Abstract
We describe a dominantly inherited β-thalassemia intermedia phenotype observed in a five-generation Portuguese family. Carriers are characterized by moderate anemia, hypochromia, microcytosis, elevated hemoglobin (Hb)A2 and HbF levels, splenomegaly, hepatomegaly, and inclusion bodies in pheripheral red blood cells after splenectomy. The molecular basis of this condition is a small deletion within the 5′ consensus splicing sequence of the second intron of the β-globin gene, IVS-II-4,5 (-AG). Reticulocyte RNA studies performed by reverse transcription-polymerase chain reaction (RT-PCR) and primer extension analysis showed three abnormally processed transcripts, which, upon sequencing, were shown to correspond to (1) skipping of exon 2, and (2) activation of two cryptic splice sites (between codons 59/60, and at IVS-II-47). In vitro translation studies of these patients' reticulocyte RNA have shown that at least one of these aberrant mRNA species is translated into an abnormally elongated peptide whose cytotoxic properties could, in part, be causing the atypical dominant mode of inheritance observed in this family. We suggest that this elongated β chain is unable to combine with an α-globin chain to form a functional Hb molecule. Its degradation would, then, exhaust the proteolytic defense mechanism of the erythroid precursors, leading to inefficient proteolysis of the free α chains in excess.
THE β-THALASSEMIAS ARE usually transmitted as autosomal-recessive disorders.1 However, some dominant forms of β-thalassemia have been identified in individuals who have inherited a single copy of an abnormal β-globin gene and a normal α-globin genotype. Thalassemia intermedia with mild anemia, jaundice, and splenomegaly was observed in these patients, as well as elevated hemoglobin (Hb)A2 and HbF levels, unbalanced α-/β-chain synthesis ratio, and presence of inclusion bodies in the erythroid precursors and peripheral red blood cells after splenectomy.2,3 The molecular basis of these dominant β-thalassemias is heterogeneous, with the majority of them being associated with mutations in the third exon, and a few located in the first and second exons of the β-globin gene4: frameshift and nonsense mutations or complex rearrangements lead to the synthesis of highly unstable truncated or elongated β-globin products.3 The main factors that determine the phenotype appear to be the length of the globin gene product, its ability to bind heme or to form functional α/β dimers and α2/β2 tetramers, and the stability of the latter in the developing erythroid precursors and in peripheral red blood cells. The continuous degradation of these nonfunctional β chains adds an extra burden to the proteolytic defense mechanism of the erythrocytic precursors, such that proteolysis of the free α chains is compromised. This leads to accumulation and precipitation of α chains to a greater extent than observed in the classical asymptomatic β-thalassemia heterozygotes. In some cases, the precipitation of the abnormal β chain is observed.3 5
Most of the mammalian genes are interrupted by introns, which are removed from mRNA precursors by the splicing machinery. At the 5′ and 3′ ends of each intron, dinucleotides GT and AG, respectively, are invariably present.6,7 Flanking these invariant dinucleotides are sequences that are fairly well conserved. Mutations within these sequences in the β-globin gene have been described that reduce, to various degrees, the efficiency of normal splicing, giving rise to abnormal globin mRNAs and producing a β+-thalassemia phenotype.8-10
Here, we describe the functional effects of a dominantly transmitted β-thalassemia determinant, detected in a large Portuguese family presenting β-thalassemia intermedia.11 The molecular basis of this condition is a deletion of nucleotides 4 and 5 of the β-globin gene IVS-II consensus donor splicing sequence, which leads, in vivo, to three abnormally spliced β-globin mRNAs and, in vitro, to an abnormal β-globin peptide. This β-globin gene mutation provides another good model for investigating the relationship between β-globin gene structure and function.
MATERIALS AND METHODS
Subjects.
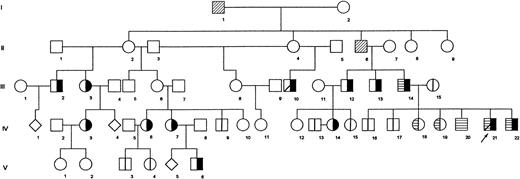
In this study, we analyzed 24 members of a five-generation Portuguese family in which β-thalassemia intermedia is inherited as a Mendelian autosomal-dominant condition (Fig 1). The disease, characterized by moderate anemia with jaundice, splenomegaly, and hepatomegaly, is transmitted vertically through different generations. The propositus (IV21) received occasional blood transfusions until 11 years of age, when he underwent a splenectomy. Before this, his spleen and liver were enlarged (9 cm and 7.5 cm below the respective costal margins). Other members of the family (IV3 and IV7) also underwent splenectomy.
Pedigree of Portuguese family shows segregation of dominantly transmitted β-thalassemia determinant. Propositus is marked by arrow. (╞) βAβthal; (⊟) αααanti3.7/αα; (│) normal tested; (▨) dead; (◊) sex unknown, individual not studied. All remaining individuals were not studied.
Pedigree of Portuguese family shows segregation of dominantly transmitted β-thalassemia determinant. Propositus is marked by arrow. (╞) βAβthal; (⊟) αααanti3.7/αα; (│) normal tested; (▨) dead; (◊) sex unknown, individual not studied. All remaining individuals were not studied.
Hematologic analysis.
Red blood cell indices were obtained with an automated cell counter. Hb analysis was performed by cellulose acetate electrophoresis at pH 8.4, isoelectrofocusing (IEF), and reverse-phase high-performance liquid chromatography (HPLC). HbA2 was quantitated by DEAE cellulose chromatography and HbF was determined by an alkali denaturation method.12 Serum iron and serum ferritin were assayed by standard methods. Globin chain synthesis was determined as previously described.13 Inclusion bodies were detected in perypheral erythrocytes after incubation with brilliant cresyl blue.14 To detect unstable Hb, the isopropanol precipitation test was performed as previously described.15
DNA analysis.
Total genomic DNA was isolated from peripheral leukocytes by a salting-out procedure,16 followed by micro phenol/chloroform extraction. β-globin haplotyping17 was performed by Southern blotting18,19 or by digestion of the appropriate polymerase chain reaction (PCR) product.20 The α-globin gene cluster was mapped by Southern blotting by usingBamHI and BglII and genomic probes specific for ζ- and α-globin genes, or by enzymatic amplification analysis using two primer sets that allow the specific amplification of the α1- and α2-globin genes.21Allele-specific oligonucleotides22 or restriction endonuclease analysis of amplified DNA were used to investigate the most frequent β-thalassemia mutations found in the Mediterranean populations.11 Both oligonucleotide and genomic probes were radioactively labeled with phosphorus 32.22,23 The sequencing of the β-globin genes from the propositus was performed on amplified double-stranded DNA by the dideoxy method24 using the Sequenase Kit Version 2.0 (US Biochemical, Cleveland, OH).
RNA analysis.
Total RNA was isolated from peripheral reticulocytes by phenol extraction of acid-precipitated polysomes as previously described.25 One microgram of total RNA was reverse-transcribed into cDNA by the random hexamer priming method using AMV reverse transcriptase (RT; Pharmacia Biotech, Uppsala, Sweden) at 42°C for 1 hour. The β-globin cDNA was then enzymatically amplified using the following primer set located within exon 1 and exon 3 of the β-globin gene: 5′-AAGTCTGCCGTTACTGCCCT-3′ (forward) and 5′-CACTTTCTGATAGGCAGCCTGC-3′ (reverse). The amplification products were then electrophoresed on a denaturant urea/formamide/polyacrylamide vertical gel and visualized directly upon ethidium bromide staining. Each fragment was extracted from the gel, reamplified with a nested pair of primers, purified, and sequenced by the dideoxy method.24 Quantitative analysis of the β-globin mRNAs was performed by primer extension. The reverse primer 5′-GTGATACTTGTGGGCCAGAT-3′, located at exon 3 of the β-globin gene, was 5′ end-labeled with [γ32P]adenosine triphosphate (ATP),22 hybridized to 1 μg of total reticulocyte RNA, and extended with RT as previously described.26 The products obtained were separated by 6% polyacrylamide gel electrophoresis under denaturing conditions. Band intensities on autoradiographs were quantitated by densitometry (Sharp Scanner JX-330; Image Master Software Phoretix, Pharmacia Biotech, Uppsala, Sweden).
Protein analysis.
One microgram of total RNA was translated in vitro27 at 30°C for 1 hour, using a micrococcal nuclease-treated rabbit reticulocyte lysate (Promega, Madison, WI) in the presence of L-[35S]methionine (Amersham, Buckinghamshire, England). The labeled translation products were separated on a TritonX-100/acid/urea 12% polyacrylamide gel28 and autoradiographed. The α-/β-globin biosynthetic ratio was determined by excising from the dried gel the newly synthesized α- and β-globin chains and quantification by liquid scintillation counting (Beckman Instruments, Fullerton, CA). As a blank control, a piece of gel of the same size was used. To determine the molecular weight of the in vitro translation products, these were excised from the Triton X-100/acid/urea-dried gel and separated on a 18% sodium dodecyl sulfate (SDS)-polyacrylamide gel.29
RESULTS
Hematologic analysis.
The patients investigated in this study showed a hypochromic, microcytic moderate anemia. The blood smear showed anisocytosis, poikilocytosis, target cells, some erythroblasts, and basophilic stippling in the erythrocytes. Separation of Hb fractions either by cellulose acetate electrophoresis or isoelectrofocusing failed to detect any abnormal Hbs. The levels of HbA2 and HbF were increased in the affected members. A marked decrease in β-globin chain synthesis was observed. Brilliant cresyl blue staining showed a large percentage of inclusion bodies in the peripheral erythrocytes of the splenectomized patients. The available hematologic and globin genotypic data in the studied family members are listed in Table1.
Available Hematologic and Globin Genotypes in Family Members Studied
| Subject . | RBC Count (×1012/L) . | Hb (g/dL) . | MCV (fL) . | MCH (pg) . | HbA2 (%) . | HbF (%) . | Reticulocytes (%) . | Ferritin (ng/mL) . | Non α/α . | Genotype . | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| α . | β . | ||||||||||
| III2 | 4.2 | 9.3-10.1 | 72 | 23.2 | 4.6 | 4.2 | — | 1,300 | — | αα/αα | βA/βthal |
| III3 | 4.2 | 7.7-9.0 | 65-66 | 20.0 | 4.2 | 1.7 | — | — | — | αα/αα | βA/βthal |
| III10 | — | 8.1 | 75 | — | 4.5 | 1.2 | — | — | — | αα/αα | βA/βthal |
| III13 | 4.3 | 9.6-11.4 | 66 | 20.7 | 4.0-4.6 | 1.8-2.1 | — | — | — | αα/αα | βA/βthal |
| III14 | 4.7 | 8.6-10.7 | 66-70 | 20.6 | 3.7 | 1.7-4.0 | 6 | 922 | — | ααα/αα | βA/βthal |
| III15 | — | 13.0 | 95 | — | 2.7 | 1.0 | — | — | 0.82 | αα/αα | βA/βA |
| IV3 | — | 8.7 | 75 | — | 3.8 | 4.8 | — | — | — | αα/αα | βA/βthal |
| IV6 | 4.6 | 9.0-10.4 | 64-68 | 21.2 | 5.0-5.4 | 0.8-0.9 | — | 241 | — | αα/αα | βA/βthal |
| IV7 | 3.9 | 8.7 | 76 | 21.9 | 3.9-5.0 | 4.5-4.7 | 4.7 | — | — | αα/αα | βA/βthal |
| IV14 | 4.1-4.4 | 9.6-9.7 | 76-78 | 21.6-23.8 | 4.7 | 3.5 | — | — | — | αα/αα | βA/βthal |
| IV16 | 4.6 | 16.2 | 101 | 35.6 | 2.8 | 0.7 | 2.5 | 120 | — | ααα/αα | βA/βA |
| IV17 | 5.0 | 17.1 | 99 | 34.5 | 1.7 | 0.3 | 0.6 | 235 | — | αα/αα | βA/βA |
| IV18 | 4.5 | 14.7 | 93 | 32.6 | 1.5 | 0.4 | 1.4 | 29.2 | — | ααα/αα | βA/βA |
| IV19 | 4.4 | 13.9 | 96 | 31.8 | 1.3 | 0.4 | 0.4 | 135 | — | ααα/αα | βA/βA |
| IV20 | 5.2 | 17.2 | 96 | 33.4 | 1.7 | 0.6 | 0.8 | 73.1 | — | ααα/αα | βA/βA |
| IV21 | 4.3 | 8.5-9.5 | 73 | 20.9 | 3.5-7.0 | 7.0 | 20.2 | 2,500 | 0.35 | ααα/αα | βA/βthal |
| IV22 | 4.8 | 7.6-9.3 | 68 | 22 | 4.0-5.8 | 4.5-7.0 | 4.5 | 175 | 0.41 | ααα/αα | βA/βthal |
| V6 | 3.3 | 7.3-7.6 | 72-76 | 23.0-23.9 | 4.8-5.1 | 4.0-5.2 | 2.2 | — | — | αα/αα | βA/βthal |
| Subject . | RBC Count (×1012/L) . | Hb (g/dL) . | MCV (fL) . | MCH (pg) . | HbA2 (%) . | HbF (%) . | Reticulocytes (%) . | Ferritin (ng/mL) . | Non α/α . | Genotype . | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| α . | β . | ||||||||||
| III2 | 4.2 | 9.3-10.1 | 72 | 23.2 | 4.6 | 4.2 | — | 1,300 | — | αα/αα | βA/βthal |
| III3 | 4.2 | 7.7-9.0 | 65-66 | 20.0 | 4.2 | 1.7 | — | — | — | αα/αα | βA/βthal |
| III10 | — | 8.1 | 75 | — | 4.5 | 1.2 | — | — | — | αα/αα | βA/βthal |
| III13 | 4.3 | 9.6-11.4 | 66 | 20.7 | 4.0-4.6 | 1.8-2.1 | — | — | — | αα/αα | βA/βthal |
| III14 | 4.7 | 8.6-10.7 | 66-70 | 20.6 | 3.7 | 1.7-4.0 | 6 | 922 | — | ααα/αα | βA/βthal |
| III15 | — | 13.0 | 95 | — | 2.7 | 1.0 | — | — | 0.82 | αα/αα | βA/βA |
| IV3 | — | 8.7 | 75 | — | 3.8 | 4.8 | — | — | — | αα/αα | βA/βthal |
| IV6 | 4.6 | 9.0-10.4 | 64-68 | 21.2 | 5.0-5.4 | 0.8-0.9 | — | 241 | — | αα/αα | βA/βthal |
| IV7 | 3.9 | 8.7 | 76 | 21.9 | 3.9-5.0 | 4.5-4.7 | 4.7 | — | — | αα/αα | βA/βthal |
| IV14 | 4.1-4.4 | 9.6-9.7 | 76-78 | 21.6-23.8 | 4.7 | 3.5 | — | — | — | αα/αα | βA/βthal |
| IV16 | 4.6 | 16.2 | 101 | 35.6 | 2.8 | 0.7 | 2.5 | 120 | — | ααα/αα | βA/βA |
| IV17 | 5.0 | 17.1 | 99 | 34.5 | 1.7 | 0.3 | 0.6 | 235 | — | αα/αα | βA/βA |
| IV18 | 4.5 | 14.7 | 93 | 32.6 | 1.5 | 0.4 | 1.4 | 29.2 | — | ααα/αα | βA/βA |
| IV19 | 4.4 | 13.9 | 96 | 31.8 | 1.3 | 0.4 | 0.4 | 135 | — | ααα/αα | βA/βA |
| IV20 | 5.2 | 17.2 | 96 | 33.4 | 1.7 | 0.6 | 0.8 | 73.1 | — | ααα/αα | βA/βA |
| IV21 | 4.3 | 8.5-9.5 | 73 | 20.9 | 3.5-7.0 | 7.0 | 20.2 | 2,500 | 0.35 | ααα/αα | βA/βthal |
| IV22 | 4.8 | 7.6-9.3 | 68 | 22 | 4.0-5.8 | 4.5-7.0 | 4.5 | 175 | 0.41 | ααα/αα | βA/βthal |
| V6 | 3.3 | 7.3-7.6 | 72-76 | 23.0-23.9 | 4.8-5.1 | 4.0-5.2 | 2.2 | — | — | αα/αα | βA/βthal |
Abbreviations: RBC, red blood cells; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin.
DNA analysis.
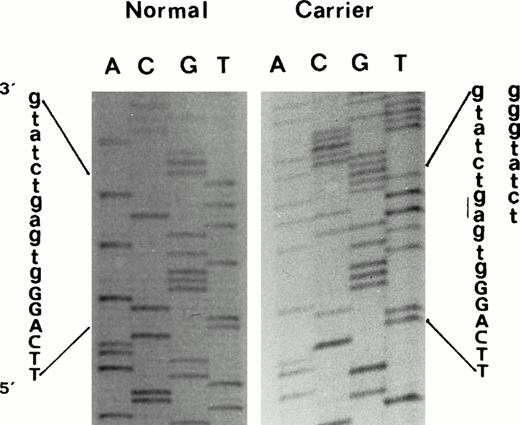
Seven of the common βthal mutations present in Mediterranean populations were not found in the propositus. However, the sequencing of the β-globin genes showed a two-nucleotide deletion (-AG) within the IVS-II 5′ splice site consensus sequence [IVS-II-4,5 (-AG)]11 (Fig 2). As this deletion eliminates a normal HinfI restriction site, its presence was easily confirmed in other affected family members by restriction digestion of the appropriate β-globin gene PCR fragment. This genetic alteration was not found in any of the nonaffected family members, or in a number of normal controls. Haplotype analysis in the β-globin gene cluster demonstrated that this β-thalassemia mutation was linked to haplotype Va.17 We also observed, in a branch of this family, the segregation of triplicated α-globin gene haplotype αααanti3.7. Three patients (III14, IV21, and IV22) were double heterozygotes for triplicated α-globin gene and the βthal mutation.
Direct sequencing of exon2/intron2 β-globin gene region of amplified double-strand DNA from propositus, using a forward primer, which shows the 2-nucleotide deletion (-AG) at position IVS-II-4,5. Deletion was confirmed by sequencing the same DNA region using an appropriate reverse primer.
Direct sequencing of exon2/intron2 β-globin gene region of amplified double-strand DNA from propositus, using a forward primer, which shows the 2-nucleotide deletion (-AG) at position IVS-II-4,5. Deletion was confirmed by sequencing the same DNA region using an appropriate reverse primer.
RNA analysis.
In an attempt to characterize the functional effect of the IVS-II-4,5 (-AG) mutation on the processing of the β-globin gene primary transcript, we analyzed the peripheral reticulocyte β-globin mRNA in the propositus and in normal controls. By RT-PCR, we detected four β-globin mRNA species of different sizes (Fig3A), which, upon sequencing, turned out to be (1) a normal β-globin mRNA; (2) an aberrant mRNA, 224 nt shorter than normal due to exon 2 skipping; (3) an aberrant mRNA, 135 nt shorter than normal, resulting from the activation of a cryptic splice site in exon 2 between codons 59/60 (AG/gtgaag); and (4) an aberrant mRNA, 45 nt longer than normal, resulting from the activation of a cryptic splice site at IVS-II-47 (TG/gttaag). These are schematically represented in Fig 3B. Quantitative analysis of the abnormal β-globin mRNAs species was performed by primer extension followed by densitometric scanning of the gel autoradiograph (Fig 3D). This failed to show any significant difference in the relative abundance of the three abnormal mRNA species.
Effect of dominantly inherited β-thalassemia mutation on β-globin expression. (A) Denaturant urea/formamide/polyacrylamide vertical gel electrophoresis of fragments obtained by RT-PCR from reticulocyte β-globin mRNAs extracted from (1) a carrier (IV7) of IVS-II-4,5(-AG) mutation (4 β-globin mRNAs species of different size were found, which, on sequencing, were shown to correspond to the normal β-globin mRNA [N], exon 2 skipping [E2S], and activation of 2 cryptic splice sites between codon 59/60 [CS 59/60] and at IVS-II-47 [CS IVS-II-47]); (2) a normal β-globin control; and (3) DNA molecular weight marker (pBR322 + BglI + HinfI). (B) Schematic representation of the 3 abnormally processed β-globin transcripts. Arrows under exons represent primers used in RT-PCR. x, stop codon position. (C) Amino acid sequence of normal human β-globin chain and predicted amino acid sequence corresponding to various aberrant β-globin transcripts. Differences in amino acid sequence are underlined. Deleted residues are indicated by dashed line. (D) Primer extension analysis of β-globin mRNA (using a reverse primer located at β-globin gene exon 3, 5′ end-labeled with [32P]dATP) in (1) a normal control; (2) a carrier of IVS-II-4,5 (-AG) mutation (showing the normal and 3 abnormal mRNA species, which, upon densitometric scanning, showed no significant difference in its relative abundance; and (3) 5′ end-labeled DNA molecular weight marker (pGEM 3 + HinfI).
Effect of dominantly inherited β-thalassemia mutation on β-globin expression. (A) Denaturant urea/formamide/polyacrylamide vertical gel electrophoresis of fragments obtained by RT-PCR from reticulocyte β-globin mRNAs extracted from (1) a carrier (IV7) of IVS-II-4,5(-AG) mutation (4 β-globin mRNAs species of different size were found, which, on sequencing, were shown to correspond to the normal β-globin mRNA [N], exon 2 skipping [E2S], and activation of 2 cryptic splice sites between codon 59/60 [CS 59/60] and at IVS-II-47 [CS IVS-II-47]); (2) a normal β-globin control; and (3) DNA molecular weight marker (pBR322 + BglI + HinfI). (B) Schematic representation of the 3 abnormally processed β-globin transcripts. Arrows under exons represent primers used in RT-PCR. x, stop codon position. (C) Amino acid sequence of normal human β-globin chain and predicted amino acid sequence corresponding to various aberrant β-globin transcripts. Differences in amino acid sequence are underlined. Deleted residues are indicated by dashed line. (D) Primer extension analysis of β-globin mRNA (using a reverse primer located at β-globin gene exon 3, 5′ end-labeled with [32P]dATP) in (1) a normal control; (2) a carrier of IVS-II-4,5 (-AG) mutation (showing the normal and 3 abnormal mRNA species, which, upon densitometric scanning, showed no significant difference in its relative abundance; and (3) 5′ end-labeled DNA molecular weight marker (pGEM 3 + HinfI).
Protein analysis.
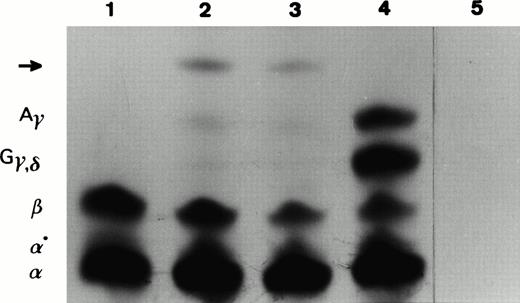
With the aim to detect the translation products of the abnormal β-globin mRNAs, we in vitro–translated patient (IV7, IV14, and V6) and normal control total reticulocyte RNA. The products obtained were separated according to their electrical charge on a TritonX100/acid/urea polyacrylamide gel. In the patients, we detected an abnormal slower-moving band in addition to the normal globins (Fig4). In vitro translation time-course experiments were performed in which aliquots were removed at regular intervals (0, 10, 20, 30, and 60 minutes) and applied to the same type of gel. No abnormal unstable peptide was observed. To characterize the abnormal peptide above by its molecular weight, we excised the pieces of dried gel that corresponded to the abnormal peptide and normal globin chains and loaded them on a 18% SDS-polyacrylamide gel. Under these experimental conditions, the abnormal slower-moving band showed a molecular weight higher than normal globins, suggesting it was the translation product of the longer abnormal β-globin mRNA (CS IVS-II-47). In vitro translation of reticulocyte RNA from a patient (IV14) showed a β-/α-globin chain synthesis ratio similar to the one observed in β-thalassemia heterozygotes: β/α = 0.53. This ratio clearly indicates a reduction of the β-globin chain biosynthetic capacity.
In vitro translation of total reticulocyte RNA isolated from peripheral blood samples of (1) a normal β-globin control, (2 and 3) a carrier (IV7, V6) of IVS-II-4,5 (-AG) mutation, (4) a newborn normal β-globin control, and (5) an in vitro translation reaction to which no exogenous RNA was added. The 35S-labeled translation products were resolved in a Triton-acid-urea polyacrylamide gel. The position of α, α°, β, Gγ, δ, andAγ globins are indicated on the left of the autoradiograph. Abnormal peptide is marked by an arrow.
In vitro translation of total reticulocyte RNA isolated from peripheral blood samples of (1) a normal β-globin control, (2 and 3) a carrier (IV7, V6) of IVS-II-4,5 (-AG) mutation, (4) a newborn normal β-globin control, and (5) an in vitro translation reaction to which no exogenous RNA was added. The 35S-labeled translation products were resolved in a Triton-acid-urea polyacrylamide gel. The position of α, α°, β, Gγ, δ, andAγ globins are indicated on the left of the autoradiograph. Abnormal peptide is marked by an arrow.
DISCUSSION
The study of naturally occurring mutations within the human β-globin gene cluster has greatly contributed to understand the mechanisms of gene expression. Most of the naturally occurring mutations and the in vitro–generated mutations that reduce the complementarity of the 5′ donor splice site with U1 SnRNP result in authentic 5′ splice-site function inactivation or reduction and several aberrant splicing events occur.30
Here, we describe a dominantly inherited β-thalassemia mutation associated with a thalassemia intermedia phenotype, located near the 5′ splice junction of intron 2 of the β-globin gene, IVS-II-4,5 (-AG), and characterize its effect on gene expression, namely, on RNA splicing. The analysis of reticulocyte RNA isolated from these patients showed three abnormally processed β-globin transcripts. The abnormal mRNA species originated by exon 2 skipping is 224 nt shorter than normal. The skipping of exon 2 results in a shift of the reading frame, which leads to a partial readthrough of the 3′ untranslated sequence of β globin mRNA until a new in-phase termination codon is encountered 11 codons downstream. If this aberrant mRNA were translated, the corresponding peptide would have an extremely altered amino acid sequence from residue 29 through to its C-terminal end (Fig 3C). Certainly, it would not be a functional globin chain. The other aberrant mRNA species, 135 nt shorter than normal, results from the activation of a cryptic splice site between codons 59/60 in exon 2 keeping the reading in frame. So, the possible peptide originated by its translation would have 45 fewer amino acid residues (59-104) than normal, corresponding to the 3′ portion of the second exon (Fig 3C). This truncated β-globin gene product is expected not to bind heme, as drastic alterations in its structure occur and therefore leave it nonfunctional. Finally, the aberrant transcript that results from the activation of a cryptic splice site in the IVS-II-47 is 45 nt longer than normal and keeps the reading frame. The β-globin chain corresponding to this abnormal mRNA species would have an insertion of 15 amino acid residues between R104 and L105 (Fig 3C), within helix-G (G5) of the β-globin chain. Four α1β1 contact points thought to be essential for dimer formation and, subsequently, for the Hb tetramer assembly are located within G-helix at positions 108, 112, 115, and 116.31 32 The introduction of 15 amino acid residues within G-helix would probably interfere with those contact points, preventing the abnormal β chain from combining with α chains to form a Hb tetramer, thus leading to ineffective erythropoiesis. However, it is possible that heme binding is responsible for the maintenance of some native secondary structure and probably this chain is less susceptible to proteolytic degradation than those without heme. The continuous degradation of these abnormal, nonfunctional β variants would add an extra burden to the proteolytic defence mechanism of the erythroid precursors, such that proteolysis of free α chains is compromised. It is therefore probable that the unstable, elongated β chain, in addition to the concomitant excess of α chains, precipitates in the differentiating red blood cell to form inclusion bodies. These, in fact, are observed in patients' blood smears after splenectomy.
Our studies performed by in vitro translation (including a time-course incubation) have shown that one of the abnormal mRNA species was translated into protein (Fig 4). We observed that the abnormal peptide has a molecular weight higher than the normal globin chains, suggesting it is the translation product of the abnormal mRNA resulting from the activation of a cryptic splice site at IVS-II-47.
The accurate and efficient selection of both 5′ and 3′ splice sites is clearly a complex process. Consensus values (CV), as a measure of complementarity to U1 snRNP, were calculated for the normal and mutated 5′ splice site sequences of the β-globin IVS-II and for the two activated cryptic splice site sequences using the method described by Shapiro and Senapathy.7 The mutation originates a drastic decrease in the score of the 5′ splice site sequence from 0.889 (GG/gtgagt) to 0.634 (GG/gtgtct). The selection of these cryptic splice sites can be understood on the basis of its higher score: 0.782 (AG/gtgaag), 135 nt upstream, and 0.739 (TG/gttaag) 45 nt downstream. Following this reasoning, selection of one 3′ splice site, out of the two possible sites lying in intron I or II, should be determined by the strength of their splice signals. The score of the 3′ splice site of the second intron is much higher than that of the first intron.33 In this way, the 3′ splice site of the second intron is powerfully selected, giving rise to exon 2 skipping, as observed in this case.
Several point mutations within the intron 5′ splicing consensus region and their effects on gene expression have been reported in a number of human disorders.34 The only other βthalmutation located in the IVS-II 5′ splice site sequence that has been characterized at functional level, IVS-II-1 (G → A), results in two abnormal β-globin mRNAs: the predominant RNA species, with an insertion of the first 47 nucleotides of the IVS-II between exons 2 and 3, and a minor species resulting from the normal first exon being directly spliced to the third.35 This suggests, in comparison to the expression effect of our mutation, that different nucleotide positions within the donor consensus splice-site sequence may play different roles in splice-site selection and in the kinetics of splicing. It is also noteworthy that, in our case, although the same cryptic splice site, at IVS-II-47, has been activated, the two-nucleotide deletion results in a different reading frame, with completely different results at the protein, cell, and organism levels.
ACKNOWLEDGMENT
We thank A. Villegas for performing the globin chain biosynthetic assay, and M.J. Peres, I. Picanço, A. Miranda, and T. Seixas for technical assistance. We also thank L. Pinho for providing further patient blood samples for analysis.
P.F. was supported by a research fellowship from Junta Nacional de Investigação Cientı́fica e Tecnológica (JNICT) and PRAXIS XXI. Supported in part by JNICT research grants.
Address reprint requests to João Lavinha, PhD, Departamento de Genética Humana, Instituto Nacional de Saúde Dr Ricardo Jorge, Avenida Padre Cruz, P-1699 Lisboa CODEX, Portugal.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

![Fig. 3. Effect of dominantly inherited β-thalassemia mutation on β-globin expression. (A) Denaturant urea/formamide/polyacrylamide vertical gel electrophoresis of fragments obtained by RT-PCR from reticulocyte β-globin mRNAs extracted from (1) a carrier (IV7) of IVS-II-4,5(-AG) mutation (4 β-globin mRNAs species of different size were found, which, on sequencing, were shown to correspond to the normal β-globin mRNA [N], exon 2 skipping [E2S], and activation of 2 cryptic splice sites between codon 59/60 [CS 59/60] and at IVS-II-47 [CS IVS-II-47]); (2) a normal β-globin control; and (3) DNA molecular weight marker (pBR322 + BglI + HinfI). (B) Schematic representation of the 3 abnormally processed β-globin transcripts. Arrows under exons represent primers used in RT-PCR. x, stop codon position. (C) Amino acid sequence of normal human β-globin chain and predicted amino acid sequence corresponding to various aberrant β-globin transcripts. Differences in amino acid sequence are underlined. Deleted residues are indicated by dashed line. (D) Primer extension analysis of β-globin mRNA (using a reverse primer located at β-globin gene exon 3, 5′ end-labeled with [32P]dATP) in (1) a normal control; (2) a carrier of IVS-II-4,5 (-AG) mutation (showing the normal and 3 abnormal mRNA species, which, upon densitometric scanning, showed no significant difference in its relative abundance; and (3) 5′ end-labeled DNA molecular weight marker (pGEM 3 + HinfI).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/2/10.1182_blood.v91.2.685/3/m_blod4020303a.jpeg?Expires=1767724496&Signature=dS1bgPWpInXQmV-KRqm59qAIQaiZrCIhIrn-x~3NLpJx3wfEp-rtvDFwcRg~hvcvt6JvyrqNd0Uo70FnmT5AMNSsoscpHdcwuWY9ZRMn~Bo7V~YklJYLwQeN3Nwt60mK0LJNTxs5CVbi4gFTX-k4UvgH8yxG8FVaByavEkeZOkYLWHCJBSKRDo~HIy4Hp48mWWHCcBvHUptHMymEUTgC4yObwgbBuL1rgIjcClfx75KuTKc9u6Yxf7sqsZvRWtiVL7S5nEy6KdJKzt1iSTOZFmvOeNeTjiCBgbhT64AnV8AvD4YJNSoIbOm2LxuLvbE1ZGAhQP6nP19-0cuJBk92fw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Effect of dominantly inherited β-thalassemia mutation on β-globin expression. (A) Denaturant urea/formamide/polyacrylamide vertical gel electrophoresis of fragments obtained by RT-PCR from reticulocyte β-globin mRNAs extracted from (1) a carrier (IV7) of IVS-II-4,5(-AG) mutation (4 β-globin mRNAs species of different size were found, which, on sequencing, were shown to correspond to the normal β-globin mRNA [N], exon 2 skipping [E2S], and activation of 2 cryptic splice sites between codon 59/60 [CS 59/60] and at IVS-II-47 [CS IVS-II-47]); (2) a normal β-globin control; and (3) DNA molecular weight marker (pBR322 + BglI + HinfI). (B) Schematic representation of the 3 abnormally processed β-globin transcripts. Arrows under exons represent primers used in RT-PCR. x, stop codon position. (C) Amino acid sequence of normal human β-globin chain and predicted amino acid sequence corresponding to various aberrant β-globin transcripts. Differences in amino acid sequence are underlined. Deleted residues are indicated by dashed line. (D) Primer extension analysis of β-globin mRNA (using a reverse primer located at β-globin gene exon 3, 5′ end-labeled with [32P]dATP) in (1) a normal control; (2) a carrier of IVS-II-4,5 (-AG) mutation (showing the normal and 3 abnormal mRNA species, which, upon densitometric scanning, showed no significant difference in its relative abundance; and (3) 5′ end-labeled DNA molecular weight marker (pGEM 3 + HinfI).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/2/10.1182_blood.v91.2.685/3/m_blod4020303b.jpeg?Expires=1767724496&Signature=ZSdaNH6C4rLSB~4SVKKIbX17eQUf7AkHAl3Hnsh2SMAZ8wPFnjCJ~JGL7Ypm1Qnt6VHa8UAcli-bBBYiYs924c27GKjF7jbO6-rZYY5bhSiCXACgvoNeX7fRmERxEfHZpI8oauYb-NRnS3HBEcSijRN1H3Ilx0ahO8-SebwBprsiJYxNdkTF~2gQnI6HLQInaPUBs5G-NSnR1lab0~fc0x7nOYaCj2oypx4xWwfaZ6sJWe2cTM9eMP1DZVPwjmv~tpl6ZTSi74--RpXs0qF0w5Gtc2kDFrotYtDHAg2R4wh01qP3T69MA93ibLdaNCJmusiFKxJ2u-YNUN7MXzwZdg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Effect of dominantly inherited β-thalassemia mutation on β-globin expression. (A) Denaturant urea/formamide/polyacrylamide vertical gel electrophoresis of fragments obtained by RT-PCR from reticulocyte β-globin mRNAs extracted from (1) a carrier (IV7) of IVS-II-4,5(-AG) mutation (4 β-globin mRNAs species of different size were found, which, on sequencing, were shown to correspond to the normal β-globin mRNA [N], exon 2 skipping [E2S], and activation of 2 cryptic splice sites between codon 59/60 [CS 59/60] and at IVS-II-47 [CS IVS-II-47]); (2) a normal β-globin control; and (3) DNA molecular weight marker (pBR322 + BglI + HinfI). (B) Schematic representation of the 3 abnormally processed β-globin transcripts. Arrows under exons represent primers used in RT-PCR. x, stop codon position. (C) Amino acid sequence of normal human β-globin chain and predicted amino acid sequence corresponding to various aberrant β-globin transcripts. Differences in amino acid sequence are underlined. Deleted residues are indicated by dashed line. (D) Primer extension analysis of β-globin mRNA (using a reverse primer located at β-globin gene exon 3, 5′ end-labeled with [32P]dATP) in (1) a normal control; (2) a carrier of IVS-II-4,5 (-AG) mutation (showing the normal and 3 abnormal mRNA species, which, upon densitometric scanning, showed no significant difference in its relative abundance; and (3) 5′ end-labeled DNA molecular weight marker (pGEM 3 + HinfI).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/2/10.1182_blood.v91.2.685/3/m_blod4020303c.jpeg?Expires=1767724496&Signature=1urOSTsCJLM~7fLdOi3~n6xT4q9Xx6eaVluhtr0quzypl-32AMmKU-0YcIS8bnvWXJPHoUaH7b5ibT-yVnjI4K~AQUfRaYhi8daFKb3bmVzIABn~hErHWkhbMHbqJcZ9bKssQwMVG8z4AJni0O2xoIIXOV0npB3~4giLdJ8v5NbMLmFNS426qDQWiUqo986SUAOEy45mlZ21kDFNCHlAMzkdzT5n9cWtg4MBVl4LwzhiO~Z8LXO8MBmsjwySt2BwaI90YRmvlLElP3ZmLR2PIv~yKRPhwxckKhnEDrpCmkawBiNbZzGaea3FwxsuII5jF-VWuCwKH5RhTR8f19GC0w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Effect of dominantly inherited β-thalassemia mutation on β-globin expression. (A) Denaturant urea/formamide/polyacrylamide vertical gel electrophoresis of fragments obtained by RT-PCR from reticulocyte β-globin mRNAs extracted from (1) a carrier (IV7) of IVS-II-4,5(-AG) mutation (4 β-globin mRNAs species of different size were found, which, on sequencing, were shown to correspond to the normal β-globin mRNA [N], exon 2 skipping [E2S], and activation of 2 cryptic splice sites between codon 59/60 [CS 59/60] and at IVS-II-47 [CS IVS-II-47]); (2) a normal β-globin control; and (3) DNA molecular weight marker (pBR322 + BglI + HinfI). (B) Schematic representation of the 3 abnormally processed β-globin transcripts. Arrows under exons represent primers used in RT-PCR. x, stop codon position. (C) Amino acid sequence of normal human β-globin chain and predicted amino acid sequence corresponding to various aberrant β-globin transcripts. Differences in amino acid sequence are underlined. Deleted residues are indicated by dashed line. (D) Primer extension analysis of β-globin mRNA (using a reverse primer located at β-globin gene exon 3, 5′ end-labeled with [32P]dATP) in (1) a normal control; (2) a carrier of IVS-II-4,5 (-AG) mutation (showing the normal and 3 abnormal mRNA species, which, upon densitometric scanning, showed no significant difference in its relative abundance; and (3) 5′ end-labeled DNA molecular weight marker (pGEM 3 + HinfI).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/2/10.1182_blood.v91.2.685/3/m_blod4020303d.jpeg?Expires=1767724496&Signature=1uSYdshChZrcMkLeAzCi3m240ffBXzh2yrivfJ5y9hnhw5vGeI8k6edI1yvm1NbnYR8zEWoc1bf1fZw0rK3G9~nxkUXcroQ619kzYoAkWV4YC8W~HW3qAJMT1ht1S7WVPrOHBLO71jW6h1dHlh8HdXl1m4X~GI0ER-YSZx-xZKMe2LudxP-tWfrJRMYJ38lXrTgFTjrjA2BsTO42A6bU1EW1uwr4xTleDC3ZthYJvyowdGz9AzeZJK2jjyGnc2gC6qmMEa9tPL2mjL-TVlUhdP7RoZbzO6Eqqm92T-w5SLiaOei5hIpJQCFBVBRXjUIz0ezV0tbolA9y-ah6WLyL4w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal