionine. Translation products were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and fluorography as described above.
Determination of ferritin content.
The intracellular concentration of H and L subunit rich ferritins was determined in aliquots of the cytoplasmic extracts used for bandshift assays using immunoassays based on rabbit polyclonal antibodies against recombinant mouse H and L ferritins. The same two proteins were used as standards. The two assays did not give cross-reactivity in the concentration range used.30
Statistical analysis.
Values are expressed as means ± standard deviation (SD). The significance of differences was evaluated with t-test using the Stata Statistical Software (Stata Corporation, College Station, TX).
RESULTS
NO modulates IRP-1 and IRP-2 activity differentially in cytokine-treated J774 cells.
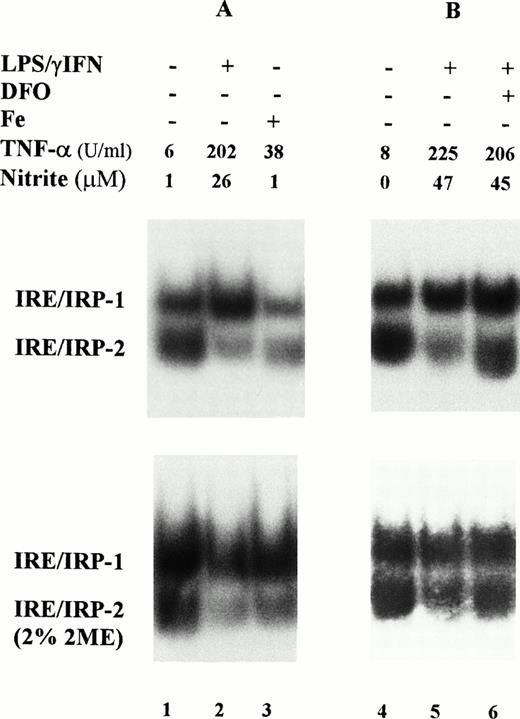
In a first series of experiments, we investigated the IRE-binding activity of IRP in cytoplasmic extracts prepared from mouse macrophage J774 cells; IRP-2 accounted for 50% or more of the total IRE binding activity in these cells (Fig 1). Stimulation with 1 μg/mL LPS plus 100 U/mL IFN-γ for 24 hours slightly enhanced IRP-1 activity, but strongly reduced that of IRP-2. Preliminary experiments (not shown) indicated no differences in response with doses ranging between 50 and 200 U/mL IFN-γ and 1 to 10 μg/mL LPS. The extent of activation of IRP-1 and of inhibition of IRP-2 was somewhat variable as shown by representative RNA-bandshift assays (Figs 1 and 2), but the pattern of response was consistent and reproducible; Table 1 summarizes the results of all experiments. As expected on the basis of previous work,3-5 binding activity of both IRP was reduced in iron-loaded cells used as a reference (Fig 1A); cytokine treatment downregulated IRP-2 activity to the same extent as iron overload. Treatment with cytokines in the presence of the iron chelator DFO prevented the decrease of IRP-2 activity (Fig 1B) suggesting that cytokines may act indirectly by increasing intracellular iron levels. Time course experiments (Fig 2 and Table 1) showed a prompt activation of IRP-1 and a progressive downregulation of IRP-2. Differences in IRP-1 binding activity were eliminated by treatment of cell extracts with 2% 2-mercaptoethanol, which completely activates IRP-1 activity in vitro,3-5 thus indicating equal loading of all samples (Figs 1 and 2, lower panels). After cytokine treatment, increased production of TNF-α, a typical marker of inflammatory response, was consistently observed.
Modulation of IRP-1 and IRP-2 activity in cytokine-treated macrophage J774 cells. J774 cells were left untreated or treated for 24 hours with cytokines (100 U/mL IFN-γ plus 1 μg/mL LPS); where indicated, the iron chelator DFO (0.05 mmol/L) was also added. Iron loading was obtained by incubating cells with 50 μg/mL ferric ammonium citrate for 24 hours (lane 3). A total of 2 μg protein of cytoplasmic extracts was analyzed for IRE-binding activity by RNA gel retardation assay with excess 32P-labeled RNA transcribed from the pSPT-fer probe containing the IRE of the ferritin H mRNA in the absence (upper panels) or presence (lower panels) of 2% 2-mercaptoethanol. TNF-α and nitrite production was assayed in the culture medium as described in Materials and Methods. TNF-α units were calculated from a standard curve with rTNF-α used as internal standard for each test.
Modulation of IRP-1 and IRP-2 activity in cytokine-treated macrophage J774 cells. J774 cells were left untreated or treated for 24 hours with cytokines (100 U/mL IFN-γ plus 1 μg/mL LPS); where indicated, the iron chelator DFO (0.05 mmol/L) was also added. Iron loading was obtained by incubating cells with 50 μg/mL ferric ammonium citrate for 24 hours (lane 3). A total of 2 μg protein of cytoplasmic extracts was analyzed for IRE-binding activity by RNA gel retardation assay with excess 32P-labeled RNA transcribed from the pSPT-fer probe containing the IRE of the ferritin H mRNA in the absence (upper panels) or presence (lower panels) of 2% 2-mercaptoethanol. TNF-α and nitrite production was assayed in the culture medium as described in Materials and Methods. TNF-α units were calculated from a standard curve with rTNF-α used as internal standard for each test.
Time course of IRP activity in cytokine-treated macrophage J774 cells. J774 cells were left untreated (lane 1) or treated for 4 (lane 2) or 24 hours (lanes 3,4) with cytokines (100 U/mL IFN-γ plus 1 μg/mL LPS); where indicated, the iNOS inhibitor NMMA (0.1 mmol/L) was also present (lane 4). A total of 2 μg protein of cytoplasmic extracts was analyzed for IRE-binding activity by RNA gel retardation assay with excess 32P-labeled probe containing IRE sequences in the absence (upper panel) or presence (lower panel) of 2% 2-mercaptoethanol. TNF-α and nitrite production was assayed in the culture medium as described in the legend to Fig 1.
Time course of IRP activity in cytokine-treated macrophage J774 cells. J774 cells were left untreated (lane 1) or treated for 4 (lane 2) or 24 hours (lanes 3,4) with cytokines (100 U/mL IFN-γ plus 1 μg/mL LPS); where indicated, the iNOS inhibitor NMMA (0.1 mmol/L) was also present (lane 4). A total of 2 μg protein of cytoplasmic extracts was analyzed for IRE-binding activity by RNA gel retardation assay with excess 32P-labeled probe containing IRE sequences in the absence (upper panel) or presence (lower panel) of 2% 2-mercaptoethanol. TNF-α and nitrite production was assayed in the culture medium as described in the legend to Fig 1.
IRP-1 and IRP-2 Activity and Ferritin Content in Cytokine-Treated J774 Cells
| . | IRP-1 (%) . | IRP-2 (%) . | FT-H (ng/mg prot) . | FT-L (ng/mg prot) . |
|---|---|---|---|---|
| Control | 100 | 100 | 218 ± 66 | 80 ± 21 |
| IFN-γ/LPS 4 h | 132 ± 14-150 | 37 ± 12-150 | — | — |
| IFN-γ/LPS 24 h | 141 ± 9-150 | 27 ± 6-150 | 489 ± 93-150 | 102 ± 47 |
| SNAP 24 h | 136 ± 11-150 | 29 ± 5-150 | 467 ± 88-150 | 100 ± 83 |
| . | IRP-1 (%) . | IRP-2 (%) . | FT-H (ng/mg prot) . | FT-L (ng/mg prot) . |
|---|---|---|---|---|
| Control | 100 | 100 | 218 ± 66 | 80 ± 21 |
| IFN-γ/LPS 4 h | 132 ± 14-150 | 37 ± 12-150 | — | — |
| IFN-γ/LPS 24 h | 141 ± 9-150 | 27 ± 6-150 | 489 ± 93-150 | 102 ± 47 |
| SNAP 24 h | 136 ± 11-150 | 29 ± 5-150 | 467 ± 88-150 | 100 ± 83 |
J774 cells were left untreated (control) or treated for 4 or 24 hours with cytokines (100 U/mL IFN-γ plus 1 μg/mL LPS) or treated with 0.5 mmol/L SNAP for 24 hours. IRP activity and ferritin content was determined as described in Materials and Methods. Values represent mean ± SD of 10 (IFN-γ/LPS) and 7 (SNAP) experiments. Values of IRP activity are given as a percentage of control.
P < .001 v controls.
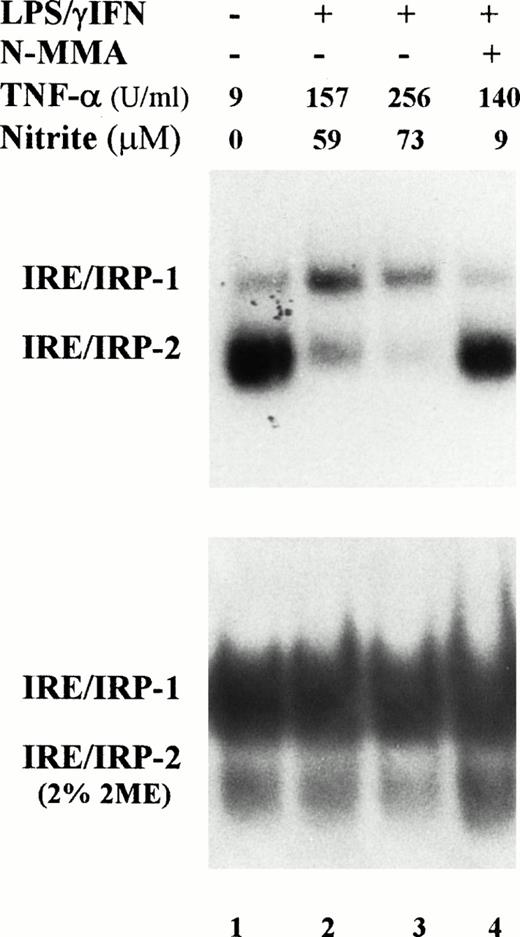
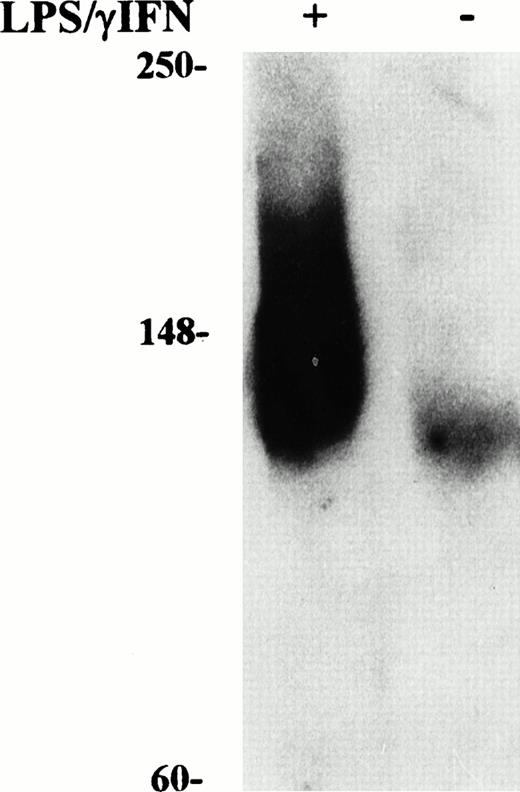
Induction of NO production by IFN-γ/LPS is well documented,2 indeed, an increased concentration of nitrite in the culture medium was found in cytokine stimulated J774 cells (Figs1 and 2). Enhanced levels of NO metabolites were the result of increased levels of iNOS, as shown by Western blot analysis of cell extracts (Fig 3). These results suggested the involvement of the NO pathway in the modulation of IRP activity. Indeed, the addition of NMMA, an iNOS inhibitor, prevented the modifications of both IRP-1 and IRP-2 activity induced by stimulation with cytokines (Fig 2, lane 4). To confirm the data indicating that NO has a role in modulation of IRP activity, we treated J774 cells with the NO donor SNAP. The addition of SNAP, but not of its inactive nonnitrosylated counterpart NAP (data not shown), had a remarkable effect on IRP activity, similar to that of treatment with IFN-γ/LPS (Fig 4A and Table1).
Western blot analysis of iNOS content in cytokine-treated macrophage J774 cells. Cytoplasmic extracts were prepared from murine J774 macrophages untreated and treated with IFN-γ/LPS for 24 hours as described in Fig 1. Equal amounts (50 μg) of denatured proteins were electrophoresed in an SDS 10% polyacrylamide gel, electroblotted to filters, and incubated with anti-iNOS antibody (1:1,000 dilution) followed by secondary antibody as described in Materials and Methods. Bands were visualized by chemiluminescence. Migration of molecular mass markers (myosin, phosphorylase B, and glutamic dehydrogenase, 250, 148, and 60 kD, respectively), loaded on the same gel, is shown on the left. The results shown are representative of six separate experiments.
Western blot analysis of iNOS content in cytokine-treated macrophage J774 cells. Cytoplasmic extracts were prepared from murine J774 macrophages untreated and treated with IFN-γ/LPS for 24 hours as described in Fig 1. Equal amounts (50 μg) of denatured proteins were electrophoresed in an SDS 10% polyacrylamide gel, electroblotted to filters, and incubated with anti-iNOS antibody (1:1,000 dilution) followed by secondary antibody as described in Materials and Methods. Bands were visualized by chemiluminescence. Migration of molecular mass markers (myosin, phosphorylase B, and glutamic dehydrogenase, 250, 148, and 60 kD, respectively), loaded on the same gel, is shown on the left. The results shown are representative of six separate experiments.
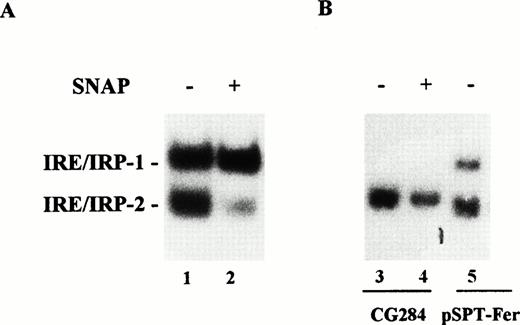
Effect of NO on IRP-1 and IRP-2 activity of macrophage J774 cells. (A) J774 cells were left untreated or treated with the NO donor SNAP (0.5 mmol/L) for 24 hours as indicated. IRP activity in cytoplasmic extracts was determined by bandshift assay using the pSPT-fer probe as described in the legend to Fig 1. The results shown are representative of seven separate experiments. (B) The mutant CG284 probe, which is specific for IRP-2, was incubated with lysates (10 μg protein) of J774 cells untreated and treated with 0.5 mmol/L SNAP for 24 hours as indicated. The pSPT-fer probe was incubated with lysates (2 μg protein) of untreated cells. Formation of RNA/protein complexes was estimated by bandshift assay as described in the legend to Fig 1. The autoradiogram shown is representative of four separate experiments.
Effect of NO on IRP-1 and IRP-2 activity of macrophage J774 cells. (A) J774 cells were left untreated or treated with the NO donor SNAP (0.5 mmol/L) for 24 hours as indicated. IRP activity in cytoplasmic extracts was determined by bandshift assay using the pSPT-fer probe as described in the legend to Fig 1. The results shown are representative of seven separate experiments. (B) The mutant CG284 probe, which is specific for IRP-2, was incubated with lysates (10 μg protein) of J774 cells untreated and treated with 0.5 mmol/L SNAP for 24 hours as indicated. The pSPT-fer probe was incubated with lysates (2 μg protein) of untreated cells. Formation of RNA/protein complexes was estimated by bandshift assay as described in the legend to Fig 1. The autoradiogram shown is representative of four separate experiments.
IRE-like sequences specifically recognized by IRP-2 have recently been described.22 An RNA probe encompassing one such mutated IRE sequence (CG284) incubated with J774 extracts formed an RNA/protein complex migrating close to the position of the faster moving complex detected by the pSPT-fer probe (Fig 4B). Binding activity was remarkably decreased in SNAP-treated cells (Fig 4B). The same results were observed in cytokine-treated cells (data not shown). The specificity of CG284 RNA for IRP-2 has previously been demonstrated by competition experiments.22
Effect of cytokine stimulation on ferritin expression.
We then investigated the effect of the concomitant increase of IRP-1 and reduction of IRP-2 on ferritin accumulation. In J774 cells treated with IFN-γ/LPS or with SNAP for 24 hours, H-rich-ferritin content was significantly enhanced, whereas L-rich ferritin concentration was somewhat increased, but the difference did not reach statistical significance (Table 1). To investigate the molecular mechanisms underlying the increase of ferritin content, we assessed ferritin synthesis: J774 cells were pulse labeled with radioactive amino acids and ferritin was immunoprecipitated and analyzed on SDS gels. A strong induction of H subunit synthesis (2.5-fold) was detected on stimulation with IFN-γ/LPS (Fig 5) and SNAP (data not shown), whereas L subunit synthesis was induced to a lower extent (1.6-fold). As previously reported,31 the mouse H subunit had a higher electrophoretic mobility than the L subunit. The finding that IRP-2 downregulation is accompanied by increased ferritin expression suggests that posttranscriptional regulation by IRP-2 plays an important role in the control of ferritin expression in this context.
Ferritin synthesis by cytokine-treated macrophage J774 cells. J774 cells untreated or treated with IFN-γ/LPS for 24 hours were incubated in the presence of 35S methionine plus cysteine (TRAN35S-LABEL) for 2 hours. Equal amounts of labeled proteins were then immunoprecipitated with antibodies against recombinant mouse H and L ferritin subunits (lanes 3 and 4); an aliquot of the supernatants containing total proteins was also loaded on the gel (lanes 1 and 2). Total proteins and immunoprecipitated ferritin chains were separated on 15% SDS/polyacrylamide gels and visualized by fluorography. The results shown are representative of four separate experiments.
Ferritin synthesis by cytokine-treated macrophage J774 cells. J774 cells untreated or treated with IFN-γ/LPS for 24 hours were incubated in the presence of 35S methionine plus cysteine (TRAN35S-LABEL) for 2 hours. Equal amounts of labeled proteins were then immunoprecipitated with antibodies against recombinant mouse H and L ferritin subunits (lanes 3 and 4); an aliquot of the supernatants containing total proteins was also loaded on the gel (lanes 1 and 2). Total proteins and immunoprecipitated ferritin chains were separated on 15% SDS/polyacrylamide gels and visualized by fluorography. The results shown are representative of four separate experiments.
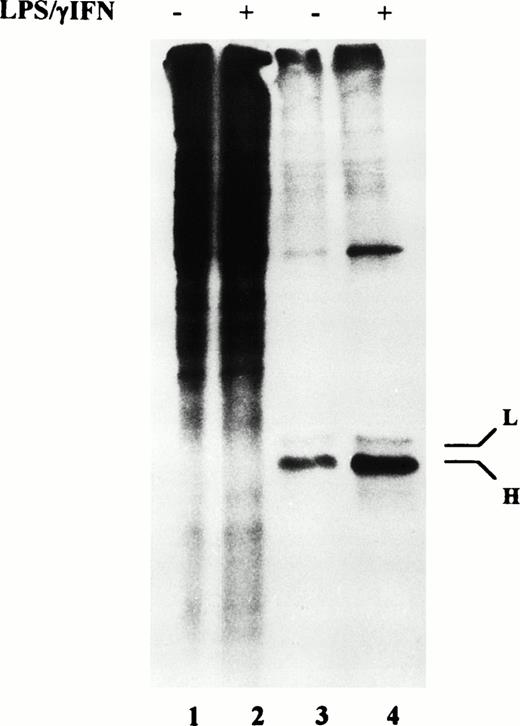
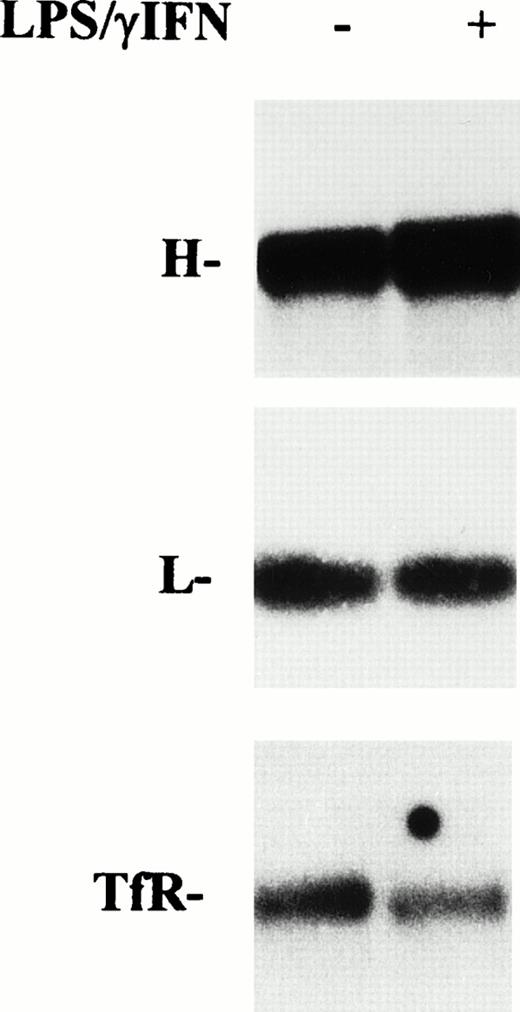
However, because modulation of mRNA coding for proteins of iron metabolism in cytokine-treated RE cells has been reported,17,18 32 a role for transcriptional mechanisms in increased ferritin synthesis cannot be ruled out. Thus, we investigated the expression of ferritin mRNAs by Northern blot analysis. As shown in Fig 6, after treatment with cytokines for 24 hours, L subunit mRNA remained at the control level, whereas H ferritin subunit mRNA was slightly increased (1.3-fold). Conversely, TfR mRNA was decreased after stimulation of cells with IFN-γ/LPS.
Effect of cytokine treatment on ferritin and transferrin receptor mRNA levels in macrophage J774 cells. Total RNA isolated from J774 cells untreated or treated with IFN-γ/LPS for 24 hours as indicated was run in denaturing agarose gels, transferred to filters, and hybridized with 32P-labeled DNA probes for H and L ferritin subunits and TfR as described in Materials and Methods. The autoradiograms shown are representative of four separate experiments.
Effect of cytokine treatment on ferritin and transferrin receptor mRNA levels in macrophage J774 cells. Total RNA isolated from J774 cells untreated or treated with IFN-γ/LPS for 24 hours as indicated was run in denaturing agarose gels, transferred to filters, and hybridized with 32P-labeled DNA probes for H and L ferritin subunits and TfR as described in Materials and Methods. The autoradiograms shown are representative of four separate experiments.
Effect of cytokine treatment on IRE-mediated translational regulation.
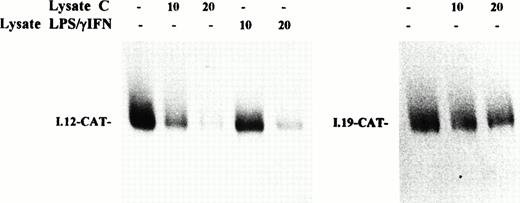
The slight increase of H chain mRNA would not completely account for the strong activation of ferritin synthesis, thus supporting the idea that the induction of ferritin expression by inflammatory cytokines is mainly controlled at the posttranscriptional level. To confirm the role of translational control in the induction of ferritin synthesis by IFN-γ/LPS, we studied the function of IRP as translational regulator by assessing the effect of the addition of cytoplasmic extracts from control or cytokine-treated J774 cells on the translation of a reporter mRNA under the control of an IRE sequence. A chimeric transcript composed of wild-type IRE sequence in front of a reporter CAT coding sequence (I.12-CAT) was translated in a cell-free system derived from wheat germ, which is devoid of endogenous IRP.33 As shown in Fig 7 (left panel), the translation of I.12-CAT was inhibited efficiently by addition of increasing amounts of IRP-containing extracts from control cells, whereas repression by extracts from stimulated cells was quantitatively less pronounced. The same result was observed in SNAP-treated cells (data not shown). The specificity of this translational control was assessed by analysis of a control mRNA; in fact, translation of I-19.CAT mRNA, which has a mutation in the IRE motif that prevents IRE/IRP interaction,24 was not significantly affected by addition of extracts from control (Fig 7, right panel) or stimulated (not shown) J774 cells. This finding indicates that cytokine-mediated downregulation of IRP-2 binding activity to an IRE sequence in bandshift assays is also reflected in an impaired functional activity in a translation repression assay.
Effect of cytokine treatment on translational repression activity of IRP in macrophage J774 cells. I-12.CAT (left panel) and I-19.CAT (right panel) mRNA, containing, respectively, the wild-type or mutated ferritin IRE sequence upstream of the CAT open reading frame, were translated in wheat germ extract in the presence of35S methionine. The effect on translational activity of the addition of increasing amounts of lysate (10 to 20 μg protein) from J774 cells untreated (lysate C) or treated with IFN-γ/LPS for 24 hours (lysate IFN-γ/LPS) was evaluated. Translation products were separated on 10% SDS/polyacrylamide gels and visualized by fluorography. The fluorogram shown is typical of four separate experiments.
Effect of cytokine treatment on translational repression activity of IRP in macrophage J774 cells. I-12.CAT (left panel) and I-19.CAT (right panel) mRNA, containing, respectively, the wild-type or mutated ferritin IRE sequence upstream of the CAT open reading frame, were translated in wheat germ extract in the presence of35S methionine. The effect on translational activity of the addition of increasing amounts of lysate (10 to 20 μg protein) from J774 cells untreated (lysate C) or treated with IFN-γ/LPS for 24 hours (lysate IFN-γ/LPS) was evaluated. Translation products were separated on 10% SDS/polyacrylamide gels and visualized by fluorography. The fluorogram shown is typical of four separate experiments.
DISCUSSION
The characterization of two IRP with structural and functional similarities, but significant differences in their mode of regulation and pattern of tissue expression has raised the question of why cells evolved two IRP. The finding that, in addition to consensus IRE, they bind a different array of selected RNA targets indicates that a possible reason for the presence of two IRP could be to enhance the versatility of the IRE/IRP regulatory system, expanding its influence to cellular processes not immediately related to intracellular iron homeostasis. However, the complexity of the system may also be increased if the two IRP respond preferentially to different stimuli. In fact, we have demonstrated preferential regulation of IRP-2 activity and consequent changes of the expression of the major proteins of intracellular iron metabolism during liver cell proliferation34 and oxidative stress.26 IRP have been shown to be also a target of NO. Whereas IRP-1 activity has been found to be induced by NO (see Weiss,2Pantopoulos,9 Drapier and Bouton,10 Richardson and Ponka,13 and Domachowske14 for review), the effect of NO on the activity of IRP-2 is less clearcut. Indeed, IRP-2 activity has been reported to be enhanced by NO in macrophages,16-18 fibroblasts,35 and the liver during the acute phase reaction,36 but not affected in hepatoma cells.37
In the present study, we showed that stimulation by cytokines weakly enhanced IRP-1 activity, but strongly reduced that of IRP-2 in the mouse macrophage cell line J774. In contrast with the present data, a coordinated increase of both IRP-1 and IRP-2 activity resulting in decreased ferritin translation has been reported in IFN-γ/LPS–treated J774 cells.17,18 We have no immediate explanation for this discrepancy, which apparently does not depend on trivial technical or experimental reasons. In fact, we used time periods and cytokine doses similar to those reported by Weiss et al.17,18 However, an evident downregulation of the fast-migrating band detected by the pSPT-fer probe, which corresponds to the IRE/IRP-2 complex,5,7 was a consistent finding in all of our experiments; in addition, the decrease of IRP-2 binding activity was confirmed by experiments using the IRP-2–specific CG284 probe. Moreover, decreased IRP-2 activity was further indicated by results obtained in a functional assay showing that the translational repressor capacity of IRP was impaired in cytokine-treated cells (see Fig 7). Evidence for the role of NO in the modulation of IRP activity in cytokine-treated J774 cells was provided by experiments in which the formation of NO by a physiologic source (ie, cytokine-stimulated iNOS, see Fig 3) was specifically prevented and by other experiments in which a compound that produces NO in solution (SNAP) was added directly to cells. As to the mechanistic aspects of the effects of NO on IRP activity, the reactivity of this compound with iron-containing proteins suggests that IRP-1 activation may depend on direct interaction of NO with the 4Fe-4S cluster.9-14 On the other hand, IRP-2, lacking the cluster, is not directly targeted by NO, which may instead downregulate the activity of this protein indirectly by increasing intracellular iron availability, as indicated by the demonstration that treatment with an iron chelator prevented the fall of IRP-2 activity (see Fig 1B).
Interestingly, the opposite modulation of IRP-1 and IRP-2 activity in stimulated J774 cells was accompanied by increased ferritin synthesis and accumulation, indicating that IRP-2 downregulation can act as the predominant effector of cellular iron homeostasis in this system. This is further demonstrated by the cytokine-dependent decrease of TfR mRNA levels found in the present study (see Fig 6) and previously.18 It should be noted that IRP-2 is particularly abundant in J774 cells in which it represents at least 50% of total IRP activity.
The prominent role of posttranscriptional mechanisms, ie, IRP-2 inactivation, in the control of ferritin induction is also supported by the finding that cytokine stimulation, in agreement with previous reports,17 18 induced only a minor increase of ferritin mRNA, which would not fully account for the increase of ferritin synthesis. The importance of posttranscriptional control in the induction of ferritin synthesis was confirmed by experiments that assessed translational regulation directly. In fact, IRP in lysates from cytokine-treated cells showed a reduced capacity to repress translation of IRE-containing transcripts in cell-free extracts. Thus, the decreased IRP-2 binding activity detected in gel retardation assays was reflected in impaired translational repression function. From a quantitative viewpoint, the slight activation of IRP-1 might counteract IRP-2 downregulation, thus explaining why induction of ferritin expression is less prominent than IRP-2 inactivation.
Although the two IRP differ in their mechanisms of regulation,5,7,8 they have been considered to be coordinately regulated7; to our knowledge, the present results are the first demonstration of an opposite response of IRP-1 and IRP-2 to a stimulus. In vitro experiments have indicated equal binding affinity and equal functional capacity of the two IRP,5,7,8 and we have shown specific modulation of IRP-2 in the absence of significant changes of IRP-1 in vivo.26,34,36 However, as IRP-1 usually accounts for the major part of IRE-binding activity, the role of IRP-2 has been generally somewhat underestimated. Here we show that when IRP-2 is abundantly expressed, as in J774 cells, modulation of its activity is sufficient to dictate intracellular iron metabolism. This also represents the first evidence that IRP-2 controls translation in living cells. The recent finding of a cell line without detectable IRP-1 supports the idea that IRP-2 can function as the sole mediator of intracellular iron metabolism.38
Diversion of iron traffic from the circulation into the storage sites within the RE system is pivotal in inducing the hypoferremia that accompanies inflammatory diseases. Previous reports,17,18which showed NO-mediated IRP activation and consequent inhibition of ferritin synthesis, are difficult to reconcile with the recognized changes of iron metabolism occurring under inflammatory conditions.2,19 The present study, by demonstrating an IRP-2–mediated increase of ferritin synthesis in cytokine-treated J774 cells, provides molecular evidence supporting previous findings in experimental models of inflammation, which showed that iron retention in RE cells was accompanied by increased ferritin expression.19 Advances in the knowledge of the molecular mechanisms underlying increased ferritin synthesis in cytokine-treated RE cells could also give insights into the pathophysiology of the anemia of chronic disease in which iron retention by macrophages limits iron availability for hematopoiesis.
ACKNOWLEDGMENT
We thank L. Kuhn for providing pSPT-fer and CG284 plasmids, K. Pantopoulos and M. Hentze for the generous gift of I-12.CAT and I-19.CAT constructs, and P. Santambrogio for determination of ferritin content.
Supported by Ministero Università e Ricerca Scientifica e Tecnologica (MURST), Consiglio Nazionale delle Ricerche (CNR), and IRCCS Ospedale Maggiore, Milano, Italy and from the UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (TDR) (to D.T.).
Address reprint requests to Dr Gaetano Cairo, Centro di Studio sulla Patologia Cellulare CNR, Via Mangiagalli 31, 20133 Milano, Italy.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal