Abstract
Protein 4.1 is a major protein of the red blood cell skeleton. It binds to the membrane through its 30-kD N-terminal domain and to the spectrin-actin lattice through its 10-kD domain. We describe here the molecular basis of a heterozygous hereditary elliptocytosis (HE) associated with protein 4.1 partial deficiency. The responsible allele displayed a greater than 70-kb genomic deletion, beginning within intron 1 and ending within a 1.3-kb region upstream from exon 13. This deletion encompassed both erythroid and nonerythroid translation initiation sites. It accounts for the largest deletion known in genes encoding proteins of the red blood cell membrane. The corresponding mRNA was shortened by 1727 bases, due to the absence of exons 2 to 12. Nevertheless, this mRNA was stable. It showed a similar pattern in lymphoblastoid cells as in reticulocytes. Differential splicing of exons within the undeleted region remained regulated in a tissue-specific manner. Exons 14, 15, and 17a were absent from both reticulocyte and lymphocyte mRNAs, whereas exon 16 was present in reticulocytes but absent from lymphocytes. Thus, differential splicing on a local scale was not dependent on the overall structure of protein 4.1 mRNA in this particular instance.
PROTEIN 4.1 IS A MAJOR component of the red blood cell (RBC) skeleton that laminates the inner surface of the plasma membrane (for review, see Lux and Palek,1Delaunay,2 and Conboy3). It contributes to the skeleton anchoring to the membrane through interactions of its 30-kD domain with band 34,5 and mainly with glycophorin C and p55.5-7 Functional studies have shown that protein 4.1 tightens the interaction of spectrin and actin via its 10-kD internal domain.8-12
Protein 4.1 arises from a single gene that maps to 1p33-p34.2.13 Multiple isoforms derive from alternative splicing.14,15 Erythroid 80-kD protein 4.1 results from two major splicing events. (1) A 17-bp motif, located in exon 2 and containing the upstream translation initiation codon, is spliced out, and (2) a 63-bp exon (exon 16) of the spectrin-actin binding domain is retained16,17 (Fig 1). Erythroid protein 4.1 is translated from the downstream initiation codon in exon 4. In nonerythroid tissues, the 17-bp motif is present and the resulting mRNA encodes a 135-kD protein 4.1, thus converting 618 bp of the erythroid 5′ untranslated region into an in-frame translatable sequence. Immunologically related forms of protein 4.1 are present in a number of tissues.18 A protein sharing a 70% homology with protein 4.1 and encoded by a distinct gene has been recently described.19
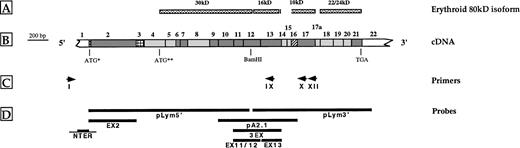
Schematic representations of protein 4.1 (A), cDNA (B), primers (C), and cDNA probes (D). (A) Structural domains of the erythroid protein 4.1, derived from limited chymotryptic cleavage. (B) (▩) Constitutive coding sequences; (▧) alternative coding sequences; (░) the 5′ end of exon 2 and exon 16 are the only motifs where alternative splicing is endowed with known physiological significance; (▦) alternative noncoding sequence; (□) untranslated regions. ATG* and ATG** indicate upstream and downstream translation initiation codons, respectively. (C) Schematic representation of the primers used in 5′ RACE experiments (see also Table 1). (D) Schematic localization of the cDNA and genomic DNA probes used in Southern blot analysis. Probes encompassing exonic (thick bar) and intronic (thin bar) regions.
Schematic representations of protein 4.1 (A), cDNA (B), primers (C), and cDNA probes (D). (A) Structural domains of the erythroid protein 4.1, derived from limited chymotryptic cleavage. (B) (▩) Constitutive coding sequences; (▧) alternative coding sequences; (░) the 5′ end of exon 2 and exon 16 are the only motifs where alternative splicing is endowed with known physiological significance; (▦) alternative noncoding sequence; (□) untranslated regions. ATG* and ATG** indicate upstream and downstream translation initiation codons, respectively. (C) Schematic representation of the primers used in 5′ RACE experiments (see also Table 1). (D) Schematic localization of the cDNA and genomic DNA probes used in Southern blot analysis. Probes encompassing exonic (thick bar) and intronic (thin bar) regions.
4.1(−) hereditary elliptocytosis (HE) designates a condition in which the RBC membrane contains a reduced (heterozygous state)20 or a usually null (homozygous state) quantity of erythroid protein 4.1. The homozygous state described so far produces a severe hemolytic picture and is exceptional.21 22
In a family with heterozygous 4.1(−) HE, we report a deletion greater than 70-kb long encompassing 10 coding exons,23 the largest deletion reported so far within genes encoding proteins of the RBC membrane. This deleted allele was designated allele 4.1 Annecy. However, the corresponding mRNA exhibited no quantitative defect, although it was truncated by 1727 bases and lacked both translation initiation codons. Lymphoblastoid cells, established from the patient's peripheral blood, contained the same mRNA truncation as reticulocytes. Differential splicing of the spared exons remained regulated in a tissue-specific manner. Thus, differential splicing on a local scale was not dependent on the overall structure of protein 4.1 mRNA in this particular instance.
MATERIALS AND METHODS
Genomic DNA studies.
The present family was screened upon during a survey of 4.1(−) HE in a small area of the French Northern Alps.24 Their HE members (mother, son, and daughter), who were typical 4.1(−) HE heterozygotes, failed to exhibit the mutation commonly encountered in their region.12 Northern blots allowed us to perceive a shortened protein 4.1 mRNA, but no further investigations were then performed.
DNA was extracted from peripheral blood leukocytes,25digested with several restriction endonucleases, and subjected to Southern blot analysis. Fragments were separated by electrophoresis through 0.8% agarose gels and transfered onto a positively charged nylon membranes (Boehringer Mannheim Corp, Indianapolis, IN). Hybridizations and washing were performed at 68°C (or 60°C for short probes) according to standard conditions suggested by the manufacturer. Among the random primed probes used, three 4.1 cDNA fragments (pLym 5′ [1.46 kb], pLym 3′ [1.28 kb], and pA2.1 [0.72 kb]) have previously been described24 (Fig1). Five polymerase chain reaction (PCR)-amplified probes (3EX, EX13, EX11/12, EX2, and NTER; Fig 1) were obtained using the following pairs of primers: VI/IX, VIII/IX, VI/VII, IV/V, and II/III, respectively (Table 1). They were cloned into pCRII (Invitrogen, San Diego, CA) and labeled by a second round of PCR with DIG-11-dUTP (Boehringer Mannheim Corp). Hybridized probes were detected with DIG Luminescent Detection kit (Boehringer Mannheim Corp) according to the manufacturer's instruction.
Oligonucleotides Used in RT or PCR Experiments
| Primer Sequence . | Location . | |
|---|---|---|
| ;I. GC (GAATTC) CTCTGAAGGTTCCAGAATCGATAG | S | Anchor-150 |
| ;II. AGAAGTGATTTGTTCGAGGGTC | S | -151 |
| ;III. GGACCCTCAGCCCTGGACC | AS | ‡ |
| ;IV. CCAGAACGCGGTCGGCCCG | S | 61-79 |
| ;V. TTCTCTGAGTTCTTCCTGAGC | AS | 680-660 |
| ;VI. AGCAGTATGAAAGTACCATCGG | S | 1558-1579 |
| ;VII. GTTTACTTGCTGTACGCTCG | AS | 1800-1781 |
| ;VIII. CTGTCGATTCGGCAGACCGA | S | 1831-1850 |
| ;IX. TCTGTGCTTTAGGGACCACTG | AS | 1953-1933 |
| ;X. GC (GAATTC) GATCTCCTCTTGACTCTTG | AS | 2180-2162 |
| ;XI. GCTCACTGATGCTGGCATGATG | AS | 2208-2187 |
| ;XII. GC (GAATTC) GGTGAGTGAGTGGATAAGCGT | AS | 2284-2264 |
| XIII. AGAGCCCACAGAAGCATGGA | S | 2012-2031 |
| ;XIV. TCACTTTTCACAGCATTGGCATTATCT | AS | 2392-2366 |
| ;XV. GACAAGAGTCAAGAGGAGATC | S | 2160-2180 |
| ;XVI. AAGGGCAGAGTTGGGGTAGG | AS | 2774-2755 |
| Primer Sequence . | Location . | |
|---|---|---|
| ;I. GC (GAATTC) CTCTGAAGGTTCCAGAATCGATAG | S | Anchor-150 |
| ;II. AGAAGTGATTTGTTCGAGGGTC | S | -151 |
| ;III. GGACCCTCAGCCCTGGACC | AS | ‡ |
| ;IV. CCAGAACGCGGTCGGCCCG | S | 61-79 |
| ;V. TTCTCTGAGTTCTTCCTGAGC | AS | 680-660 |
| ;VI. AGCAGTATGAAAGTACCATCGG | S | 1558-1579 |
| ;VII. GTTTACTTGCTGTACGCTCG | AS | 1800-1781 |
| ;VIII. CTGTCGATTCGGCAGACCGA | S | 1831-1850 |
| ;IX. TCTGTGCTTTAGGGACCACTG | AS | 1953-1933 |
| ;X. GC (GAATTC) GATCTCCTCTTGACTCTTG | AS | 2180-2162 |
| ;XI. GCTCACTGATGCTGGCATGATG | AS | 2208-2187 |
| ;XII. GC (GAATTC) GGTGAGTGAGTGGATAAGCGT | AS | 2284-2264 |
| XIII. AGAGCCCACAGAAGCATGGA | S | 2012-2031 |
| ;XIV. TCACTTTTCACAGCATTGGCATTATCT | AS | 2392-2366 |
| ;XV. GACAAGAGTCAAGAGGAGATC | S | 2160-2180 |
| ;XVI. AAGGGCAGAGTTGGGGTAGG | AS | 2774-2755 |
The location of nucleotides is given within the sequence published by Conboy et al.15
Abbreviations: S, sense strand; AS, antisense strand.
Sequence complementary to the anchor used in the Amplifinder kit (Clontech).
‡Genomic sequences isolated from the W27 clone23; they are located upstream and downstream from exon 1, respectively.23
Amplification of up to 20-kb DNA fragments was performed by PCR using the Advantage Tth polymerase mix/Genomic PCR kit (Clontech Laboratories Inc, Palo Alto, CA). One or two rounds of 10 to 35 cycles were performed as follows: denaturation at 95°C for 20 seconds and annealing-extension at 70°C for 20 minutes.
mRNA studies.
Total reticulocyte RNA was obtained from peripheral blood as previously described.24 Lymphoblastoid cell lines were kindly established by Prof G. Lenoir (International Agency for Research on Cancer, Lyon, France) from blood sample of proband I.2. Total RNA was extracted from cultured cells according to Chomczynski and Sacchi.26
Reverse transcription (RT) and PCR were performed essentially as previously referred to22 using sequence specific primers (Table 1). The 30 to 35 cycles of amplification were performed as follows: denaturation at 94°C for 45 seconds, annealing at 56°C for 30 seconds or at 60°C for 45 seconds, extension at 72°C for 60 seconds. 5′ Rapid amplification of cDNA ends (5′RACE) was performed with the 5′-Amplifinder Race kit (Clontech) following the manufacturer's protocol. The PCR products were analyzed by electrophoresis on 3% or 4% NuSieve GTG agarose gels (FMC BioProducts, Rockland, ME) and cloned into pUC 18 EcoRI/BAP (Pharmacia, Uppsala, Sweden) or pCRII (Invitrogen) vector. Nucleotide sequencing was performed on double-strand DNAs by the dideoxynucleotide method, with the Sequenase kit (US Biochemical, Cleveland, OH), using universal M13/pUC primers.
Northern blot analysis was performed on reticulocyte and lymphoblastoid cell RNA using random primed pLym 5′ and pLym 3′ probes, as previously described.24
RESULTS
A shortened protein 4.1 mRNA.
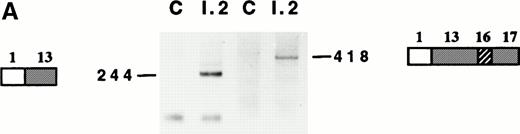
5′RACE was performed to characterize the shortened protein 4.1 mRNA revealed by Northern blot analyses.24 Either random hexamers or primer XI in RT experiments and two pairs of primers (I/XII) or (I/X) in PCR experiments (Table 1 and Fig 1) were used. The reactions yielded a major 522-bp fragment (not shown) and a 418-bp fragment, respectively, corresponding to the abnormal 4.1 allele (Fig 2A). Because the size of the amplified fragment would exceed 1 kb (maximum size recommended by the manufacturer) in the normal allele, no PCR products were expected from these amplifications in control mRNA. Indeed, no PCR products were obtained from the control (Fig 2A). 5′RACE using an upstream reverse primer within exon 13 (primer IX, Fig 1 and Table 1) showed a similar pattern, with a major 244-bp band in the proposita (Fig 2A). The amplified product suggested the presence of the 3′ region of exon 13, encompassing primer IX sequence, on the abnormal mRNA.
Reticulocyte mRNA analysis. (A) 5′RACE on reticulocyte mRNA, using the pairs of primers I/IX (left side) and I/X (right side). C, control; I.2, proposita. The PCR products are schematically represented and their sizes in basepairs are shown. For schematic representation of exons, see Fig 1. (B) Use of internal cryptic splicing sites within exon 13. Sequencing analysis performed on RT-PCR products derived from normal12 or 4.1 Annecy (this report) mRNAs showed discrete use of cryptic acceptor sites within exon 13. The several sequences obtained from 4.1 Annecy tend to suggest a different pattern of use of these sites. () Major site; (—) secondary site; (---) site not used. Intronic and exonic sequences are represented in lowercase and capital bolded letters, respectively.
Reticulocyte mRNA analysis. (A) 5′RACE on reticulocyte mRNA, using the pairs of primers I/IX (left side) and I/X (right side). C, control; I.2, proposita. The PCR products are schematically represented and their sizes in basepairs are shown. For schematic representation of exons, see Fig 1. (B) Use of internal cryptic splicing sites within exon 13. Sequencing analysis performed on RT-PCR products derived from normal12 or 4.1 Annecy (this report) mRNAs showed discrete use of cryptic acceptor sites within exon 13. The several sequences obtained from 4.1 Annecy tend to suggest a different pattern of use of these sites. () Major site; (—) secondary site; (---) site not used. Intronic and exonic sequences are represented in lowercase and capital bolded letters, respectively.
The 418-bp and 244-bp fragments were cloned and 8 positive clones (4 of each size) were sequenced. They all showed an abnormal sequence with exon 1 3′ end joined to exon 13 5′ end, suggesting a large deletion of about 1727 bp (Fig 2B). This deletion encompasses 10 coding exons (exons 2 through 12, exon 3 being a non coding sequence23) and therefore removes both upstream and downstream translation initiation codons (Fig 1). To confirm the 5′RACE results, and knowing that the deletion spares exon 1 sequences, we performed a standard RT-PCR experiment using primers IV and IX, designed within exon 1 and exon 13, respectively. Again, a specific PCR product was obtained from the patient's RNA only. Nucleotide sequencing of the 167-bp corresponding band confirmed the large deletion observed in 5′RACE experiments (not shown).
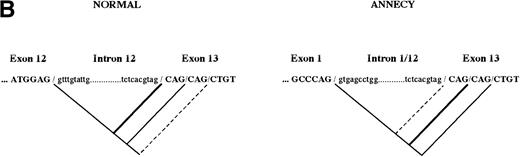
In addition to the large deletion, nucleotide sequencing of the 5′RACE clones showed two discrete changes at the new exon 1/exon 13 boundary (Fig 2B). None of the 8 clones actually showed an intact junction with three repeated CAG codons. Instead, 5 clones showed two of these repeats, and the three remaining clones contained only one CAG codon. Exon 1/intron 1 boundary sequence does not display an alternative donor splicing site with a consensus context. Therefore, it most likely participates to the splicing reaction with the wildtype splicing site. However, the succession of three AG dinucleotides at the intron 13/exon 13 junction offers three potential acceptor splicing sites. It is therefore plausible that the skipping of CAG codons must derive from the use of internal cryptic acceptor sites within exon 13 (Fig 2B). In a previous work, we brought up the use of the first internal cryptic site in intron 12 splicing.12 Sequencing analysis performed here suggests that splicing of the chimeric intron 1/12 preferentially uses the first internal cryptic site, and secondarily the very downstream cryptic site, which is virtually not used in normal intron 12 splicing.
Deleted protein 4.1 mRNA in lymphoblastoid cells.
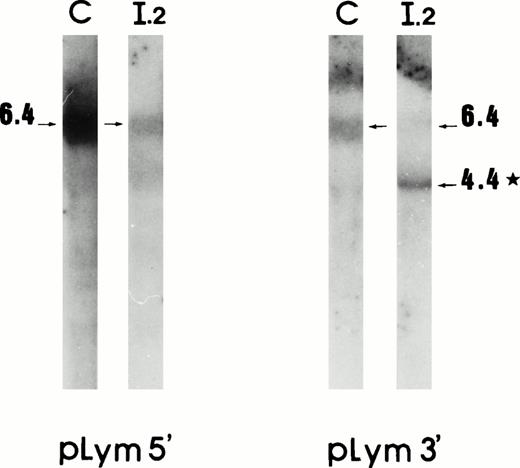
Lymphoblastoid cell mRNAs originating from the proposita and from a control were investigated. Northern blot analysis using pLym 5′ showed a single 6.4-kb band in both the control and the patient. As in reticulocyte 4.1 mRNA, pLym 3′ showed a second major 4.4-kb band, in the proposita (Fig 3). Thus, the mutant mRNA in lymphoblastoid cells was also stable and presented a similar 2-kb deletion.
mRNA deletion in lymphoblastoid cells. Northern blot analysis using cDNA probes pLym5′ and pLym3′. The band sizes are given in kilobases. *Additional band present in the proposita. C, control; I.2, proposita.
mRNA deletion in lymphoblastoid cells. Northern blot analysis using cDNA probes pLym5′ and pLym3′. The band sizes are given in kilobases. *Additional band present in the proposita. C, control; I.2, proposita.
RT-PCR experiments using primers IV and IX showed a 167-bp fragment similar to that found in reticulocyte mRNA; the Taq I digestion provided a rapid mean to probe the nature of this fragment. These data suggest that lymphoblastoid cells also presented the same 1727-bp mRNA deletion, which connected exon 1 to exon 13.
A large genomic deletion encompassing 11 exons.
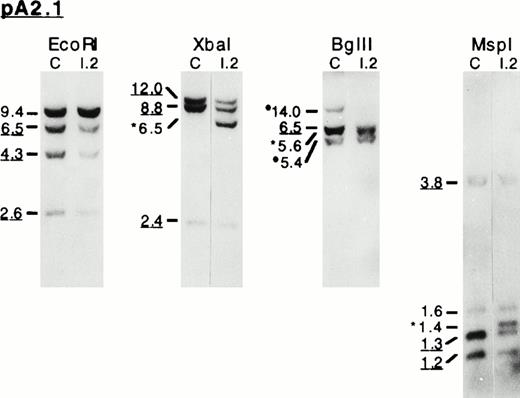
To test the hypothesis of a gene rearrangement responsible for the mRNA deletion, we first performed a Southern blot analysis of genomic DNA from patients I.2 and II.1 and from the unaffected parent I.1 using a pair of restriction endonucleases Msp I and Xba I. Hybridization with either pLym 3′ or pA2.1 (Fig 1) showed an additional band in both patients occurring at the expense of a normal fragment; pLym 5′ probe did not detect this particular fragment (not shown). These findings suggested that the responsible genomic abnormality would involve the 5′ half of the gene and would end within a genomic region encompassed by pA2.1 sequence.
Genomic DNAs were then further investigated by Southern blot analysis using pA2.1 probe and EcoRI, Xba I, Bgl II, orMsp I restriction enzymes. In 4.1(−) HE patients, several bands presented a diminished signal. On Xba I, Bgl II, and Msp I restriction patterns, additional abnormal-sized bands were obtained (Fig 4A). These experiments, together with the mRNA studies, strongly suggested that the 4.1(−) allele bears a large deletion at its 5′ half.
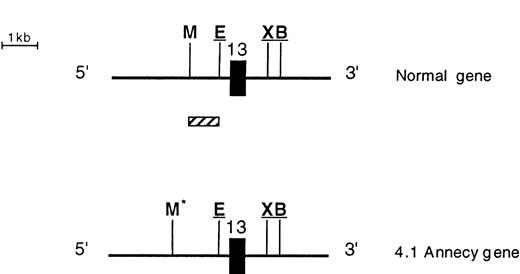
Southern blot analysis of the 3′ endpoint genomic deletion. (A) Southern blot analysis of genomic DNA from control (C) and proposita (I.2), using pA2.1 cDNA probe (see also Fig 1). Restriction enzymes used are indicated. Band sizes (in kilobases) are given. Underlined sizes correspond to half intensity bands; *abnormal bands; •polymorphic site c.24 (B) Partial restriction map, around exon 13, of normal 4.1 gene (top) and shortened allele 4.1 Annecy (bottom). X, B, M, and E represent restriction sites ofXba I, Bgl II, Msp I, and EcoRI enzymes, respectively. Underlined sites had been previously localized (Baklouti et al23 and unpublished data). Localization of Msp I site on the normal gene stemmed from results shown on (A). *Msp I site originating from intron 1 region. (░) The approximately 1-kb region containing the 3′ endpoint of the genomic deletion.
Southern blot analysis of the 3′ endpoint genomic deletion. (A) Southern blot analysis of genomic DNA from control (C) and proposita (I.2), using pA2.1 cDNA probe (see also Fig 1). Restriction enzymes used are indicated. Band sizes (in kilobases) are given. Underlined sizes correspond to half intensity bands; *abnormal bands; •polymorphic site c.24 (B) Partial restriction map, around exon 13, of normal 4.1 gene (top) and shortened allele 4.1 Annecy (bottom). X, B, M, and E represent restriction sites ofXba I, Bgl II, Msp I, and EcoRI enzymes, respectively. Underlined sites had been previously localized (Baklouti et al23 and unpublished data). Localization of Msp I site on the normal gene stemmed from results shown on (A). *Msp I site originating from intron 1 region. (░) The approximately 1-kb region containing the 3′ endpoint of the genomic deletion.
The 3′ endpoint of the deletion was delineated by Southern blot analysis using shorter cDNA probes: 3EX, EX11/12, and EX13 (Fig 1). The abnormal-sized fragments detected with pA2.1 were consistently shown by EX13 probe, which is an exon 13-specific probe, suggesting that the 3′ endpoint must be at the vicinity of this exon. These data, together with our previous genomic clone mapping (Baklouti et al23 and unpublished data), allowed us to locate the 3′ endpoint within a 1-kb intronic region situated 250 bp upstream from exon 13 (Fig 4B).
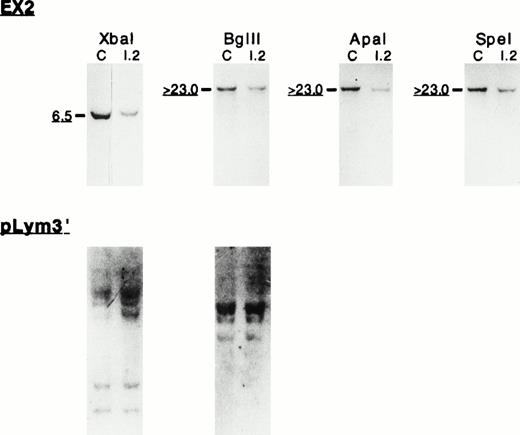
To define the size and the 5′ endpoint of the genomic deletion, we analyzed the genomic region encompassing exons 1 and 2. Southern blot using EX2 cDNA probe (Fig 1) on genomic DNA digested withXba I, Bgl II, Apa I, or Spe I (Fig 5) showed half intensity signals in 4.1(−) HE carriers. The comparable signal intensity between the control and the proposita, using pLym3′ cDNA probe, argues against a DNA loading difference and strongly suggests that the reduced signal observed with EX2 probe is related to the gene deletion. These results indicated that the genomic deletion encompassed all the coding exons absent from the mRNA (exons 2 to 12). We further investigated the genomic region surrounding exon 1 using NTER cDNA probe (Fig 1) and fairly rare cutters (BamHI, EcoRV, and Kpn I). The patterns obtained did not show any specific differences between the controls and 4.1(−) HE patients (not shown), suggesting that no additional event such as an inversion or a translocation had occurred during the deletional event.
Southern blot analysis of the 5′ endpoint genomic deletion. (Top) Southern blot analysis of genomic DNA from control (C) and proposita (I.2), using EX2 cDNA probe (see also Fig 1). Notice the reduced signal intensity (underlined sizes bands) consistently found in the patient with EX2 probe. Restriction enzymes used are indicated. Band sizes (in kilobases) are given. (Bottom) Hybridization of the sameXba I/Bgl II membrane with pLym3′ probe that covers the nondeleted region of 4.1 Annecy gene.
Southern blot analysis of the 5′ endpoint genomic deletion. (Top) Southern blot analysis of genomic DNA from control (C) and proposita (I.2), using EX2 cDNA probe (see also Fig 1). Notice the reduced signal intensity (underlined sizes bands) consistently found in the patient with EX2 probe. Restriction enzymes used are indicated. Band sizes (in kilobases) are given. (Bottom) Hybridization of the sameXba I/Bgl II membrane with pLym3′ probe that covers the nondeleted region of 4.1 Annecy gene.
We finally attempted to capture the genomic region containing both the 5′ and the 3′ deletion endpoints using a long range PCR and primers IV and IX (Table 1). The PCR experiments, using conditions for up to the 20-kb long fragment, failed to amplify the deleted 4.1 gene. These data, together with the absence of additional bands on Southern blots using either EX2 or NTER probes, as explained above, suggested that the 5′ endpoint must lie within intron 1, several kilobases away from both exon 1 and exon 2. Moreover, the exon-intron organization of protein 4.1 gene study23 presented a gap in the delineation of some intronic sequences: among others, intron 1 appears to be much larger than 25 kb. Therefore, exons 1 and 13 would be separated by at least 70 kb.
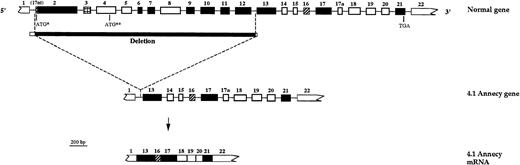
In conclusion, 4.1 Annecy allele most likely results from a genomic deletion that spans over 70 kb (Fig 6).
Schematic representation of the genomic deletion. (Top) Schematic out-of-scale representation of protein 4.1 gene. Tentative boundaries of the genomic deletion are depicted. (▪) Genomic deletion encompassing exons 2 to 12. (□) 5′ and 3′ endpoints located downstream from exon 1 and in a 1-kb stretch upstream from exon 13, respectively. (Middle) Schematic representation of 4.1 Annecy gene. (Bottom) Representation of the shortened 4.1 Annecy mRNA. For schematic representation of exons, see Fig 1.
Schematic representation of the genomic deletion. (Top) Schematic out-of-scale representation of protein 4.1 gene. Tentative boundaries of the genomic deletion are depicted. (▪) Genomic deletion encompassing exons 2 to 12. (□) 5′ and 3′ endpoints located downstream from exon 1 and in a 1-kb stretch upstream from exon 13, respectively. (Middle) Schematic representation of 4.1 Annecy gene. (Bottom) Representation of the shortened 4.1 Annecy mRNA. For schematic representation of exons, see Fig 1.
Erythroid and lymphoid expression of alternatively spliced exons.
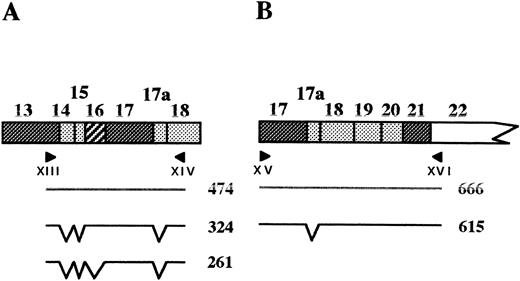
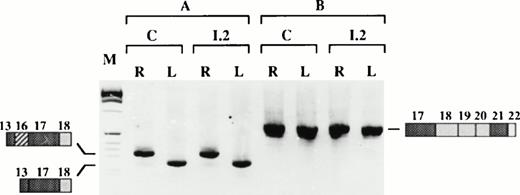
The RT-PCR and 5′RACE experiments described above clearly showed that the chimeric intron, made of the 5′ part of intron 1 and the 3′ part of intron 12, is correctly spliced leading to a junction of exon 1 and exon 13. We next asked whether the large genomic deletion affects the tissue-specific splicing of the remaining exons. Two overlapping RT-PCR experiments were performed on reticulocyte and lymphoblastoid cell RNAs using primers designed within the nondeleted cDNA sequence (Fig 7). These experiments showed that exons 14 and 15, tandemly present in brain,23,27 were absent from normal and shortened mRNAs in both cell types. Exon 17a, recently described as a muscle specific exon,23 was likewise absent in all cases. Similarly, exon 16 splicing still follows a tissue-specific regulation, regardless of the presence or absence of the upstream region (exons 2 to 12) on the pre-mRNA: Exon 16 was indeed present in reticulocyte mRNA, but absent from lymphocyte mRNA (Fig 7).
Tissue-specific splicing of exons downstream from the deletion. (Top) Schematic representation of the PCR strategy; the expected PCR products and their sizes in basepairs are shown. (Bottom) RT-PCR on reticulocyte (R) or lymphoblastoid cell (L) mRNA, using primers XIII/XIV (A) or primers XV/XVI (B). C, control; I.2, proposita; M, size marker.
Tissue-specific splicing of exons downstream from the deletion. (Top) Schematic representation of the PCR strategy; the expected PCR products and their sizes in basepairs are shown. (Bottom) RT-PCR on reticulocyte (R) or lymphoblastoid cell (L) mRNA, using primers XIII/XIV (A) or primers XV/XVI (B). C, control; I.2, proposita; M, size marker.
DISCUSSION
The present protein 4.1 allele, allele Annecy, provides the first known instance of a very large deletion affecting a gene encoding an RBC membrane protein (for review, see Delaunay2). It carries a deletion longer than 70 kb. The 5′ endpoint of the deletion is situated in the intronic sequence 3′ of exon 1. The 3′ endpoint is located within a 1-kb region of the intronic sequence finishing 250 bp 5′ upstream to exon 13. The extent of this deletion cannot be assessed with greater accuracy, because the sizes and sequences of the introns of protein 4.1 gene are still to be defined.23 The mRNA that arises from this allele 4.1 Annecy in erythroid cells carries a 1727-bp deletion, corresponding to 10 coding exons (exons 2 to 12; exon 3 is a noncoding sequence). This deletion includes both the upstream and the downstream translation initiation codons.
Following alleles 4.1 Madrid,22 4.1 Aravis,12and 4.1 Lille,28 allele 4.1 Annecy is the fourth 4.1(−) HE allele the molecular basis of which is defined at the gene level. The scarcity of protein 4.1 gene mutations elucidated so far contrasts with the profusion of HE generating mutations inSPTA1 and SPTB genes, encoding α- and β-spectrin chains, respectively. This is presumably accounted for by the lack of information at the protein level (protein 4.1 is just manifested by its absence) and the knowledge, only recently acquired,23 of the protein 4.1 gene structure.
The nucleotide sequence of mutated 4.1 mRNA Annecy is defined by a deletion spanning exon 2 to exon 12 region and removing 1727 bases. Exon 1 and exon 13 are joined, implying that the splicing had normally occurred between these 2 exons. However, 3 nucleotides were missing in the mutated sequence (exon 1)GCCCAGCAGCTG(exon 13) as compared with the expected joined sequences (exon 1)GCCCAG/CAGCAGCTG(exon13). This stemed from the use of a cryptic site within exon 13. Such an alternative splicing has already been observed, although as a minor event, in normal mRNA.12 Sequencing analysis showed still another minor species generated, presumably by the skipping of a second CAG triplet (exon 1)GCCCAGCT(exon 13) (Fig 2B), and the use of the remaining AG cryptic site of the repetitive CAG sequence. These data suggest that the sequence of exon 1 is identical to that in the normal 4.1 mRNA sequence and that the 5′ end of exon 13 is submitted to extra splicing events due to repetitive CAG sequences.
Among all 4.1(−) alleles described to date, only those alterations whose expression is confined in the RBCs have been found at the homozygous state; this is the case for allele 4.1 Madrid, mutated in the erythroid translation initiation codon22 and the Algerian 4.1 allele lacking three exons, including the one that carries the downstream codon.29 Use of the upstream AUG codon allows protein 4.1 expression in nonerythroid tissues. However, allele 4.1 Annecy is the only variant lacking both translation initiation sites. One would speculate that such a variant would not exist at the homozygous state.
Large deletions within functional genes usually jeopardize the cytoplasmic mRNA accumulation either because of an altered transcription initiation, defective splicing, RNA transport, or in-frame premature stop codon. We and others have shown that the alteration of the translation initiation site often destabilizes mRNA22,30-32 (for review, see Cooper33). Shortened mRNA 4.1 Annecy, although it potentially encodes for nonfunctional protein(s), remained stable in the cytoplasm. It did so not only in nucleated cells (lymphoblastoid cells), but also in enucleated cells (reticulocytes), long after the nucleus to cytoplasm transport.
We and others have shown that protein 4.1 pre-mRNA is endowed with complex alternative splicing in a tissue and developmental stage-specific manners.14-17 In mRNA 4.1 Annecy, the splicing pattern of exons downstream from the deletion is similar to splicing in normal protein 4.1 mRNA. These findings, consistent with our recent functional studies (Baklouti et al34 and unpublished data), suggest that cis-elements involved in exon recognition must lie within targeted exons and their close vicinity intronic sequences.
In this work, we have defined the alteration of allele 4.1 Annecy, which is responsible for 4.1(−) HE. We accurately defined the 1727-bp deletion at the mRNA level. This mRNA was stable and presented the same feature in lymphoblastoid cells. At the gene level, we delineated the first very large genomic deletion encountered in RBC membrane protein genes, extending over 70 kb, starting and ending quite far from the corresponding exons within introns 1 and 12, respectively. Miniallele 4.1 Annecy nevertheless allowed the occurrence of a splicing that connected exons 1 and 13 into a stable and shortened mRNA.
ACKNOWLEDGMENT
The authors thank family PE for their kind cooperation. We thank Prof G. Lenoir and M. Vuillaume (International Agency for Research on Cancer) for obtaining the lymphoblastoid cell line and Dr J.P. Magaud for growing the cells.
Supported by the Association Française contre les Myopathies, the Centre National de la Recherche Scientifique (URA 1171), the Université Claude Bernard Lyon I, the Conseil de la Région Rhône-Alpes, the Institut National de la Santé et de la Recherche Médicale (CRE 930405), the Programme Hospitalier de Recherche Clinique (1995-1997), and the Institut Pasteur de Lyon.
Address reprint requests to N. Dalla Venezia, PhD, CNRS UMR 5641, Laboratoire de Génétique, Domaine Rockefeller, 8 Avenue Rockefeller, 69373 Lyon Cedex 08, France.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal