Abstract
Recently, we and other groups reported in acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) a strong correlation between cytogenetic rearrangements leading to 17p deletion, a typical form of dysgranulopoiesis combining pseudo-Pelger-Huët hypolobulation and small vacuoles in neutrophils, and p53 mutation. To gain further insight into this “17p-syndrome,” we studied 17 cases of AML and MDS with 17p deletion by whole chromosome painting (WCP) and fluorescence in situ hybridization (FISH) with probes spanning the 17p arm, including a p53 gene probe. Cytogenetically, 15 patients had unbalanced translocation between chromosome 17 and another chromosome (chromosome 5 in nine cases and unidentified chromosome -add 17p- in three cases), one patient had monosomy 17, and one had i(17q). All rearrangements appeared to result in 17p deletion. Sixteen patients had additional cytogenetic rearrangements. WCP analysis confirmed the cytogenetic interpretation in all cases and identified one of the cases of add 17p as a t(17;22). WCP also identified chromosome 17 material on a marker or ring chromosome in two cases of t(5;17). FISH analysis with 17p markers made in 16 cases showed no deletion of the 17p markers studied in the last two patients, who had no typical dysgranulopoiesis; p53 mutation analysis in one of them was negative. In the 14 other cases, FISH showed a 17p deletion of variable extent but that always included deletion of the p53 gene. All 14 patients had typical dysgranulopoiesis, and all but one had p53 mutation and/or overexpression. These findings reinforce the morphologic, cytogenetic, and molecular correlation found in the 17p- syndrome and suggest a pathogenetic role for inactivation of tumor suppressor gene(s) located in 17p, especially the p53 gene.
MYELODYSPLASTIC syndromes (MDSs) are clonal bone marrow stem cell disorders characterized by ineffective hematopoiesis leading to blood cytopenias and by a high incidence of progression to acute myeloid leukemia (AML).1,2 Nonrandom chromosomal abnormalities resulting in 17p deletion are seen in 3% to 4% of AML and MDS cases.3-6 They include unbalanced translocations between 17p and another chromosome, mainly chromosome 5, and less often monosomy 17, i(17q), or del 17p. Most of those patients have several other cytogenetic abnormalities, and about 30% of the cases are therapy-related.7 We found a strong correlation in AML and MDS between 17p deletion and a typical form of dysgranulopoiesis combining pseudo-Pelger-Huët hypolobulation and the presence of small vacuoles in granulocytes.6,7 We also found a strong correlation between 17p deletion and p53 mutation, suggesting that MDS and AML with 17p deletion constitute a new morphologic-cytogenetic-molecular “entity” in those disorders.8,9 Our results confirmed a previous report that found a correlation between 17p deletion (mainly through i(17q)) and pseudo-Pelger-Huët hypolobulation in blast crisis chronic myeloid leukemia (CML),10 a situation where p53 mutation is also a frequent event, generally correlated to 17p deletion.11Finally, the correlation between 17p deletion and pseudo-Pelger-Huët hypolobulation in MDS was recently confirmed by another group.12
In our description of MDS and AML with 17p deletion, all chromosomal studies were made by conventional cytogenetic analysis. To study more precisely the chromosomal segments involved in those rearrangements, we studied by whole chromosome painting (WCP) and fluorescence in situ hybridization (FISH) 17 cases of AML and MDS where 17p deletion was observed by conventional cytogenetics.
SUBJECTS AND METHODS
Patients.
Seventeen cases of MDS and AML where conventional cytogenetics showed chromosome abnormalities leading to 17p deletion, diagnosed at our institution between 1987 and 1996 according to French-American-British criteria,13 and for whom adequate material was available were studied.
Their characteristics at diagnosis are shown in Table1. Six patients had AML, and 11 had MDS. Nine cases were therapy-related. A review of bone marrow slides was made for the analysis of myelodysplastic features, particularly dysgranulopoiesis. Conventional cytogenetic analysis was performed on bone marrow cells, after short-term (24 hours) culture without stimulation. Chromosomes were identified by RHG and GTG banding and classified according to the International System for Human Cytogenetic Nomenclature.14 Hematologic and cytogenetic findings for three of the patients were included in a previous report.7
Hematologic and Cytogenetic Findings in 17 MDS and AML Patients With 17p Deletion
| Patient No. . | Sex/Age . | Therapy Related . | FAB Type . | “Typical” Dysgranulopoiesis-151 . | Conventional Cytogenetic Findings . |
|---|---|---|---|---|---|
| t(5;17) translocation | |||||
| 1 | F/63 | No | M1AML | Yes | 43,XX,del(2)(p13),−3,del(5)(q11q23),der(5)t(5;17)(q23;q11),del(6)(p21), −7,add(9)(p13),add(12)(p11),del(15)(q15),−17 [17] /46,XX [1] |
| 2* | M/40 | No | RAEB-T | Yes | 42,XY,−3,−4,−5,−7,−11,−12,−16, der(17)t(5;17)(p11;p11),−20,+4mar [22] /46,XY [4] |
| 3 | M/71 | Yes | RAEB | Yes | 45,XY,−5, r(7), der(17)t(5;17)(p11;p12),+mar1 [7] /45,idem,del(3)(p13) [3] /46,XY [3] |
| 4 | M/68 | Yes | RAEB | Yes | 44,X,−Y,−3,−5,der(5)t(3;5)(p13;q31),der(17)t(5;17)(p11;p11),−18,+r, +mar [20] |
| 5 | M/75 | Yes | M2AML | Yes | 45,XY,−5, der(17)t inv(5;17)(p11q14;p11) [15] /46,XY [2] |
| 6 | M/82 | Yes | RAEB-T | Yes | 45,XY,−5, der(17)t(5;17)(p11;p11) [9] /46,idem,+8 [3] /45,XY,−5,add(16)(q21),−17,der(18)t(17;18)(q11;q23),+mar [1] |
| 7* | F/67 | Yes | RAEB | Yes | 44,XX,−5,−7, der(17)t(5;17)(p11;p11) [11] |
| 8 | M/57 | No | RAEB | No | 43-44,XY,add(2)(p11),−5,add(5)(p1?),der(5)t(5;17)(q13;q11),−7,−13,−17, del(20)(q12),−21,+(0-1)r,+mar1,+mar2 [7] |
| 9 | M/69 | No | RAEB | No | 44-46,XY, der(5)t(5;17)(q11;q11),del(7)(q21q34),+i(8)(q10), dup(13)(q11p13), ?add(16)(q11),−17,−(0-1)18,−21,+r [15] /46,XY [1] |
| Rearrangement between 17p and another chromosome | |||||
| 10 | M/62 | Yes | M5AML | Not evaluable | 44-46,XY,(0-1)add(2)(q35),der(7)t(1;7)(p11;p21),+(0-1)i(8)(q10), t(9;11)(p21;q23),−12,−(0-1)13,der(17)t(12;17)(q11;p11), (0-1)add(22)(p11),+(0-1)mar,(1-2)dmin(cp7) [16] |
| 11* | M/76 | No | RAEB-T | Yes | 43-45,XY,3,add(5)(p?),del(5)(q14q32),+11,add(11)(q23), der(12)t(12;17)(p12;q11),del(13)(q13q14),−17,−22,+mar,ace [cp4] |
| 12 | F/81 | No | M2AML | Yes | 41-42,XX,del(4)(q13),−5,der(9)t(9;10)(p21;q11),−12,t(13;15)(p10;p10), −15,add(16)(q11),der(17)t(17;21)(p11;q11),?i(21)(q10),−21,−22,add(22)(p11), +(1-3)mar [18] /46,XX [1] |
| 13 | F/75 | Yes | M2AML | Yes | 44-45,XX,del(5)(q11q34),−17, add(17)(p13), −18,t(?21;22)(q22;q11), mar(15),+mar1 [8] |
| 14 | F/71 | No | RAEB | Yes | 46-49,XX,del(5)(q13q33),−7,del(9)(q23q32), add(17)(p11), −18, (0-1)del(20)(q11),−21,+22,+(2-4)mar [11] /46,XX,del(5)(q13;q33) [1] /46,XX [2] |
| 15 | M/47 | Yes | RAEB | Yes | 44-45,XY,−(0-1)7,add(12)(p12),−(0-1)15, add(17)(p11), add(21)(q13), +(0-1)mar [22] |
| Monosomy 17 and i(17q) | |||||
| 16 | M/55 | Yes | M2AML | Yes | 45,XY,del(5)(q13,q33),del(11)(p12),add(15)(q23),−16,−17,−20, +mar1,+mar( cp7) [13] |
| 17 | M/68 | No | RAEB | Yes | 46,XY,i(17)(q10) [14] /46,XY [3] |
| Patient No. . | Sex/Age . | Therapy Related . | FAB Type . | “Typical” Dysgranulopoiesis-151 . | Conventional Cytogenetic Findings . |
|---|---|---|---|---|---|
| t(5;17) translocation | |||||
| 1 | F/63 | No | M1AML | Yes | 43,XX,del(2)(p13),−3,del(5)(q11q23),der(5)t(5;17)(q23;q11),del(6)(p21), −7,add(9)(p13),add(12)(p11),del(15)(q15),−17 [17] /46,XX [1] |
| 2* | M/40 | No | RAEB-T | Yes | 42,XY,−3,−4,−5,−7,−11,−12,−16, der(17)t(5;17)(p11;p11),−20,+4mar [22] /46,XY [4] |
| 3 | M/71 | Yes | RAEB | Yes | 45,XY,−5, r(7), der(17)t(5;17)(p11;p12),+mar1 [7] /45,idem,del(3)(p13) [3] /46,XY [3] |
| 4 | M/68 | Yes | RAEB | Yes | 44,X,−Y,−3,−5,der(5)t(3;5)(p13;q31),der(17)t(5;17)(p11;p11),−18,+r, +mar [20] |
| 5 | M/75 | Yes | M2AML | Yes | 45,XY,−5, der(17)t inv(5;17)(p11q14;p11) [15] /46,XY [2] |
| 6 | M/82 | Yes | RAEB-T | Yes | 45,XY,−5, der(17)t(5;17)(p11;p11) [9] /46,idem,+8 [3] /45,XY,−5,add(16)(q21),−17,der(18)t(17;18)(q11;q23),+mar [1] |
| 7* | F/67 | Yes | RAEB | Yes | 44,XX,−5,−7, der(17)t(5;17)(p11;p11) [11] |
| 8 | M/57 | No | RAEB | No | 43-44,XY,add(2)(p11),−5,add(5)(p1?),der(5)t(5;17)(q13;q11),−7,−13,−17, del(20)(q12),−21,+(0-1)r,+mar1,+mar2 [7] |
| 9 | M/69 | No | RAEB | No | 44-46,XY, der(5)t(5;17)(q11;q11),del(7)(q21q34),+i(8)(q10), dup(13)(q11p13), ?add(16)(q11),−17,−(0-1)18,−21,+r [15] /46,XY [1] |
| Rearrangement between 17p and another chromosome | |||||
| 10 | M/62 | Yes | M5AML | Not evaluable | 44-46,XY,(0-1)add(2)(q35),der(7)t(1;7)(p11;p21),+(0-1)i(8)(q10), t(9;11)(p21;q23),−12,−(0-1)13,der(17)t(12;17)(q11;p11), (0-1)add(22)(p11),+(0-1)mar,(1-2)dmin(cp7) [16] |
| 11* | M/76 | No | RAEB-T | Yes | 43-45,XY,3,add(5)(p?),del(5)(q14q32),+11,add(11)(q23), der(12)t(12;17)(p12;q11),del(13)(q13q14),−17,−22,+mar,ace [cp4] |
| 12 | F/81 | No | M2AML | Yes | 41-42,XX,del(4)(q13),−5,der(9)t(9;10)(p21;q11),−12,t(13;15)(p10;p10), −15,add(16)(q11),der(17)t(17;21)(p11;q11),?i(21)(q10),−21,−22,add(22)(p11), +(1-3)mar [18] /46,XX [1] |
| 13 | F/75 | Yes | M2AML | Yes | 44-45,XX,del(5)(q11q34),−17, add(17)(p13), −18,t(?21;22)(q22;q11), mar(15),+mar1 [8] |
| 14 | F/71 | No | RAEB | Yes | 46-49,XX,del(5)(q13q33),−7,del(9)(q23q32), add(17)(p11), −18, (0-1)del(20)(q11),−21,+22,+(2-4)mar [11] /46,XX,del(5)(q13;q33) [1] /46,XX [2] |
| 15 | M/47 | Yes | RAEB | Yes | 44-45,XY,−(0-1)7,add(12)(p12),−(0-1)15, add(17)(p11), add(21)(q13), +(0-1)mar [22] |
| Monosomy 17 and i(17q) | |||||
| 16 | M/55 | Yes | M2AML | Yes | 45,XY,del(5)(q13,q33),del(11)(p12),add(15)(q23),−16,−17,−20, +mar1,+mar( cp7) [13] |
| 17 | M/68 | No | RAEB | Yes | 46,XY,i(17)(q10) [14] /46,XY [3] |
*Patients no. 2, 7, 11 had been reported in reference no. 7 as patients no. 7, 1 and 29, respectively.
Combining pseudo Pelger Huët hypolobulation and vacuoles in >5% neutrophils.
WCP.
WCP was performed in the 17 cases on metaphase spreads with cyanine-3 (Cy-3)–labeled WCP chromosome 17 probe and with fluorescein isothiocyanate (FITC)-labeled WCP probes for chromosomes 5, 7, 12, 18, 21, and 22 (Biovation, Aberdeen, UK). Hybridization was performed according to the manufacturer's protocol.
Slides were counterstained in Vectashield anti-fading medium (Vector Laboratories, Burlingame, CA) containing 4,6-diamino-2-phenylindole-dihydrochloride ([DAPI] 0.2 μg/mL).
FISH analysis with chromosome 17 markers, including the p53 gene.
To determine the extent of 17p deletion and whether the p53 gene located in 17p13.1 was deleted, we performed FISH analysis, in 16 of 17 cases with 3 yeast artificial chromosomes (YACs), 961 F10, 904 B5, and 914 C7, localized in 17p11.2, 17p11.2, and 17p12,15respectively, and with two probes specific for p53 and Miller-Dieker syndrome (MD) genes localized in 17p13.1 and 17p13.3, respectively. A chromosome 17 centromeric probe (D17Z1) and, in patients with t(5;17), a chromosome 5 centromeric probe (D5Z2) were also used. YACs were obtained from the Centre d'Etude du Polymorphisme Humain (Paris, France, courtesy of Dr D. Le Paslier), D17Z1 and p53 probes were from Vysis (Woodcreek Drive, IL), and MD and D5Z2 probe were from Oncor (Gaithersburg, MD).
Total yeast DNA was labeled by nick-translation with biotin-16-dUTP (Boehringer, Mannheim, Germany), and unincorporated nucleotides were removed on G50 Sephadex columns. The metaphase slides were first treated with RNase A and pepsin as described by Wiegnant et al.16 Hybridization was performed as described by Chaffanet et al.17 Briefly, the slides were denatured and hybridized at 37°C with 1 μg labeled probe in the presence of 30 μg human Cot 1 DNA (Boehringer) and 50 μg salmon sperm DNA (Sigma, St Louis, MO). Detection of the hybridized YAC probes was performed by indirect immunofluorescence using FITC-conjugated avidin (Vector Laboratories) in combination with biotinylated goat anti-avidin (Vector Laboratories). Vectashield anti-fading medium (Vector Laboratories) containing propidium iodide (0.4 μg/mL) was applied to the slides to stain double-stranded DNA.
D5Z2, D17Z1, p53, and MD probes were hybridized according to the conditions of the manufacturer. D17Z1 and p53 probes were directly labeled by Spectrum green and Spectrum orange, respectively. D5Z2 and MD probes were labeled by digoxigenin and detected by the FITC-conjugated antidigoxigenin (Oncor). DNA was counterstained with DAPI (0.2 μg/mL).
Fluorescence signals were visualized on a Zeiss Axioskop epifluorescence microscope (Zeiss, Oberkochen, Germany) equipped with a double- or triple-bandpass filter. Fluorescence images were captured using the Quips Smart Capture FISH Imaging Software (Vysis). For each slide, 15 metaphases were analyzed, on average.
Analysis of p53 mutations.
Detection of p53 mutations was made on DNA extracted from bone marrow cells by single-stranded conformation polymorphism (SSCP) analysis of exons 5 to 8 of the p53 gene and/or immunocytochemical analysis of p53 protein on bone marrow slides as previously reported.8,18 In our experience, overexpression of p53 by this method in MDS and AML is always associated with a p53 gene missense mutation.18
RESULTS
Interpretation of Chromosome Rearrangements by Conventional Cytogenetics, WCP, and FISH Analysis With Centromeric Probes
t(5;17) Translocation
Cytogenetically 9 cases (no. 1 to 9) had an unbalanced t(5;17) translocation with a breakpoint in 17p11 in 5 cases, in 17p12 in 1 case, and in 17q11 in 3 cases; the breakpoint on chromosome 5 was in 5p11 in 5 cases and in 5q11, 5q13, and 5q23 in 1 case each (Figs 1 and2).In the remaining patient with t(5;17) (no. 5), the translocation was associated with an inv(5)(p11q14). Cytogenetically, the derivative chromosome appeared to have a chromosome 17 centromere (der 17) in 6 cases and a chromosome 5 centromere (der 5) in 3 cases.
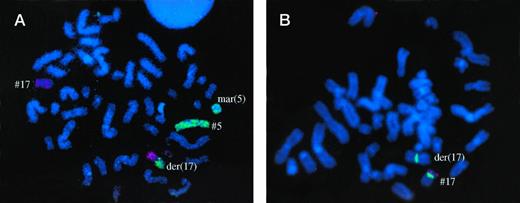
Patient no. 3, with typical der(17) t(5;17) by GTG banding and WCP, and loss of one p53 allele. Some chromosome 5 material was found on a marker chromosome.
Patient no. 3, with typical der(17) t(5;17) by GTG banding and WCP, and loss of one p53 allele. Some chromosome 5 material was found on a marker chromosome.
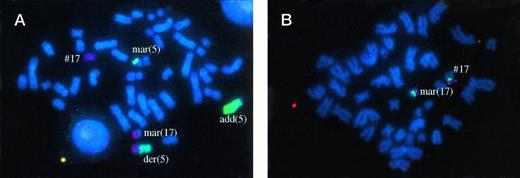
Patient no. 8, with der(5) t(5;17) by GTG banding and WCP. Non identified chromosome material was translocated on the other chromosome 5 (add(5)). Some chromosome 5 material was also found on a marker chromosome (mar(5)). Another marker chromosome contained chromosome 17 material which included the p53 gene (fig 2B) and other markers studied on 17p. This marker (mar(17)) also contained chromosome 7 material. Therefore, this patient apparently had no visible deletion of 17p material.
Patient no. 8, with der(5) t(5;17) by GTG banding and WCP. Non identified chromosome material was translocated on the other chromosome 5 (add(5)). Some chromosome 5 material was also found on a marker chromosome (mar(5)). Another marker chromosome contained chromosome 17 material which included the p53 gene (fig 2B) and other markers studied on 17p. This marker (mar(17)) also contained chromosome 7 material. Therefore, this patient apparently had no visible deletion of 17p material.
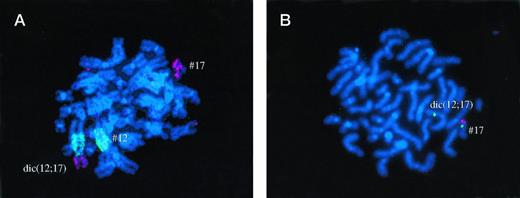
Patient no. 11, where RHG banding suggested presence of der(12) t(12;17). FISH analysis showed that chromosome 17 centromere was also present on the derivative chromosome, which was therefore a dic (12;17). One p53 allele was deleted.
Patient no. 11, where RHG banding suggested presence of der(12) t(12;17). FISH analysis showed that chromosome 17 centromere was also present on the derivative chromosome, which was therefore a dic (12;17). One p53 allele was deleted.
WCP confirmed involvement of chromosomes 5 and 17 in all cases. FISH analysis with centromeric probes for chromosomes 5 and 17, performed in 8 of the 9 cases to show the presence or absence of those centromeres in the derivative t(5;17) chromosome, confirmed conventional cytogenetics results in 7 cases. However, in patient no. 4, FISH analysis showed the presence of chromosome 5 centromere and the absence of chromosome 17 centromere on the derivative chromosome, contrary to conventional cytogenetics, demonstrating that it was a der(5) and not a der(17).
In patient no. 9 with t(5;17), WCP showed that the ring chromosome contained chromosome 17 material. In patient no. 8, one marker was shown to contain chromosome 17 and chromosome 7 material and the other to contain chromosome 5 material (Fig 2A). In patient no. 3, chromosome 5 material was also present on a marker chromosome (Fig 1A).
Rearrangement Between 17p and Another Chromosome
t(12;17).
Two patients cytogenetically had t(12;17) but with different breakpoints on chromosomes 12 and 17: der(17)t(12;17)(q11;p11) in patient no. 10 and der(12)t(12;17)(p12;q11) in patient no. 11. (Fig3A). WCP confirmed the involvement of chromosomes 12 and 17 and the presence of a chromosome 17 centromere in patient no. 10. However, in patient no. 11, both chromosome 12 and 17 centromeres were present on the derivative chromosome, which was therefore a dic(12;17) (Fig 3A).
t(17;21).
One patient (no. 12) cytogenetically had der(17)t(17;21)(p11;q11), an interpretation confirmed by WCP.
t(17;18).
In one of the patients with t(5;17) (no. 6), one mitose with der(18)t(17;18)(q11;q23), also resulting in 17p deletion, was observed. FISH analysis confirmed the cytogenetic interpretation.
add 17p.
In three patients (no. 13, 14, and 15), the chromosomal segment involved in the translocation with 17p was not identified by conventional cytogenetic analysis. In the three cases, WCP showed that chromosome 5 was not involved. In patients no. 13 and 14 but not patient no. 15, the derivative chromosome was shown to carry a chromosome 17 centromere. No WCP analysis for chromosomes other than no. 5 could be performed in patient no. 15 due to lack of available slides. WCP analysis could be made for chromosomes 5, 18, 21, and 22 in patients no. 13 and 14 and also for chromosomes 7 and 12 in patient no. 14. No involvement of those chromosomes in the translocation was found in patient no. 14. In patient no. 13, the translocation could be identified as der(17)t(17;22). Patient no. 13, in addition to a translocation involving the 17p13 band, had lost the nonrearranged chromosome 17 copy (Table 2).
WCP, FISH, and p53 Analysis in 17 MDS and AML Patients With 17p Deletion
| Patient No. . | WCP . | FISH Analysis . | P53 Analysis . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| D17Z1 Centromere 17 . | 961F10 17p11.2 D17S922 D17S921 D17S839 D17S955 . | 904B5 17p11.2 D17S799 D17S936 . | 914C7 17p12 D17S954 . | p53 17p13.1 . | MD 17p13.3 D17S379 . | p53 Mutation (SSCP) . | p53 Overexpression (immunocytochemistry) . | ||
| t(5;17) translocation | |||||||||
| 1 | der(5)t(5;17) | − | − | − | − | − | − | ND | ND |
| 2 | der(17)t(5;17) | + | + | + | − | − | − | Exon 5 | ND |
| 3 | der(5)t(5;17) | + | + | − | − | − | − | ND | Yes |
| 4 | der(5)t(5;17) | − | − | − | − | − | − | Exon 7 | Yes |
| 5 | der(17)t(5;17) | + | − | − | − | − | − | No | No |
| 6* | der(17)t(5;17) | + | − | − | − | − | − | Exon 5 | Yes |
| der(18)t(17;18) | − | ND | ND | ND | ND | ND | |||
| 7 | der(?)t(5;17) | ND | ND | ND | ND | ND | ND | ND | ND |
| 8 | der(5)t(5.17) | − | − | − | − | − | − | No | No |
| mar(7;17) | + | + | + | + | + | + | |||
| 9 | der(5)t(5;17) | − | − | − | − | − | − | ND | ND |
| r(17) | + | + | + | + | + | + | |||
| Rearrangement between 17p and another chromosome | |||||||||
| 10 | der(17)t(12;17) | + | + | + | − | − | − | Exon 8 | Yes |
| 11 | dic(12;17) | + | + | + | + | − | − | Exon 8 | Yes |
| 12 | der(17)t(17;21) | + | + | + | + | − | − | ND | Yes |
| 13 | der(17)t(17;22) | + | + | + | + | + | − | Exon 7 | Yes |
| −17 | − | − | − | − | − | − | |||
| 14 | add(17) | + | + | + | + | − | − | ND | Yes |
| 15 | add(17) | − | − | − | − | − | − | Exon 7 | ND |
| Monosomy 17 and i(17q) | |||||||||
| 16 | −17 | − | − | − | − | − | − | Exon 5 | Yes |
| 17 | idic(17)(p11) | ++ | − | − | − | − | − | ND | ND |
| Patient No. . | WCP . | FISH Analysis . | P53 Analysis . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| D17Z1 Centromere 17 . | 961F10 17p11.2 D17S922 D17S921 D17S839 D17S955 . | 904B5 17p11.2 D17S799 D17S936 . | 914C7 17p12 D17S954 . | p53 17p13.1 . | MD 17p13.3 D17S379 . | p53 Mutation (SSCP) . | p53 Overexpression (immunocytochemistry) . | ||
| t(5;17) translocation | |||||||||
| 1 | der(5)t(5;17) | − | − | − | − | − | − | ND | ND |
| 2 | der(17)t(5;17) | + | + | + | − | − | − | Exon 5 | ND |
| 3 | der(5)t(5;17) | + | + | − | − | − | − | ND | Yes |
| 4 | der(5)t(5;17) | − | − | − | − | − | − | Exon 7 | Yes |
| 5 | der(17)t(5;17) | + | − | − | − | − | − | No | No |
| 6* | der(17)t(5;17) | + | − | − | − | − | − | Exon 5 | Yes |
| der(18)t(17;18) | − | ND | ND | ND | ND | ND | |||
| 7 | der(?)t(5;17) | ND | ND | ND | ND | ND | ND | ND | ND |
| 8 | der(5)t(5.17) | − | − | − | − | − | − | No | No |
| mar(7;17) | + | + | + | + | + | + | |||
| 9 | der(5)t(5;17) | − | − | − | − | − | − | ND | ND |
| r(17) | + | + | + | + | + | + | |||
| Rearrangement between 17p and another chromosome | |||||||||
| 10 | der(17)t(12;17) | + | + | + | − | − | − | Exon 8 | Yes |
| 11 | dic(12;17) | + | + | + | + | − | − | Exon 8 | Yes |
| 12 | der(17)t(17;21) | + | + | + | + | − | − | ND | Yes |
| 13 | der(17)t(17;22) | + | + | + | + | + | − | Exon 7 | Yes |
| −17 | − | − | − | − | − | − | |||
| 14 | add(17) | + | + | + | + | − | − | ND | Yes |
| 15 | add(17) | − | − | − | − | − | − | Exon 7 | ND |
| Monosomy 17 and i(17q) | |||||||||
| 16 | −17 | − | − | − | − | − | − | Exon 5 | Yes |
| 17 | idic(17)(p11) | ++ | − | − | − | − | − | ND | ND |
*In patient no. 6, no t(17;18) cells could be analyzed by FISH.
Abbreviation: ND, not done.
Monosomy 17 and i(17q).
The two last patients had monosomy 17 (no. 16) and i(17)(q10) (no. 17), respectively. In patient no. 16, WCP confirmed monosomy 17. In patient no. 17, WCP and FISH analysis showed that the patient actually had an idic(17)(p11), as two signals for the chromosome 17 centromere probe were present.
Complex Cytogenetic Rearrangements
Sixteen patients had one or several additional chromosomal abnormalities, including the presence of unidentified ring or marker chromosome(s) in 12 cases. In particular, with the exception of patients no. 10, 15, and 17, cytogenetic rearrangements resulted in loss of part or all of the long arm of one chromosome 5. Patient no. 1 also had deletion of the long arm of the other chromosome 5.
FISH Analysis of 17p Segment
FISH analysis of 17p with three YACs and the p53 and MD probes was performed in 16 of 17 cases. Only one signal for some or all of the 17p probes was seen in 14 cases, and two signals for all 17p probes in the two remaining cases, patients no. 8 and 9. In those last two patients where, as before, WCP showed chromosome 17 material on a marker and ring chromosome, respectively, FISH analysis therefore strongly suggested that this material corresponded to all 17p material translocated from the derivative (5;17), although 17p microdeletions, undetectable with the probes used, could not be excluded.
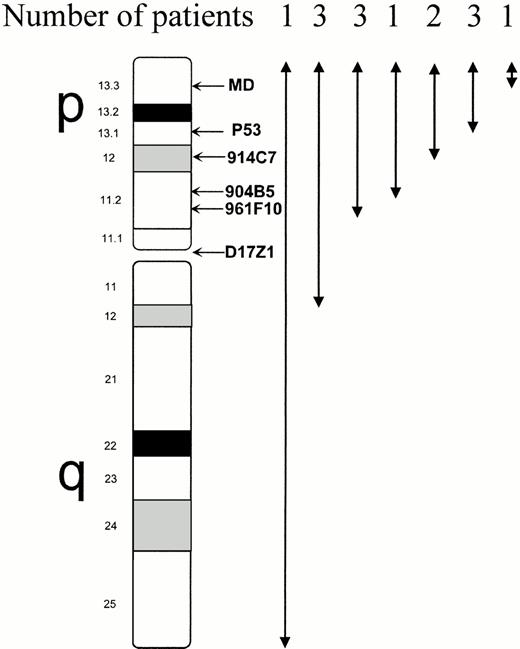
Among the 14 other patients, only one signal was found for all 5 17p probes in 7 cases; four of them also had loss of chromosome 17 centromere (D17Z1) signal, including the patient with monosomy 17 and 3 patients who cytogenetically had a breakpoint in 17q11. In the remaining 7 cases, only one signal was seen: for the 4 17p distal probes in 1 case, for the 3 distal probes in 2 cases, and for the 2 distal probes (p53 and MD) in three cases (Fig4); in the last patient (no. 13), the deletion involved MD probe only. (In this patient, in fact, the four proximal 17p markers showed only one signal, whereas MD showed no signal. This was due to the presence of both monosomy 17 and der(17) t(17;22)(p13;q?) leading to del 17p13).
Extent of 17p deletion analyzed by FISH using 5 markers spanning 17p and a chromosome 17 centromeric probe (D17Z1) in the 14 patients with confirmed 17p deletion. The deletion involved the p53 gene in all cases except one, but this patient (patient no. 13) had also monosomy 17, and therefore loss of one p53 allele. The proximal breakpoint was in 17q in 3 patients, as shown by deletion of D17Z1 probe, and located in 17q11 by conventional cytogenetic analysis. (FISH results in patients no. 8 and 9, where 17p deletion was not confirmed, are excluded from this figure).
Extent of 17p deletion analyzed by FISH using 5 markers spanning 17p and a chromosome 17 centromeric probe (D17Z1) in the 14 patients with confirmed 17p deletion. The deletion involved the p53 gene in all cases except one, but this patient (patient no. 13) had also monosomy 17, and therefore loss of one p53 allele. The proximal breakpoint was in 17q in 3 patients, as shown by deletion of D17Z1 probe, and located in 17q11 by conventional cytogenetic analysis. (FISH results in patients no. 8 and 9, where 17p deletion was not confirmed, are excluded from this figure).
FISH analysis was generally in agreement with cytogenetic analysis for determination of the breakpoint on chromosome 17. However, in patient no. 3, FISH analysis showed the breakpoint on chromosome 17 to be in rather 17p11 rather than 17p12, in patient no. 4 and no. 15 in 17q rather than 17p11, and in patient no. 12 and no. 14 in 17p12 rather than 17p11.
All 14 patients with loss of 17p material analyzed by FISH had only one signal for the p53 probe.
p53 Mutations and Myelodysplastic Features
Analysis of the p53 mutation by SSCP and/or analysis of p53 expression by immunocytochemistry could be made in 13 cases. In our experience in AML and MDS, p53 overexpression by immunochemistry on bone marrow slides is always associated with a p53 missense mutation.18 A mutation and/or overexpression of p53 was found in 11 cases, who all had 17p monosomy and loss of one p53 allele by FISH. No p53 mutation or overexpression was found in patient no. 8, who had no 17p deletion and no p53 deletion by FISH analysis, and in patient no. 5, although he had 17p deletion and loss of one p53 allele by FISH. In the other patient without 17p and p53 deletion by FISH (no. 9), analysis of p53 mutation and expression could not be made.
Dysgranulopoiesis could be assessed in 16 cases, as bone marrow blastic infiltration was massive in patient no. 10 and no mature granulocytes were present. Typical pseudo-Pelger-Huët hyposegmentation and small vacuoles in greater than five percent of the granulocytes was seen in 14 patients: 13 of them had 17p deletion and deletion of one p53 allele (the remaining patient was not studied by FISH), 10 had p53 mutation and/or overexpression, one had no p53 mutation, and three were not studied for p53. The remaining two patients (no. 8 and 9) had no dysgranulopoiesis and, as seen before, no 17p deletion by FISH and (in patient no. 8) no p53 mutation or overexpression.
Finally, no dysmegakaryopoiesis typical of that observed in the “5q-syndrome” (including the presence of large monolobulated megakaryocytes) was seen in any of the 10 patients where megakaryocytes were visible on marrow smears.
DISCUSSION
Relatively large numbers of cytogenetic rearrangements leading to 17p deletion have been reported in AML and MDS.19 They include monosomy 17, isochromosome 17q, and unbalanced translocations between chromosome 17 and another chromosome. The most frequent other chromosome involved in the unbalanced translocation was chromosome 5, followed by chromosomes 7, 12, 18, and 21, whereas other chromosomes were rarely implicated.20-25 However, with rare exceptions,25 data were only obtained by conventional cytogenetics and no WCP or other FISH analyses were performed.
We therefore studied WCP and FISH analysis with probes specific for 17p in 17 cases of AML and MDS with 17p deletion by conventional cytogenetics and where adequate material was available.6This analysis seemed important to perform because in our experience and in the literature AML and MDS with 17p deletion generally have complex karyotypes, with unidentified chromosome markers that could contain 17p material, and also because the chromosomal segment translocated on 17p sometimes cannot be identified.6 19
In the patients where conventional cytogenetics suggested unbalanced t(5;17), t(12;17), t(17;18), or t(17;21) translocation leading to 17p deletion, WCP confirmed the translocation between chromosome 17 and chromosome 5, 12, 18, and 21, respectively. WCP also confirmed conventional cytogenetic findings in one patient with monosomy 17. However, combination of WCP and FISH analysis with centromeric probes for chromosome 17 and partner chromosomes modified the interpretation of cytogenetic data in a few cases. In particular, one of the patients thought to have t(12;17) by conventional cytogenetics was shown to have dic(12;17), one patient classified as i(17q) by conventional cytogenetics was shown to have idic(17)(p11), and one patient classified as der(17)t(5;17) was reclassified as der(5)t(5;17).
In the three cases where the chromosome segment translocated to 17p could not be identified by conventional cytogenetics, WCP showed that it corresponded to chromosome 22q material in one case (no. 13). In another patient (no. 14), the partner chromosome could not be identified, but it was not chromosome 5, 7, 12, 18, 21, or 22. In the last case (no. 15), only chromosome 5 could be analyzed by WCP, ruling out t(5;17).
WCP and FISH analysis showed that in two cases of t(5;17) (patients no. 8 and 9), no 17p material appeared to be deleted. In those two cases, WCP showed chromosome 17 material on a marker chromosome and a ring chromosome, respectively. The presence of two copies of all 17p markers studied, including p53, was confirmed in both patients by FISH analysis. None of those two patients had typical dysgranulopoiesis, and p53 mutation was not found in the assessable case. By contrast, WCP and FISH analysis confirmed 17p deletion and loss of one p53 allele in the remaining patients. All had typical dysgranulopoiesis, and all but one of the assessable cases had p53 mutation and/or overexpression (which in our experience is synonymous with the presence of a p53 missense mutation in MDS and AML).18 These findings reinforce our previous results showing the very strong correlation in MDS and AML between 17p deletion, a typical form of dysgranulopoiesis, and p53 mutation.7 They bring further support to the description of MDS and AML with 17p deletion as a specific morphologic-cytogenetic-molecular entity.7 12
Of note, although in all but three patients cytogenetic rearrangements led to 5q deletion, no dysmegakaryopoiesis typical of the 5q− syndrome was seen. On the other hand, the typical dysmegakaryopoiesis of the 5q− syndrome has been mainly observed in cases of isolated del(5q) and is less frequent when del(5q) is associated with other chromosome changes, as in the cases reported here.26 Also of note was the presence of t(9;11)(p21;q23) in addition to t(12;17) in one patient (no. 10). This patient had previously received anthracycline-based chemotherapy for lymphoma and had M5AML, suggesting a typical case of therapy-related AML occurring after agents targeting topoisomerase II.27t(12;17) unbalanced translocation associated with the p53 mutation in this patient was possibly a secondary genetic event. Alternatively, as a relatively high proportion of AML and MDS cases with cytogenetic rearrangements leading to 17p deletion are therapy-related, antineoplastic drugs could have provoked several distinct genetic lesions, resulting in both t(9;11) and t(12;17) translocation.
FISH analysis with YACs spanning 17p confirmed the variability of the breakpoints and extent of the deletion of 17p, already suggested by conventional cytogenetics. Even in t(5;17) cases, the breakpoint on chromosome 17 was variable: the most frequent breakpoints were in 17p11, but breakpoints also occurred in 17p12, 17p13, and 17q. In the two cases with t(12;17), likewise, the breakpoint and extent of deleted 17p material was different in each patient. However, all but one of the patients with loss of 17p material had a breakpoint proximal to the p53 gene, which was deleted in all cases. The exception was patient no. 13, where the 17p breakpoint occurred between p53 and MD marker, situated in p13.3. However, this patient has also lost one p53 allele, through loss of the other chromosome 17 copy. Although no FISH analysis of chromosome 5 and chromosome 12 breakpoints was performed in this study, cytogenetic results showed, as for 17p, variable breakpoints on these two chromosomes. These results confirm previously published cytogenetic data in unbalanced translocations resulting in 17p deletion, which found breakpoints in 17p11, 17p12, or 17q11 and rarely in 17p13, and also variable breakpoints on partner chromosomes.19
Precise analysis of larger numbers of unbalanced translocations involving 17p and different chromosomes may in the future identify discrete breakpoint clusters on chromosome 17, where genes involved in leukemogenesis could be located, as in the example of the MLLgene in 11q23 or TEL gene in 12p13. However, the variable extent of 17p deletion we observed in this study rather suggests the presence of tumor suppressor gene(s) on 17p, inactivated by the deletion. The p53 gene could be a good candidate, or one of the candidate genes. Indeed, in patients with deletion of 17p material, deletion of one p53 allele was found in all cases and mutation of the non deleted p53 allele in all but one case. A pathogenetic role of p53 inactivation in neoplasia has been shown, especially in experiments of gene transfer of a normal p53 gene in tumor cells with p53 inactivation, where loss of tumorigenicity was found.28 In the case of hematologic malignancies, it is strongly suspected that p53 inactivation plays a role in the progression of CML to AML,11 of chronic lymphocytic leukemia to Richter's syndrome, and of low grade NHL to large cell lymphoma.29 In MDS and AML, 17p deletion is associated with advanced disease, low response to chemotherapy, and short survival.7,8 30 In those patients, p53 inactivation could play a role in disease progression and also explain the presence of complex cytogenetic abnormalities, as p53 inactivation can generate gene instability.
We confirmed in this study the close correlation between typical dysgranulopoiesis combining pseudo-Pelger-Huët hypolobulation and small vacuoles in neutrophils, 17p deletion, and p53 mutation. However, the relationship between this typical dysgranulopoiesis and p53 mutation in MDS and AML with 17p deletion is unclear. We had previously shown that these dysgranulopoietic features did not correspond to images of apoptosis.7 Furthermore, one would expect p53 inactivation to result in reduction rather than increase in the number of apoptotic cells. Whether this typical dysgranulopoiesis results from inactivation of one of several genes involved in granulopoiesis and situated on 17p (and therefore lost by 17p deletion) will have to be further explored.
Fig 1C, 2C, 3C. Partial karyotypes showing rearrangements of chromosome 17 (arrow).
Fig 1A, 2A, 3A. Whole chromosome painting (WCP) with cyanine-3–labeled probe for chromosome 17 and FITC-labeled probes for chromosome 5 (Fig 1A and 2A), or chromosome 12 (Fig 3A). Fig 1B, 2B, 3B. FISH analysis with a Spectrum green–labeled probe for chromosome 17 centromere (D17Z1) and with a Spectrum orange–labeled probe for the p53 gene.
ACKNOWLEDGMENT
The authors are indebted to Anne-Sophie Legrain and Dominique Pruvot for expert technical assistance.
V.S. and C.P. contributed equally as first authors of this work.
Supported by the Fondation contre la Leucémie, the Association de Recherche sur le Cancer, and the Centre Hospitalier Universitaire de Lille.
Address reprint requests to Pierre Fenaux, MD, Service des Maladies du Sang, C.H.U., 1 Place de Verdun, 59037 Lille, France.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal