Abstract
The FAC protein encoded by the gene defective in Fanconi anemia (FA) complementation group C binds to at least three ubiquitous cytoplasmic proteins in vitro. We used here the complete coding sequence ofFAC in a yeast two-hybrid screen to identify interacting proteins. The molecular chaperone GRP94 was isolated twice from a B-lymphocyte cDNA library. Binding was confirmed by coimmunoprecipitation of FAC and GRP94 from cytosolic, but not nuclear, lysates of transfected COS-1 cells, as well as from mouse liver cytoplasmic extracts. Deletion mutants of FAC showed that residues 103-308 were required for interaction with GRP94, and a natural splicing mutation within the IVS-4 of FAC that removes residues 111-148 failed to bind GRP94. Ribozyme-mediated inactivation of GRP94 in the rat NRK cell line led to significantly reduced levels of immunoreactive FAC and concomitant hypersensitivity to mitomycin C, similar to the cellular phenotype of FA. Our results demonstrate that GRP94 interacts with FAC both in vitro and in vivo and regulates its intracellular level in a cell culture model. In addition, the pathogenicity of the IVS-4 splicing mutation in the FAC gene may be mediated in part by its inability to bind to GRP94.
THE AUTOSOMAL RECESSIVE disorder Fanconi anemia (FA) frequently leads to bone marrow failure, leukemia, and other cancers at an early age.1,2 Hence, it is regarded as an important genetic model of preleukemia and cancer susceptibility. Cellular manifestations of FA include chromosomal instability, enhanced sensitivity to bifunctional cross-linking agents (eg, mitomycin C [MMC]) and to oxygen,3 cell cycle defects characterized by delays in the G2 phase of the cell cycle (Seyschab et al4 and the references therein) and a predisposition to apoptosis.5-8 Although the molecular basis of these abnormalities is not well understood, we and others have challenged earlier views9 on the pathogenesis of FA and suggested that it does not necessarily involve a primary defect in DNA repair.10-12
At least eight distinct complementation groups of FA have been defined by somatic cell fusions,13-16 three of which map to different chromosomal loci.13,17,18 The genes for complementation groups A (FAA)19,20 and C (FAC)21 have been cloned, and mutations consisting of deletions, a splicing defect, as well as nonsense and missense mutations have been identified (for examples, see Whitney et al,22 Verlander et al,23 Gibson et al,24 and Lo Ten Foe et al25). However, the predicted polypeptide sequences of FAA and FAC have not shed light on their biological functions. The 163-kD FAA protein contains a putative nuclear localization signal,19,20 but otherwise it has little homology to known proteins, including FAC. It may play a role in DNA repair.26 Although fibroblasts from certain FA patients have elevated rates of homologous recombination, the significance of this observation for particular complementation groups and for the proximal effects of FA proteins are unknown.27 The FAC protein is considerably smaller (63 kD),10,11,21 and cytoplasmic residence is obligatory for its ability to correct the hypersensitivity of FA group C (FA-C) cells to bifunctional alkylators.12
We have shown that at least three ubiquitous cytoplasmic proteins interact with FAC.28 One of the interacting proteins is the microsomal enzyme NADPH:cytochrome c (P-450) reductase. The amino-terminal region of FAC binds to the cytosolic, membrane-proximal domain of the reductase and attenuates its catalytic activity (H. Youssoufian et al, manuscript submitted). We postulate that, in the absence of FAC, excess reactive metabolites of MMC and perhaps other compounds that are subject to bioreductive modification could damage DNA and compromise chromosomal stability.
In this study, we show that FAC binds directly to a member of the glucose-regulated family of proteins, GRP94.29-31 This abundant, stress-inducible glycoprotein with homology to the molecular chaperone HSP90 is localized primarily in the lumen of the endoplasmic reticulum (ER). There is also evidence for a transmembrane form of GRP94 that spans the ER membrane and extends into the cytoplasm.32-34 GRP94 has been shown to bind to both normal and mutant proteins,32-36 including p185erbB2 and a mutant form of herpes simplex virus-1 glycoprotein B. GRP94 may act in tandem with GRP78 on partially folded intermediates of the nascent Ig chains.35 It has also been implicated in a variety of other cellular processes,29 particularly intracellular Ca2+homeostasis.37 This association with FAC establishes a novel, albeit indirect, function for GRP94 in the maintenance of chromosomal stability in mammalian cells.
MATERIALS AND METHODS
Cell culture.
NRK cells stably transfected with GRP78-promoter constructs (gift of Dr Amy Lee, University of Southern California, Los Angeles)38 driving a GRP94 ribozyme (NRK-Ribo) and a vector control (NRK-Vect) were maintained in Dulbecco's Modified Essential Medium (DMEM; GIBCO-BRL, Grand Island, NY) containing 10% fetal bovine serum (FBS) and G418 (400 μg/mL). COS-1, HSC536 (FA-C lymphoblastoid cells), and HSC536-FAC (HSC536 cells transfected with wild-type FAC) were maintained as described previously.11 12
Plasmid constructions and cDNA libraries.
Mammalian expression vectors encoding wild-type FAC cDNA and a fusion cDNA of FAC with the constant region of the IgG1 heavy chain (FAC-Ig) have been described previously.28 As a control for FAC-Ig, seven residues from the amino-terminal region of FAC were fused in-frame with the IgG1 epitope in the vector pED6 to generate ΔFAC-Ig. Full-length FAC cDNA was also fused in-frame downstream of the DNA-binding domain of GAL4 in the vector pGBT (Clontech, Palo Alto, CA) to generate pGBT-FAC. Carboxy-terminal deletion mutants of FAC and an interstitial deletion that removes exon 4 (the product of the IVS4+4 mutation, which can generate a 111-bp, in-frame deletion of exon 4)22 were derived by polymerase chain reaction (PCR) and cloned into either pGBT or pBD-GAL4Cam (Strategene, La Jolla, CA). Inserts in both expression vectors were under the control of the ADH1 promoter. cDNA libraries from human B-lymphocyte, fetal liver, bone marrow, and HeLa cells cloned as fusion genes with the GAL4 transcriptional activation domain in the vector pACT were obtained from a commercial source (Clontech). A 2.4-kb EcoRI fragment obtained from pACT-GRP94 that contains the entire coding region of human GRP94 was cloned into pcDNA3 (Invitrogen, San Diego, CA) for expression into COS-1 cells. Human factor VIII cDNA in the vector pMT2 was used in cotransfection studies.
Yeast two-hybrid screen.
The yeast strain HF7c was transformed initially with pGBT-FAC by the lithium acetate procedure (MATCHMAKER Two Hybrid System; Clontech) and grown on synthetic minimal medium (SD) in the absence of Trp. As expected, colonies appearing on the plate lacked transcriptional activity. Next, pooled colonies were transformed with cDNA libraries (200 μg) and selected on SD plates in the presence of 50 fmol/L 3-aminotriazole, but lacking Leu, Trp, and His. Colonies that grew in the absence of these amino acids were tested for their ability to activate the lacZ reporter gene. Plasmids recovered from such LacZ+ colonies were used to transform Escherichia coli, which were selected on M9 plates in the absence of Leu to obtain activation-domain plasmids.
Analysis of yeast two-hybrid inserts.
The inserts obtained from the yeast two-hybrid screen were characterized by DNA sequencing using Sequenase Version 2.0 and analyzed by the BLAST program of the National Center for Biotechnology Information database.
β-Galactosidase activity.
Yeast transformants were assayed for β-galactosidase expression using a filter assay.39 Briefly, yeast cells were transferred onto Whatman filters (Whatman, Maidstone, UK), permeabilized in liquid nitrogen, and placed on Whatman No. 1 filter papers that had been soaked in Z buffer (60 mmol/L NaHPO4, 40 mmol/L NaH2PO4, 10 mmol/L MgCl2, 50 mmol/L β-mercaptoethanol) containing 1 μg/mL 5-bromo-4-chloro-3-indolyl-β-D-galactoside at 30°C. Color developed between 5 minutes and 10 hours and was scored by visual inspection.
Cell transfections.
COS-1 cells at 40% to 60% confluence in 100-mm dishes were transfected with plasmid DNA at a concentration of 1 μg/mL by the diethylaminoethyl ether (DEAE)-dextran procedure as before.28 Cells were then cultured in DMEM-10% FBS for 16 hours, detached with trypsin, and replated in fresh medium. NRK-Vect and NRK-Ribo cells were transiently transfected with Lipofectamine (GIBCO-BRL) for 5 hours with 20 μg of total plasmid DNA according to the suggestions of the manufacturer. After 16 hours, 2 × 106 cells detached with trypsin were replated on 60-mm dishes for ribozyme induction.
Immunoprecipitation and immunoblotting.
After 48 hours (for transfected COS-1 cells) or 72 hours (for transiently transfected NRK cells), the monolayer was washed with cold phosphate-buffered saline and detached by scraping, and cytosolic extracts were prepared by incubation of the cell pellets in hypotonic buffer (20 mmol/L HEPES, pH 7.9, 10 mmol/L NaCl, 10% glycerol, 0.5 mmol/L EDTA, protease inhibitors). Nuclei were pelleted by centrifugation at 600g for 5 minutes, and the supernatant was centrifuged at 100,000g for 30 minutes to obtain a cytosolic extract. Soluble liver extracts from C57BL/6 mice were prepared by homogenization in ice-cold buffer (20 mmol/L HEPES, pH 7.9, 150 mmol/L NaCl) containing protease inhibitors. Nuclei and cellular debris were pelleted by centrifugation at 600g for 10 minutes. The final concentration of all of the extracts was adjusted to 20 mmol/L HEPES (pH 7.9), 50 mmol/L NaCl, 5% glycerol, 0.25 mmol/L EDTA, and 0.1% Nonidet P-40. In some experiments, the concentration of NaCl or EDTA was increased to 150 mmol/L and 2 mmol/L, respectively. Extracts of transfected cells were incubated with the anti-FAC antibody typically for 2 hours and with protein A-agarose beads for 1 hour at 4°C. Liver extracts were incubated with either anti-FAC antibody or with a rat monoclonal antibody specific for GRP94 (StressGen Biotechnologies Corp, Victoria, British Columbia, Canada), and immunocomplexes were precipitated with a mixture of immobilized protein A and protein G. Beads were washed three times with binding buffer over 30 minutes and boiled in 1× Laemmli buffer containing 200 mmol/L dithiothreitol, and the proteins were resolved by denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (NEN, Boston, MA) by electroblotting. After blocking in TBST buffer (50 mmol/L Tris, pH 8.3, 150 mmol/L NaCl, 0.05% Tween-20) containing 5% nonfat milk, filters were incubated sequentially with the primary antibodies (1 μg/mL) for 2 hours and appropriate secondary antibodies conjugated to horseradish peroxidase (HRP; GIBCO-BRL) for 45 minutes, which were then washed and visualized by chemiluminescence (ECL; Dupont, Wilmington, DE).
Determination of factor VIII level.
COS-1 cells transfected with pMT2-factor VIII were incubated for 36 hours in DMEM-10% FBS containing aprotinin. The conditioned medium was clarified of cellular debris by centrifugation at 600g for 5 minutes, and factor VIII antigen level was determined by enzyme-linked immunosorbant assay using the monoclonal antibodies ESH2 (American Diagnostica, Greenwich, CN) and factor VIII (Genetics Institute, Cambridge, MA) for capture and detection, respectively. Antigen levels were confirmed by the COATEST chromogenic assay (Chromogenix/DiaPharma, Franklin, OH) using purified recombinant factor VIII as standard.
Determination of cell survival.
For measurements of cell survival, 2 × 105exponentially growing cells were exposed to MMC (0 to 250 ng/mL; Sigma Chemical Co, St Louis, MO) for 5 days, and viable cells were counted by trypan blue exclusion.
RESULTS
Identification of GRP94-FAC interaction by yeast two-hybrid assay.
To identify proteins that interact with FAC, we used the yeast two-hybrid system,39 a genetic strategy that can identify in vivo protein-protein interactions by exploiting the ability of fusion proteins to reconstitute a transcriptional activator. Plasmids encoding FAC cDNA fused with the DNA-binding domain of GAL4 (pGBT-FAC) and libraries of cDNAs (human B-lymphocyte, bone marrow, fetal liver, and HeLa cells) fused to the transcriptional activation domain of GAL4 (in pACT) were used to cotransform HF7c yeast cells. Double transformant colonies were screened and selected under His, Trp-, and Leu-nutrient conditions. Viable colonies were then assayed for LacZ expression. Using full-length FAC as bait, we identified 15 positive clones: 2 clones from the B-lymphocyte library, 3 from bone marrow, 4 from fetal liver, and 6 from HeLa cells. Both clones from the B-lymphocyte library conferred FAC-dependent expression of LacZ in yeast cells (Fig 1A). Sequence analysis of these clones showed the full-length coding region of human GRP94 (a 782-amino acid polypeptide with a 21-amino acid signal peptide), in-frame with the GAL4 DNA-activation domain.
Interaction of GRP94 with FAC by yeast two-hybrid analysis. (A) Structure of full-length FAC and carboxy-terminal truncated mutants (amino acid residues are residues indicated) and (B) an in-frame protein product of the IVS4+4 mutation that deletes exon 4 sequences were fused in-frame, downstream of the DNA-binding domain of GAL4 in the vector pGBT or pBDGAL4Cam. Transcriptional activation ofLacZ in yeast transformed with these constructs as well as with pACT-GRP94 that contains the entire coding region of human GRP94 fused to the GAL4 transcriptional activation domain was assessed by a filter color assay. The rapidity and intensity of color development was scored by visual inspection as shown: −, white color after 12 hours; +, blue color after 1 to 12 hours; ++, dark blue color or blue color after less than 1 hour.
Interaction of GRP94 with FAC by yeast two-hybrid analysis. (A) Structure of full-length FAC and carboxy-terminal truncated mutants (amino acid residues are residues indicated) and (B) an in-frame protein product of the IVS4+4 mutation that deletes exon 4 sequences were fused in-frame, downstream of the DNA-binding domain of GAL4 in the vector pGBT or pBDGAL4Cam. Transcriptional activation ofLacZ in yeast transformed with these constructs as well as with pACT-GRP94 that contains the entire coding region of human GRP94 fused to the GAL4 transcriptional activation domain was assessed by a filter color assay. The rapidity and intensity of color development was scored by visual inspection as shown: −, white color after 12 hours; +, blue color after 1 to 12 hours; ++, dark blue color or blue color after less than 1 hour.
GRP94-binding domain of FAC.
To further delineate the amino acid sequences that are important for the interaction of FAC with GRP94, we constructed carboxy-terminal truncation mutants of FAC and subcloned into pGBT9 to generate fusion proteins with the DNA-binding domain of GAL4 (Fig 1A). Yeast cells were cotransformed with FAC constructs in pGBT9 and full-length GRP94 in pACT. Fusion polypeptides containing residues 1-467 or 1-308 of FAC bound to GRP94, as indicated by the transcriptional activation ofLacZ. By contrast, a fusion protein containing FAC residues 1-103 did not bind GRP94. Thus, residues 103-308 of FAC interact with GRP94.
A common natural mutation in FAC affects the splice donor sequence of intron 4 (called IVS4+4 allele)22 and gives rise to two different truncated products. A severe truncation of carboxy-terminal sequences results by the partial removal of exon 4, use of a downstream cryptic splice donor site, and frameshift. The second, and potentially more interesting, mutation gives rise to complete in-frame deletion of exon 4 involving codons 111 to 148. Because the sequences removed by this mutation are included within the GRP94 binding domain, we tested the possibility that the in-frame protein generated by the IVS4+4 allele is defective in its interaction with GRP94. Indeed, when this fragment was fused to the DNA-binding domain of GAL4 and introduced into yeast cells with pACT-GRP94, there was no evidence of LacZ transactivation (Fig 1B). By contrast, wild-type FAC in pGBT9 cotransformed with pACT-GRP94 resulted in strong transactivation. These results suggest that the pathogenesis of FA in the setting of the IVS4+4 mutation may result, at least in part, through a deficient interaction of the mutant FAC protein with GRP94.
FAC and GRP94 can be coimmunoprecipitated.
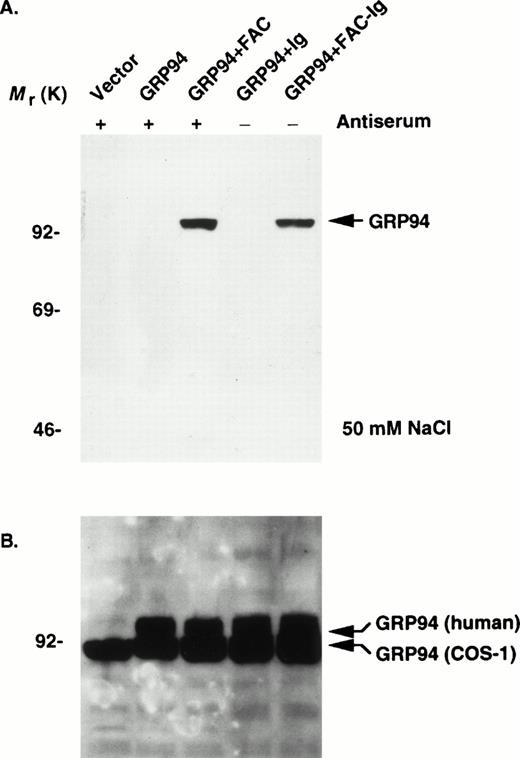
The endogenous expression of FAC in most mammalian cells is very low.11 To look for an interaction between FAC and GRP94 in mammalian cells and characterize binding parameters, we used two complementary strategies. First, we overexpressed GRP94 in COS-1 cells singly or by cotransfection with either wild-type human FAC or a functional isoform fused to the heavy chain of human IgG1 (FAC-Ig). After 48 hours, we performed sequential immunoprecipitation and immunoblotting experiments. Because FAC is localized primarily in the cytoplasm,10,11 we prepared cytoplasmic lysates from COS-1 cells transfected with pED6-FAC and immunoprecipitated with anti-FAC antibodies and protein A-agarose. Immunocomplexes resolved by SDS-PAGE were then immunoblotted and probed with a rat monoclonal antidody specific for GRP94. Lysates containing FAC-Ig were immunoprecipitated directly with protein A-agarose. In both cases, the expected 94-kD GRP94 protein coimmunoprecipitated with FAC and FAC-Ig (Fig 2A). Immunoblotting of the total lysates showed abundant expression of the human GRP94 in cells transfected with pcDNA3-GRP94. The anti-GRP94 antibody also cross-reacted with a 90-kD species in both transfected and nontransfected COS-1 cells, which probably represents the endogenous monkey GRP94 (Fig 2B). FAC-GRP94 complexes were not detected in cells transfected with the empty vector, with GRP94 alone, or with ΔFAC-Ig. Thus, it is possible that the human FAC fails to interact with the monkey homologue. Stable complexes were formed in 50 mmol/L NaCl and in the absence of EDTA, but these complexes were disrupted when the concentration of NaCl and EDTA were raised to 150 mmol/L and 2 mmol/L, respectively (data not shown). Similar immunoprecipitations from mock-transfected or ΔFAC-Ig–transfected cells showed no evidence of interactions with GRP94. Moreover, under similar conditions, the 78-kD heat shock protein (BiP; also called Ig heavy chain binding protein) did not coimmunoprecipitate with FAC (data not shown). These data demonstrate that functionally competent forms of FAC associate with GRP94 in vivo. The sensitivity of FAC-GRP94 interaction to NaCl and EDTA concentrations suggests several possibilities, including a low-affinity interaction in vivo and stabilization of the complex by divalent cations. Second, we attempted to obtain evidence for FAC-GRP94 interaction in normal tissues by sequential immunoprecipitation of protein complexes and immunoblotting. As described previously, our anti-FAC antibody recognizes the murine fac protein,40which is expressed in higher amounts in the liver than in most other adult tissues (H. Youssoufian, unpublished observations). Both anti-FAC and anti-GRP94 antibodies were capable of immunoprecipitating GRP94 from cytoplasmic extracts (Fig 3). The anti-GRP94 antibody precipitated a greater amount of GRP94 than the anti-FAC antibody. These observations demonstrate that fac-GRP94 complexes are normally present in the cytoplasm of mammalian cells.
FAC binds to GRP94 in vivo in transfected cells. (A) COS-1 cells transfected with the indicated constructs (vector, pED6; remainder as described in the Materials and Methods) were processed after 48 hours, and cytoplasmic extracts were prepared in a final concentration of 50 mmol/L NaCl. Extracts were immunoprecipitated with either anti-FAC antiserum and protein A-agarose sequentially (+) or with protein A-agarose (−) only, boiled, resolved by SDS-PAGE, and probed for GRP94 expression by immunoblotting. (B) Analysis of transfected COS-1 cell lysates directly by immunoblotting and probing with anti-GRP94 antibody. The endogenous COS-1 GRP94 appears slightly smaller than the transfected human GRP94 cDNA (shown as bent arrows).
FAC binds to GRP94 in vivo in transfected cells. (A) COS-1 cells transfected with the indicated constructs (vector, pED6; remainder as described in the Materials and Methods) were processed after 48 hours, and cytoplasmic extracts were prepared in a final concentration of 50 mmol/L NaCl. Extracts were immunoprecipitated with either anti-FAC antiserum and protein A-agarose sequentially (+) or with protein A-agarose (−) only, boiled, resolved by SDS-PAGE, and probed for GRP94 expression by immunoblotting. (B) Analysis of transfected COS-1 cell lysates directly by immunoblotting and probing with anti-GRP94 antibody. The endogenous COS-1 GRP94 appears slightly smaller than the transfected human GRP94 cDNA (shown as bent arrows).
Interaction of murine fac with GRP94 in vivo. Liver cytoplasmic extracts from C57B/6 mice were subjected to immunoprecipitation with either polyclonal anti-FAC antibody or monoclonal anti-GRP monoclonal antibody. The anti-FAC antibody can bind efficiently protein A, whereas the rat anti-GRP94 antibody binds well to protein G, but not to protein A. Thus, to increase the recovery of immune complexes and minimize quantitative differences that may result from such interactions, immune complexes were precipitated with a mixture of protein A-agarose and protein G-agarose. After electrotransfer to PVDF membranes, the expression of GRP94 was detected using anti-GRP94 antibody (1 μg/mL), HRP-conjugated goat antirat IgG, and EC. Lane 1, 50 μg extract analyzed directly without prior immunoprecipitation; lane 2, 250 μg extract incubated with protein A-agarose and protein G-agarose without primary antibodies; lane 3, 250 μg extract immunoprecipitated with anti-FAC antibody (4 μg in 250 μL extract) and subsequently with protein A-agarose and protein G-agarose; 250 μg extract immunoprecipitated with anti-GRP94 (4 μg in 250 μL extract) antibody and subsequently with protein A-agarose and protein G-agarose. The positions of murine GRP94 and rabbit IgG heavy chain are shown (arrow).
Interaction of murine fac with GRP94 in vivo. Liver cytoplasmic extracts from C57B/6 mice were subjected to immunoprecipitation with either polyclonal anti-FAC antibody or monoclonal anti-GRP monoclonal antibody. The anti-FAC antibody can bind efficiently protein A, whereas the rat anti-GRP94 antibody binds well to protein G, but not to protein A. Thus, to increase the recovery of immune complexes and minimize quantitative differences that may result from such interactions, immune complexes were precipitated with a mixture of protein A-agarose and protein G-agarose. After electrotransfer to PVDF membranes, the expression of GRP94 was detected using anti-GRP94 antibody (1 μg/mL), HRP-conjugated goat antirat IgG, and EC. Lane 1, 50 μg extract analyzed directly without prior immunoprecipitation; lane 2, 250 μg extract incubated with protein A-agarose and protein G-agarose without primary antibodies; lane 3, 250 μg extract immunoprecipitated with anti-FAC antibody (4 μg in 250 μL extract) and subsequently with protein A-agarose and protein G-agarose; 250 μg extract immunoprecipitated with anti-GRP94 (4 μg in 250 μL extract) antibody and subsequently with protein A-agarose and protein G-agarose. The positions of murine GRP94 and rabbit IgG heavy chain are shown (arrow).
Modulation of intracellular levels of FAC with GRP94.
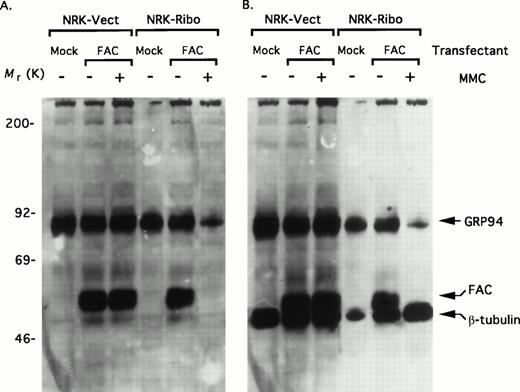
Does the interaction of FAC with GRP94 have any functional consequence? To assess this possibility, we took advantage of a cell line in which the expression of GRP94 can be conditionally inhibited by a targeted ribozyme that can cleave the GRP94 mRNA.38 The ribozyme is driven by the stress-inducible GRP78 promoter. Under steady-state conditions, the activity of the promoter is minimal; consequently, the expression of the ribozyme is also limited. However, under stress conditions, the activation of the promoter can induce high levels of the ribozyme, which degrades the GRP94 mRNA. Rat NRK cells stably expressing GRP78 promoter constructs were transiently transfected with pED6-FAC. After 48 hours, cells were either untreated (steady-state) or treated with 100 nmol/L MMC for 24 hours (stress), and total cell lysates were assayed for GRP94 and FAC expression simultaneously. Lysates of NRK-Vect cells probed simultaneously for GRP94 and FAC (Fig 4A) and subsequently for β-tubulin as a loading control (Fig 4B) showed no differences in the levels of FAC and GRP94 in the presence or absence of MMC. Although the lack of an effect by MMC on FAC expression is consistent with similar previous observations,11 the absence of induction in GRP94 levels seemed incongruent. A likely explanation may be an incomplete induction of the endogenous GRP94 promoter by MMC. This possibility is consistent with previous observations that the GRP94 promoter is generally weaker than the related GRP78 promoter.38 By contrast, MMC treatment of NRK-Ribo cells that presumably induces the targeted ribozyme lead to profound reductions in both the level of GRP94 and of FAC. Indeed, after MMC treatment, the transfected FAC was no longer detectable by immunoblotting. By contrast, there were no significant changes in the expression of β-tubulin, indicating that the reduced levels of FAC and GRP94 do not result from a nonspecific cytotoxic effect of MMC on these cells. These results demonstrate that the intracellular level of FAC is closely coupled to the level of GRP94.
Ribozyme-mediated inactivation of GRP94 causes severe reduction in the level of FAC. (A) NRK-Vect and NRK-Ribo cells transfected with pED6 (mock) or pED6-FAC (FAC) were either untreated or treated with 100 nmol/L MMC for 24 hours. Cytosolic lysates were then assayed for expression of FAC and GRP94 simultaneously by immunoblotting. (B) The same immunoblot as in Fig 3A was reprobed with an antibody against β-tubulin.
Ribozyme-mediated inactivation of GRP94 causes severe reduction in the level of FAC. (A) NRK-Vect and NRK-Ribo cells transfected with pED6 (mock) or pED6-FAC (FAC) were either untreated or treated with 100 nmol/L MMC for 24 hours. Cytosolic lysates were then assayed for expression of FAC and GRP94 simultaneously by immunoblotting. (B) The same immunoblot as in Fig 3A was reprobed with an antibody against β-tubulin.
GRP94 inactivation in vivo.
We also performed experiments to discount the possibility that reduced level of GRP94 in ribozyme-activated cells results from a spurious effect on vector-encoded cDNA. First, Northern analysis of cytoplasmic RNA from NRK-Ribo cells transfected with pED6-FAC showed no differences in the steady-state levels of FAC mRNA in MMC-treated or untreated cells (data not shown). Thus, a transcriptional effect on FAC expression would appear to be unlikely. Second, we identified another molecular target of GRP94 that is regulated by protein-protein interactions and analyzed its expression concomittantly with FAC. We reasoned that such an independent assay for the intracellular function of GRP94 could further minimize the possibility of an artifact caused by MMC or the ribozyme on the observed variations in the levels of FAC. Previous studies have shown that secretion of the human coagulation protein factor VIII from transfected Chinese Hamster Kidney cells is enhanced by antisense inhibition of GRP78.41 Because GRP78 and GRP94 can work in tandem,35 we tested the effect of GRP94 levels on factor VIII secretion. In pilot studies, we cotransfected pcDNA3-GRP94 and pMT2-factor VIII in COS-1 cells and noted a significant reduction in the secretion of factor VIII with GRP94 coexpression (data not shown). Next, we cotransfected NRK-Vect and NRK-Ribo cells with pED6-FAC and pMT2-factor VIII. It should be noted that pED6 and pMT2 are closely related vectors, and any potential effect of the ribozyme on the vector is most likely comparable. After 24 hours, cells were exposed to 100 nmol/L MMC continuously for 36 hours and the conditioned media was assayed for factor VIII activity. There was a significant increase in factor VIII activity in the media of NRK-Ribo cells treated with MMC, but not in NRK-Vect cells (Fig 5A). Third, an alternative possibility is that FAC regulates the intracellular level of GRP94. In this case, the level of GRP94 should be different in parental FA-C cells compared with complemented cells. Immunoblot analysis of total cellular lysates from HSC536 and HSC536-FAC cells showed no significant variations in the level of GRP94 (Fig 5B). There was also little variation in GRP94 levels in cells of other FA groups and in non-FA cells (data not shown). Thus, the alternative possibility that FAC regulates the expression of GRP94 is not supported by these data.
GRP94 expression in vivo. (A) Enhanced factor VIII secretion by inactivation of GRP94. NRK-Vect and NRK-Ribo cells transfected with pMT2-factor VIII were analyzed for their ability to secrete factor VIII in the presence or absence of MMC. (B) Invariant GRP94 levels in parental and complemented FA-C cells. Lysates of parental HSC536 and HSC536-FAC cells were analyzed by immunoblotting with anti-GRP94 antibody. Each lane contained 10 μg protein as determined by the Bradford assay.
GRP94 expression in vivo. (A) Enhanced factor VIII secretion by inactivation of GRP94. NRK-Vect and NRK-Ribo cells transfected with pMT2-factor VIII were analyzed for their ability to secrete factor VIII in the presence or absence of MMC. (B) Invariant GRP94 levels in parental and complemented FA-C cells. Lysates of parental HSC536 and HSC536-FAC cells were analyzed by immunoblotting with anti-GRP94 antibody. Each lane contained 10 μg protein as determined by the Bradford assay.
GRP94 inactivation causes hypersensitivity to MMC.
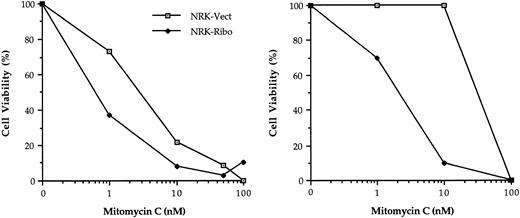
The hallmark of FA cells is their hypersensitivity to bifunctional cross-linking agents. By conditional inactivation of FAC, we reasoned that NRK-Ribo cells should display an enhanced sensitivity to MMC. NRK-Vect and NRK-Ribo cells were treated with MMC and viable cells were counted after 5 days. The results of two separate experiments are shown (Fig 6). NRK-Ribo cells were significantly more sensitive to the cytotoxic effects of MMC than NRK-Vect cells. The difference in the absolute values between the two experiments may reflect unequal levels of ribozyme activation by MMC. These results demonstrate that inactivation of GRP94 leads to a cross-linker-sensitive phenotype similar to that of FA cells.
GRP94 inactivation induces sensitivity to MMC. NRK-Vect or NRK-Ribo cells treated with the indicated concentrations of MMC were analyzed after 5 days for the presence of viable cells by trypan blue staining. Two independent experiments are shown. Values represent the mean of triplicates.
GRP94 inactivation induces sensitivity to MMC. NRK-Vect or NRK-Ribo cells treated with the indicated concentrations of MMC were analyzed after 5 days for the presence of viable cells by trypan blue staining. Two independent experiments are shown. Values represent the mean of triplicates.
DISCUSSION
In this study, we demonstrate that FAC-GRP94 immunocomplexes can be detected in mammalian cells, and residues 103-308 of FAC are essential for this interaction. At first glance, the abundance of heat-shock factors and their frequent isolation from yeast two-hybrid screens, possibly through interactions with misfolded domains, might raise concern that the interaction of FAC with GRP94 is of dubious biological significance. However, both structural mapping studies and functional evidence show that, in this instance, this possibility is unlikely. First, we demonstrate that GRP94 can form protein complexes in normal mouse liver cells. Relatively large amounts of extract were required for this experiment, because the expression of both human FAC and murine fac is low in most mammalian cells. Our ability to demonstrate such an interaction in normal cells gives credence to the data from heterologous systems. Second, the identification of a specific binding domain on FAC makes it less likely that the observed interaction results are artifactual. FAC-GRP94 complexes are stable at relatively low salt concentrations, but dissociate in both higher salt concentrations and in the presence of EDTA. These observations suggest that the interaction of FAC with GRP94 may be transient or of low-affinity and subject to regulation by divalent cations. Third, our preliminary experiments fail to show an interaction between GRP94 and FAA (H. Youssoufian, unpublished observation, November 1997), which also contains a large number of leucine residues and other hydrophobic segments. Fourth, and most important, the use of a powerful genetic system in mammalian cells demonstrates that the FAC-GRP94 biochemical interactions are functionally relevant. Ribozyme-mediated inactivation of GRP94 reduces the level of GRP94 protein, which in turn greatly reduces the level of immunoreactive FAC. Concomittant with these reductions is an acquired sensitivity of the host cells to MMC. Taken together, our data support the conclusion that GRP94 most likely interacts with FAC in vivo and regulates its intracellular level. A plausible interpretation might be that GRP94 prevents the premature degradation of FAC.
A wealth of biochemical data has defined a broad spectrum of biological activities for GRP94.29-38 Consistent with its role in antigenic recognition,29 GRP94 acts as a peptide acceptor and participates in the loading of MHC class I molecules. These peptides are usually bound tightly and can only be eluted by relatively harsh conditions, such as acid extraction.34 As a molecular chaperone, GRP94 also binds to a number of mutant and wild-type proteins,29-32,35,36 but the available evidence indicates that such interactions are of considerably lower affinity. This also appears to be the case for the interaction between GRP94 and FAC. However, the abundance of GRP94 and the rapid turnover of FAC with a half-life of approximately 45 minutes42 could make even low-affinity interactions physiologically tenable. Thus, it is plausible that any disruption of FAC-GRP94 interaction could lead to a rapid degradation of FAC. This interpretation is consistent with our inability to detect immunoreactive FAC after even partial inactivation of GRP94. It is also similar to the interaction between p185erbB2 and GRP94,32 whereby disruption of the complex by ansamycins leads to a dramatic reduction in the half-life of p185erbB2 from 9 hours to 2 hours and hastens its degradation. The latter role of GRP94 is similar to its postulated effect on FAC.
It is noteworthy that the 205-amino-acid central domain of FAC that binds to GRP94 contains seven heptameric sequences that represent a characteristic alternating array of hydrophobic residues and conform to the general motif [Hy(W/X)HyXHyXHy]. This motif is thought to mediate interactions between peptides and BiP.43 Given that BiP and GRP94 act in tandem on folding peptides, it is possible that both chaperones use similar motifs for protein interactions. One of these heptamers (GLGYAPI) is located within the relatively small (37 amino acids in length) exon 4 of FAC, suggesting that this site may be particularly important for FAC-GRP94 interaction.
The subcellular localization of GRP94 is somewhat controversial. Although a large fraction of GRP94 is thought to reside in the lumen of the ER, there is also evidence that a fraction of this protein spans the ER membrane.31 In the latter model, the domain organization of GRP94 predicts a relatively short, amino-terminal intraluminal domain that contains two potential glycosylation sites, a single transmembrane domain, and a carboxy-terminal cytoplasmic domain. Given the known cytoplasmic location of FAC, we suggest that its association is most likely with the membrane-bound isoform of GRP94. By contrast, the intraluminal form of GRP94 is unlikely to interact with FAC. The absence of a signal peptide in FAC and the inability ofN-glycanase to affect the mobility of FAC immunoprecipitates (H. Youssoufian, unpublished observation, November 1994) make it highly unlikely that FAC passes through the ER. Thus, only a fraction of GRP94 molecules would be expected to associate with FAC. This is perhaps consistent with our immunoprecipitation results (Fig 3) in which anti-GRP94 antibody precipitates a considerably larger fraction of GRP94 than anti-FAC antibody. However, other explanations (eg, a difference in the avidity of the two antibodies) may also apply. Given the topological organization of GRP94, an additional prediction of our data is that FAC associates with the carboxy-terminal domain of GRP94. We plan to test this prediction and identify specific residues on GRP94 that are critical for FAC-GRP94 interaction.
Does GRP94 simply serve as a scaffold for FAC? Although the answer is not clear, a number of observations about the stress response of mammalian cells as well as their vulnerability to DNA damage and apoptosis open the possibility of a broader array of effects. GRP94 is induced by interferon,44 which is increasingly implicated in the pathogenesis of FA. For example, a recent study has demonstrated that hematopoietic progenitor cells from fac-knockout mice have an enhanced sensitivity to interferon-γ.8,45 In addition, failure to signal a stress response by GRP94 may predispose lymphoid cells to apoptosis.46 Whether this response is related to the lower apoptotic threshhold of FA cells5-8 remains to be determined. It should be possible to dissect these molecular pathways by manipulating GRP94 levels in cells using molecular techniques (Little and Lee38 and this study) or drugs.32
At present, we are unable to explain completely several of our own observations. For example, why should human FAC fail to interact with the monkey homologue of GRP94? The latter gene has not been cloned and its evolutionary relatedness to the human gene is unknown. Such sequence conservation may be less than expected, because the human and monkey proteins differ in size by approximately 4 kD (Fig 2). Also, although our data demonstrate that the effect of MMC on the NRK-Ribo cells is most likely mediated by induction of the ribozyme promoter, we cannot exclude other mechanistic possibilities. In several models of detoxification, both heat shock (a classic inducer of GRP94) and MMC appear to regulate the expression of target genes, such as DT-diaphorase, by modulating the activity of NF-κB.47Hence, there may be secondary effects of NF-κB activation that could play a role in this process. MMC can also induce heat-shock proteins in nonmammalian systems, but its effect is not necessarily equivalent for all such proteins.48 Thus, it is not surprising that MMC induces the GRP78 promoter in the NRK-Ribo cells, but not the promoter of GRP94. Nevertheless, additional experiments will be required to characterize these phenomena.
In conclusion, our studies demonstrate that a central domain of FAC binds to GRP94, which adds to our knowledge of the domain structure of FAC. The carboxy-terminus of FAC is thought to bind to cdc2,49 and the amino-terminus binds NADPH cytochrome P-450 reductase (Youssoufian et al, manuscript submitted). This modular organization of FAC should facilitate future analysis of its physical structure. Finally, the interaction between FAC and GRP94 suggests a novel approach to cancer chemotherapy. Disruption of this complex in vivo may sensitize cancer cells to MMC and related agents.
ACKNOWLEDGMENT
The authors thank Drs Amy Lee and Randall J. Kaufman for their gifts of cell lines and reagents, Sally Tetrault for technical assistance, and Dr Neal Young for stimulating discussions.
Supported in part by grants from the National Institutes of Health (HL52138 to H.Y. and PO1 HL42443 to R.J.W.) and the Aplastic Anemia Foundation of America (to H.Y.).
Address reprint requests to Hagop Youssoufian, MD, Department of Molecular and Human Genetics, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal