Abstract
We have isolated several T-cell clones from lymphocytes infiltrating a human major histocompatibility class (MHC) II negative cutaneous T-cell lymphoma (CTCL). We describe here two of these clones, TC5 and TC7, with, respectively, a CD4+CD8dim+ and CD4+CD8− phenotype. Both clones mediated a specific MHC class I–restricted cytotoxic activity toward the fresh autologous tumor cells, and autologous tumor cell lines previously established with interleukin-2 (IL-2) and IL-7 from the skin and from the blood. Analysis of the T-cell receptor (TCR) Vβ gene expression showed that the tumor cells, which were shown to have a trisomy 7 by fluorescent in situ hybridization, expressed Vβ7/Jβ2.3, Vβ13/Jβ2.5, and Vβ22/Jβ2.5 rearrangements. Phenotypic analysis using specific anti-Vβ monoclonal antibodies indicated that only Vβ13 could be detected on the cell membrane of the tumor cells. Analysis of the TCR Vβ gene expression of the clones showed that TC5 and TC7 expressed a unique TCR-Vβ transcript, corresponding, respectively, to Vβ5/Jβ2.3 and Vβ17/Jβ2.7 gene segments. To determine whether these reactive T lymphocytes were present in vivo, we used specific primers corresponding to TC5- and TC7-Vβ TCR transcripts. The results showed that both cytotoxic T-cell clones were present at the lesional skin site and amplified in vitro. TC7 was found in the patient peripheral blood invaded by tumoral cells, whereas TC5 was not, indicating that the repertoire of the reactional lymphocytes differs in the blood and at the tumor site. These results show for the first time the presence of reactive T lymphocytes with CD4 or double-positive phenotype infiltrating a CTCL. These findings raise the question of the role of these antitumoral effector T cells in the tumor growth.
THE CONTRIBUTION of the immune system to host defense against solid tumors has been extensively investigated during the last few years.1,2 In humans, the most widely studied model to support the concept of antitumor immunity is malignant melanoma.3-10 Several studies have described T-cell clones or T-cell lines developed either from tumor-infiltrating lymphocytes or from peripheral blood (PB) exhibiting major histocompatibility complex (MHC)-restricted cytotoxic activity against autologous tumor cells. Subsequently, these highly specific, mainly CD8+ T-cell clones were used as tools to identify the recognized tumor antigens encoded by three families of genes, namely, MAGE, BAGEand GAGE.11-13 These genes are frequently expressed in a wide range of tumor types, including lung carcinoma and breast tumors.14,15 In normal tissue these gene products have only been observed in testis and placenta and appear to represent tissue-specific antigens.16 More recently, melanoma-reactive T-lymphocytes recognizing the products of mutated genes were isolated. This finding suggests that certain proteins contain common mutations that give rise to nonself T-cell epitopes which could be really tumor-specific.17
In the present report we studied the immune response against cutaneous T-cell lymphomas (CTCL). CTCLs are a heterogeneous group of lymphoproliferative disorders.18,19 Mycosis fungoides is clinically characterized by slowly progressing erythematous patches and plaques, and histologically by infiltration of the epidermis and dermis by clonally derived malignant lymphocytes with a mature CD3+CD4+ phenotype.19-22 A more aggressive form of CTCL occurs when the malignant cells become nonepidermotropic and is associated with extra-cutaneous involvement. Sézary syndrome is an erythrodermic form of CTCL with blood involvement. Pleomorphic small and medium CTCL is a rare form of CTCL characterized clinically by the occurrence of cutaneous nodules and tumors, and histologically by a nonepidermotropic lymphoid infiltrate.23 It has been reported that lesional tumor-infiltrating lymphocytes (TIL) in CTCL contain a mixture of various nonneoplastic lymphoid cells, including CD4+ and CD8+ lymphocytes.24 The role of these TIL in CTCL remains to be fully elucidated.24,25 Two-color immunohistologic analysis has shown that the majority of infiltrating CD8+ cells express MHC class II antigens, and may be functional cytotoxic T lymphocytes (CTL), as they express TIA-1, an RNA-binding protein that can induce nuclear fragmentation and may be the cytolytic granule factor responsible for causing apoptosis in the target cells.26 Phenotypic studies are not able to address the antigen specificity of these CD8+ cells, and it is possible that they are bystander cells, nonspecifically recruited into CTCL lesions, which do not exhibit any cytotoxic activity against the autologous tumor cells. Therefore, the main objective of the present report was to isolate and study TIL in CTCL to determine whether these lymphocytes are reactive against tumor cells, and whether they can also be detected in the blood invaded by tumor cells. For this purpose, we developed T-cell clones from lymphocytes infiltrating a T-cell receptor (TCR) Vβ13+ CD4+ MHC class II− CTCL. We successfully isolated autologous tumor-specific cytotoxic CD4+CD8dim+and CD4+ T-cell clones. We show that these T-cell clones exhibit MHC class I–restricted cytotoxicity. Comparison of the TCR transcripts expressed by these T-cell clones showed the presence of these reactive T lymphocytes at the tumor site, their expansion during the in vitro coculture in the presence of cytokines, and the presence of one only of these clones in the patient PB invaded by the same tumoral cells. Our findings show for the first time that an MHC class I–restricted tumor-specific reactive CD4+ T-cell clone, which could be isolated from CTCL-infiltrating reactive TIL, was also present in the blood, whereas a CD4+CD8dim+tumor-specific T-cell clone was only present in the tumoral site of an MHC class II–negative CTCL.
MATERIALS AND METHODS
Patients.
Patient Cou was an 81-year-old man with a mycosis fungoides initially presenting as disseminated infiltrated patches and plaques and no extra-cutaneous involvement. After a 5-year follow-up, this mycosis fungoides evolved into a pleomorphic large T-cell lymphoma presenting as disseminated cutaneous tumors and 30% atypical lymphocytes in the peripheral blood. Patient Zia was a 45-year-old man with a pleomorphic small and medium T-cell lymphoma presenting as disseminated cutaneous tumors and no extra-cutaneous or blood involvement. Patient Lak was a 50-year-old woman with a Sezary syndrome defined by the presence of erythroderma, disseminated lymphadenopathy, and 40% atypical lymphocytes in the PB. Patients were not previously treated with chemotherapy. Skin and blood samples were taken after informed consent. Ethical committee approval for the study was obtained.
TIL cultures and clones.
TIL from patients Cou and Zia were obtained from tumor fragments mechanically dispersed into single-cell suspensions and cultured in 12-well plates (Becton Dickinson, Lincoln Park, NJ) in culture medium consisting of RPMI 1640 (GIBCO, Paisley, UK), 2 mmol/L L-glutamine, penicillin (100 U/mL), streptomycin (100 μg/mL), 10% heat-inactivated human serum, 25 U/mL recombinant interleukin-2 (rIL-2; kindly provided by EuroCetus, Amsterdam, The Netherlands), and rIL-7 (10 ng/mL) (kindly provided by Sanofi Recherche, Labège, France). For patient Lak, the cocultures of nonmalignant lymphocytes and tumor lymphocytes were done with mononuclear cells isolated by the technique of Ficoll-Isopaque (Pharmacia Fine Chemicals, Piscataway, NJ) density gradient centrifugation. The various cocultures were tested at day 10 for their ability to exhibit specific cytotoxic activity against the cells from the tumor specimen previously frozen. At day 10 of the cocultures, the lymphocytes from patient Cou were further cloned by a limiting dilution method in which the cells were plated at a concentration of 0.3 cells per well into round-bottomed 96-well plates (Greiner, Nürtingen, Germany). Plates were previously fed with irradiated allogeneic PB lymphocytes (PBL) from two healthy donors (5 × 104 cells per well) in complete medium containing 25 U/mL of rIL-2 and 1 mg/mL phytohemagglutinin (PHA; Wellcome, Beckenham, UK). Cultures were fed every 3 days with rIL-2–containing medium and the growing cloned cells were expanded as previously described.27
Tumor cell lines.
Fresh CTCL tumor cells were obtained from mechanically dispersed tumor fragments (at the same time as isolation of TIL). One portion of the tumor specimen was rapidly frozen in liquid nitrogen awaiting RNA and DNA extraction. Another portion was frozen in human serum plus 10% dimethyl sulfoxide for later use in cell mediated cytotoxicity assays. To develop the tumoral line Cou-LS, the remaining cells were cultured in 24-well tissue-culture plates (Becton Dickinson) at a concentration of 105 cells/mL of RPMI medium containing 10% heat-inactivated human serum, 25 U/mL of rIL-2, and 10 ng/mL of rIL-7. The cultures were fed two to three times per week with fresh medium and split at a 1:2 ratio when necessary. After 2 months of culture most cells were tumoral cells and were maintained in culture for more than 2 years. The line Cou-LB was developed from Cou PBL and cultured as the line Cou-LS. The fresh and cultured CTCL from patient Cou were HLA-A1,A2 and HLA-B5(51),B35. The clonal origin of the growing cell lines was systematically tested by analyzing their clonal reactivity with an anti-TCR Vβ13 monoclonal antibody (MoAb) and by TCR Vβ trancripts analysis.
MoAbs and phenotypic analysis.
MoAbs such as anti-CD3, anti-CD4, anti-CD8, anti-HLA class II, anti-HLA class I, and TCR αβ were produced locally. Other MoAbs were obtained through the exchanges of the Vth international workshop on the differentiation antigens.28 Most anti-TCR Vβ MoAbs were purchased from Immunotech-Coulter (Marseille, France), whereas only the anti-TCR Vβ13 MoAb was from BIOadvance (Emerainville, France). The MoAb B1.23.2, which reacts with monomorphic determinants shared by HLA-B, HLA-C and only a few HLA-A alleles, and the MoAb L243, reactive with monomorphic determinants shared by HLA-DR, HLA-DP, and HLA-DQ, were kindly provided by Dr P. Le Bouteiller (INSERM, Toulouse, France). These MoAbs were used as ascites fluid and, when needed, coupled to fluorescein isothiocyanate (FITC) or biotin. Phenotypic analysis was performed using a single argon flow cytometer analyzer (Epics XL; Coulter, Miami, FL). Indirect immunofluorescence assays were performed using an FITC-conjugated goat anti-mouse Ig from Caltag Laboratories (San Francisco, CA) or a phycoerythrin (PE)-labeled goat anti-mouse Ig from Immunotech (Marseille, France). For two-color immunofluorescence experiments, cells were treated as already described.29
Proliferation assays.
The proliferative response of the tumor cell lines to various cytokines was determined by measuring the [3H]-thymidine incorporation (cpm) of 50 × 103 responder cells. These tests were carried out in 96-well round-bottomed plates in 0.2 mL of culture medium containing 10% inactivated human serum. The cytokines used to test the proliferation of the T-cell lines, ie, IL-2 (25 U/mL), IL-4 (1,000 U/mL), and IL-7 (10 ng/mL), were kindly provided by Sanofi Recherche (Labège, France). After 54 hours, the various culture wells were individually pulsed with 1 mCi (=37 kBq) of [3H]-thymidine and obtained 18 hours later. [3H]-thymidine incorporation was measured in a microplate scintillation counter (Topcount; Packard Instrument Co, Meriden, CT).
Cytotoxicity assays.
Cytotoxicity assays were performed according to a standard51Cr-release method. Effector cells were TIL cultures and TIL-derived T-cell clones. Cryopreserved, noncultured tumor cells, and cultured tumor lines were used as target cells. Assays at various effector to target cell (E:T) ratios with 5 × 10351Cr-labeled target cells/well were performed in triplicate, using 96-well V-bottomed microtiter plates. The final culture volume was 200 μL per well. After 4 hours of culture, plates were spun and 100 μL of supernatant was removed from each well and counted in a gamma-counter for the determination of 51Cr release. The percentage of lysis was determined as previously described.29 For blocking experiments, anti-CD4 MoAb was added to effector cells for 30 minutes at room temperature and the cells were washed before mixing them with 51Cr-labeled target cells. In contrast, anti-HLA class I MoAbs W6/32 and B1.23.2 were added to the 51Cr-labeled target cells for 30 minutes at 4°C and then the cells were washed before using them. When anti-CD3 MoAb was used, 1 μg/mL of purified CD3X3 MoAb was added into culture well with the effector and target cells during the whole assay.
Complementarity determining region 3 (CDR3) size analysis of Vβ transcripts using polymerase chain reaction (PCR).
To study the Vβ transcripts expressed by T-cell clones and tumor cell lines, a run-off methodology was used.30 The Vβ and Cβ-specific primers and the procedure used for CDR3 size analysis have been reported previously.31 Briefly, tumoral and nontumoral tissue samples (0.2 to 0.5 g tissue or 5 × 106 cells) were resuspended in 6 mol/L guanidium thiocyanate buffer. Total RNA was then purified by CsCl gradient centrifugation. For PBL, total RNA was extracted using a modified guanidium thiocyanate phenol/chloroform method (RNAzol B method). cDNA was prepared by standard method using reverse transcriptase (RT) and an oligo-dT primer. cDNA copies of 0.1 μg RNA were amplified in 40 cycles Vβ/Cβ PCR in 50 μL and aliquots (2 μL) were copied in 1- to 5-cycle runoff reactions primed with fluorescent (ABI fluorophore Fam)-labeled oligonucleotides specific for Cβ or Jβ fluorophores. Runoff products were then subjected to electrophoresis on an ABI sequencer (Applied Biosystems, Foster City, CA) in the presence of fluorescent size markers and analyzed with the 672 Genescan software (Perkin Elmer SA, Courbevoie, France).
Semiquantitative PCR analysis.
TCR Vβ gene segment usage was determined using a semiquantitative PCR analysis as described previously.30 cDNA amplifications were performed over 30 cycles with the fluorescent Cβ primer and the same panel of Vβ primers than for the CDR3 size analysis. The intensities of the different peaks present in all Vβ subfamilies were added, and the percentages of each Vβ subfamily were calculated and represented as histograms.
Directed sequencing of PCR products.
PCR products were purified using Qiagen columns (Qiaquick PCR purification kit; Qiagen, Hilden, Germany), and resuspended in 20 μL of sterile water. The purified products were directly sequenced in both directions with a PRISM ready reaction DyeDeoxy Terminator cycle sequencing kit and a 373A DNA sequencer (Applied Biosystems).
Genomic analysis.
High-molecular-weight DNA was extracted by a standard proteinase K digestion and a phenol/chloroform extraction. Two microliters of the DNA samples were subjected to PCR in a 50-μL reaction volume with a Vβ-specific primer and Jβ primer. The PCR cycles were followed by a final 10-minute elongation at 72°C. An aliquot of each amplification reaction was visualized on ethidium bromide–stained 2% agarose gel.
Fluorescent in situ hybridization (FISH).
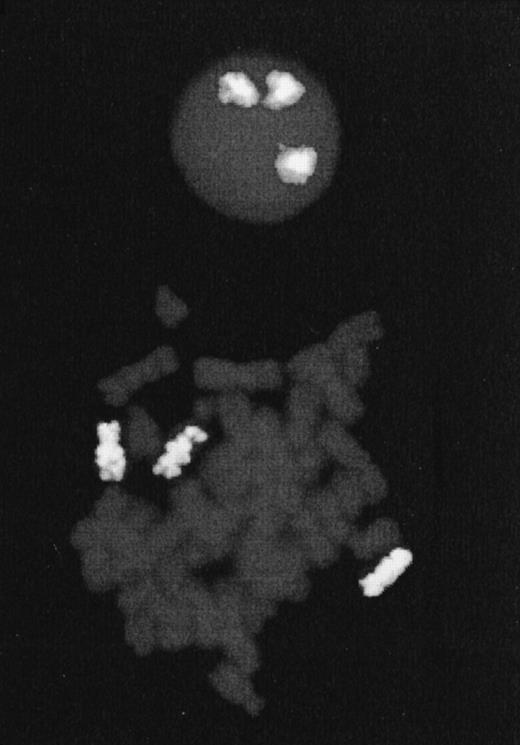
After adjunction of colcemid to exponentially growing cells, metaphase spreads were prepared with standard procedures of hypotonic treatment, methanol/acetic fixation, and dropped on chilled slides. Whole chromosome probe specific for chromosome 7 was of commercial origin (Oncor, Gaithersburg, MD) and was labeled with digoxigenin. FISH was performed following the manufacturer's instructions, and digoxigenin-labeled DNA was detected using anti-digoxigenin-tetramethylrhodamine isothiocyanate (TRITC) antibodies (Boehringer Mannheim, Mannheim, Germany). Chromosomes were counterstained with DAPI (4,5-diamino-2-phenylindole). Fifty mitoses were analyzed and showed in paradiploid cells a homogenous trisomy 7 with no translocation.
RESULTS
CTCL suspensions showed specific cytotoxic activities against autologous tumor cells.
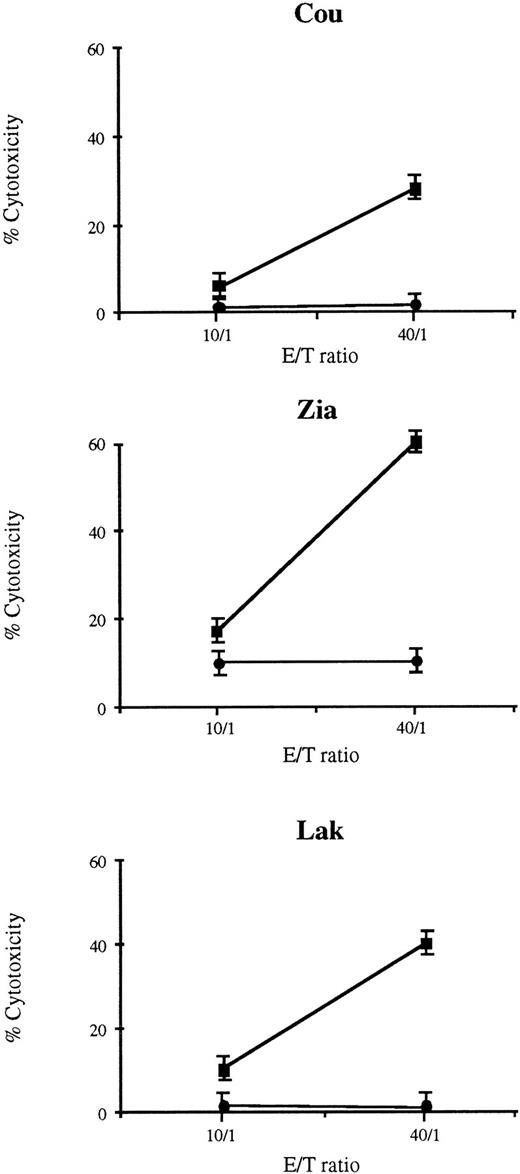
Single-cell suspensions obtained from skin tumor fragments of patients Cou and Zia and PBL from patient Lak were cultured in vitro for 10 days with culture medium containing 10% inactivated human serum supplemented with rIL-2 and rIL-7. Approximately 90% of cells were CD4+, and this percentage of CD4+ cells was not modified after 10 days of culture with IL-2/IL-7. The cultured T lymphocytes were then tested for their ability to mediate cytotoxic activity against a suspension of non-cultured autologous tumor CD4+ T lymphocytes. The results presented in Fig 1 indicate that cocultures from each patient exhibited cytotoxic activity against the previously frozen autologous tumoral cells. The lytic activity of these effector cells were reproducibly significant at the respective highest effector/target ratio tested and decreased with lower effector/target ratios. No lytic activity could be detected against allogeneic tumor cells. As can be seen from the representative experiments shown in Fig 1, the cytotoxic effector cells from Lak were unable to kill allogeneic tumor cells from patient Cou and, conversely, the cytotoxic lymphocytes from Cou failed to kill the allogeneic tumor cells of Lak.
Cytotoxic activity of TIL from three patients with CTCL. TIL were cultured with IL-2 and IL-7, and were tested at day 10 for their ability to exhibit specific cytotoxic activity against the cells of the tumor specimen previously frozen. For the three patients, the TIL cultures exhibited a specific cytotoxic activity against autologous tumor T-cells (▪) whereas no cytotoxic activity was found on allogeneic tumor T cells (•). Results are expressed as the mean of triplicates ± SD.
Cytotoxic activity of TIL from three patients with CTCL. TIL were cultured with IL-2 and IL-7, and were tested at day 10 for their ability to exhibit specific cytotoxic activity against the cells of the tumor specimen previously frozen. For the three patients, the TIL cultures exhibited a specific cytotoxic activity against autologous tumor T-cells (▪) whereas no cytotoxic activity was found on allogeneic tumor T cells (•). Results are expressed as the mean of triplicates ± SD.
Establishment of IL-7/IL-2–dependent lymphoma cell lines.
To maintain in long-term cultures the tumor cells, we subsequently developed a T-cell line from the tumor fragment of patient Cou. This CTCL line (Cou-LS) was generated by culturing the tumoral skin cells with rIL-2 and rIL-7. The phenotype of the growing line was TCRαβ+CD4+CD8−MHC class I+ and MHC class II−, and was the same as that of the fresh tumor cells directly isolated from the skin (see below). The proliferative response to cytokines of the tumor cell line Cou-LS is shown in Table 1. IL-7, IL-4, and to a lesser extent IL-2 were growth factors for this tumor cell line. In addition, a synergistic effect of IL-2 and IL-7 was observed for the growth of this cell line, whereas no additive effect was observed with IL-4 and IL-7. Similar proliferative results were obtained with a tumor T-cell line developed from the blood, named Cou-LB (data not shown).
Proliferative Response to Cytokines of the Tumor Cell Line Cou-LS
| Culture Conditions . | Proliferative Response . |
|---|---|
| Medium | 115 ± 31* |
| IL-2 | 1,923 ± 481 |
| IL-4 | 9,094 ± 441 |
| IL-7 | 19,983 ± 3,512 |
| IL-2 + IL-7 | 38,619 ± 5,064 |
| IL-4 + IL-7 | 18,703 ± 1,006 |
| Culture Conditions . | Proliferative Response . |
|---|---|
| Medium | 115 ± 31* |
| IL-2 | 1,923 ± 481 |
| IL-4 | 9,094 ± 441 |
| IL-7 | 19,983 ± 3,512 |
| IL-2 + IL-7 | 38,619 ± 5,064 |
| IL-4 + IL-7 | 18,703 ± 1,006 |
*Results are expressed as mean cpm ± SD for tritiated thymidine incorporated by 50 × 103 cells during the last 18 hours of a 3-day culture.
Characterization of the TCR transcripts in Cou-LS tumor cell line.
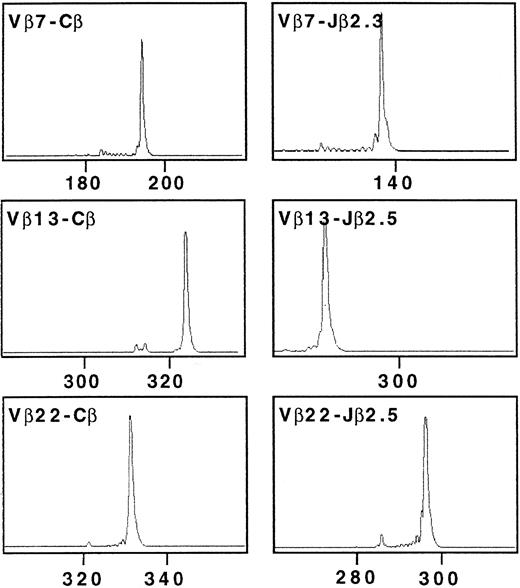
The TCRβ gene segment usage by Cou-LS tumor cell line was determined using an RT-PCR approach and a panel of previously described Vβ and Jβ primers. TCR β chain structure determination of Cou-LS tumor cell line showed three TCR β transcripts corresponding to Vβ7/Jβ2.3, Vβ13/Jβ2.5, and Vβ22/Jβ2.5 (Fig 2). These results suggested a trisomy of chromosome 7, which was further evidenced by FISH, showing that Cou-LS is a diploid cell line with homogenous trisomy 7 and no translocation (Fig 3). It must be noted that this chromosomal abnormality was not secondary to the culture with cytokines, as the three TCR β transcripts were also found in the tumor fragment and in the PBL of the patient. Direct sequencing of the junctional region corresponding to each of these TCR β transcripts indicated that only the Vβ13/Jβ2.5 transcript was in frame. The sequence of the CDR3 of the Vβ13/Jβ2.5 transcript was: (Vβ13.2) TGT GCC AGC AG … C CCC AGC GGG CGG AAA … CAA GAG (Jβ2.5).
Complementarity-determining region 3 (CDR3) size analysis of TCR Vβ7, Vβ13, and Vβ22 transcripts in Cou-LS tumor cell line analyzed with fluorescent Cβ, Jβ2.3, and Jβ2.5. cDNA made from total RNA extracted from Cou-LS were amplified in a PCR reaction primed by Vβ7/Vβ13/Vβ22 and Cβ-specific primers. The unlabeled amplification products were elongated with nested fluorescent Cβ, Jβ2.3, and Jβ2.5. Aliquots were subjected to electrophoresis and analysis on an automated sequencer. Cou-LS tumor cell line showed three TCR β transcripts corresponding to Vβ7/Jβ2.3, Vβ13/Jβ2.5, and Vβ22/Jβ2.5.
Complementarity-determining region 3 (CDR3) size analysis of TCR Vβ7, Vβ13, and Vβ22 transcripts in Cou-LS tumor cell line analyzed with fluorescent Cβ, Jβ2.3, and Jβ2.5. cDNA made from total RNA extracted from Cou-LS were amplified in a PCR reaction primed by Vβ7/Vβ13/Vβ22 and Cβ-specific primers. The unlabeled amplification products were elongated with nested fluorescent Cβ, Jβ2.3, and Jβ2.5. Aliquots were subjected to electrophoresis and analysis on an automated sequencer. Cou-LS tumor cell line showed three TCR β transcripts corresponding to Vβ7/Jβ2.3, Vβ13/Jβ2.5, and Vβ22/Jβ2.5.
FISH performed on Cou-LS cells with whole chromosome paint for chromosome 7. To the top a nucleus with three chromosomal domains; to the bottom a mitosis with three labeled chromosomes 7.
FISH performed on Cou-LS cells with whole chromosome paint for chromosome 7. To the top a nucleus with three chromosomal domains; to the bottom a mitosis with three labeled chromosomes 7.
TCR β gene rearrangement analysis in the skin tumor.
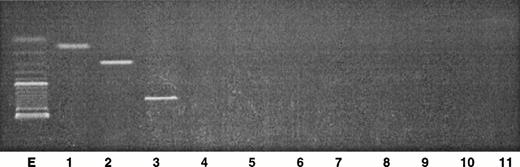
To discriminate the nonneoplastic cells from the tumor clone within the initial skin tumor cells, PCR analysis was performed on genomic DNA extracted from the fresh skin tumor. The results clearly show that the three rearrangements Vβ7/Jβ2.3, Vβ13/Jβ2.5, and Vβ22/Jβ2.5 were amplified at the genomic level (Fig4). It must be noted that the size corresponding to the Vβ22/Jβ2.5 rearrangement corresponded to a higher molecular weight, compared with the expected size obtained from the corresponding cDNA (Fig 2). The two other transcripts corresponded to the expected size. As control, amplification of Vβ5 and Vβ8 primers with Jβ1.1, Jβ1.2, Jβ2.1, and Jβ2.7 gave negative results. These primers were retrospectively chosen according to the results obtained with PCR analysis on cDNA, indicating a single peak.
Analysis of the PCR products obtained from genomic DNA in the skin tumor. Two microliters of the DNA sample were amplified with a 30-cycle PCR, using primers specific for Vβ genes in combination with Jβ primers. The PCR products were analyzed by electrophoresis through a 2% agarose gel. Lane E, size markers (100 bp); lane 1, Vβ7/Jβ2.3; lane 2, Vβ13/Jβ2.5; lane 3, Vβ22/Jβ2.5; lane 4, Vβ5/Jβ1.1; lane 5, Vβ5/Jβ1.2; lane 6, Vβ5/Jβ2.1; lane 7, Vβ5/Jβ2.7; lane 8, Vβ8/Jβ1.1; lane 9, Vβ8/Jβ1.2; lane 10, Vβ8/Jβ2.1; lane 11, Vβ8/Jβ2.7.
Analysis of the PCR products obtained from genomic DNA in the skin tumor. Two microliters of the DNA sample were amplified with a 30-cycle PCR, using primers specific for Vβ genes in combination with Jβ primers. The PCR products were analyzed by electrophoresis through a 2% agarose gel. Lane E, size markers (100 bp); lane 1, Vβ7/Jβ2.3; lane 2, Vβ13/Jβ2.5; lane 3, Vβ22/Jβ2.5; lane 4, Vβ5/Jβ1.1; lane 5, Vβ5/Jβ1.2; lane 6, Vβ5/Jβ2.1; lane 7, Vβ5/Jβ2.7; lane 8, Vβ8/Jβ1.1; lane 9, Vβ8/Jβ1.2; lane 10, Vβ8/Jβ2.1; lane 11, Vβ8/Jβ2.7.
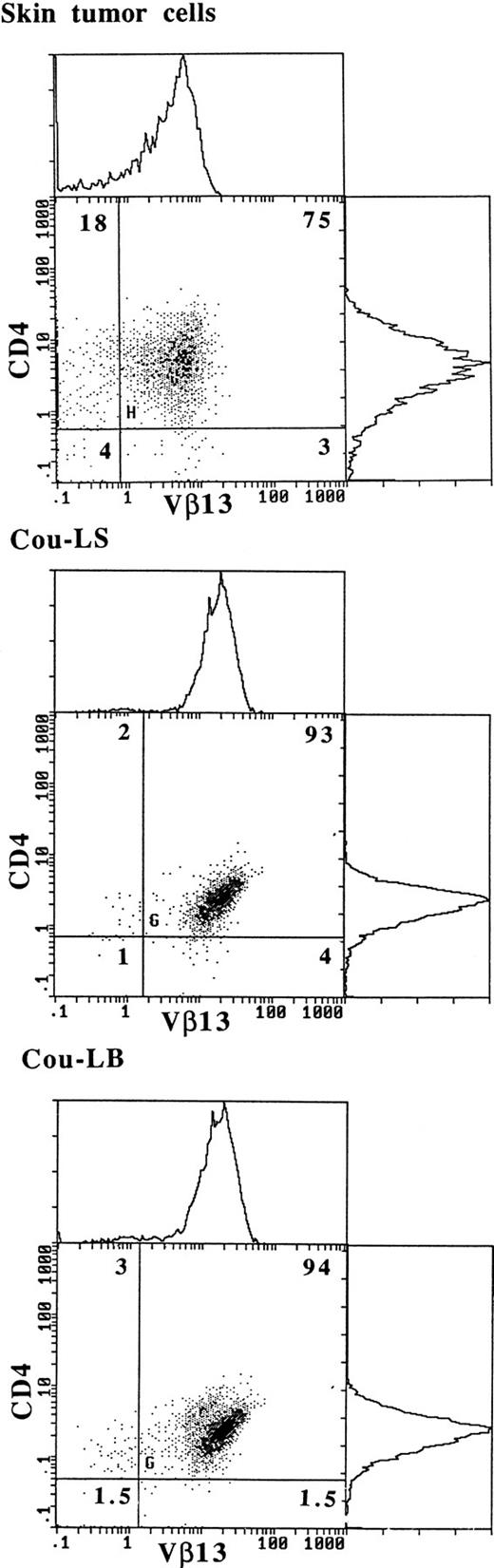
Vβ13 is expressed on the membrane of fresh tumor cells and of both T-cell lines.
To confirm that TCR Vβ13 was expressed by tumor cells, two-color immunofluorescence analyses were performed on fresh skin tumor cells and on both T-cell lines Cou-LS and Cou-LB (Fig 5). The results show that within the initial tumor, 80% of the CD4+ lymphocytes expressed Vβ13. It must be noted that a minority of the Vβ13+population was CD4−. In addition, the Vβ13− reactive lymphocytes were composed mainly of CD4+ lymphocytes. Both long-term T-cell lines derived respectively from the skin and from the blood had a CD4+Vβ13+ phenotype. The PBL from the patient contained 30% of CD4+Vβ13+ tumoral cells (data not shown).
Expression of Vβ13 on fresh skin tumor cells and on both cultured T-cell lines Cou-LS and Cou-LB. Double immunostaining flow-cytometric analysis was performed, using a PE-conjugated anti-CD4 MoAb, and an anti-Vβ13 MoAb plus an FITC-conjugated goat anti-mouse Ig.
Expression of Vβ13 on fresh skin tumor cells and on both cultured T-cell lines Cou-LS and Cou-LB. Double immunostaining flow-cytometric analysis was performed, using a PE-conjugated anti-CD4 MoAb, and an anti-Vβ13 MoAb plus an FITC-conjugated goat anti-mouse Ig.
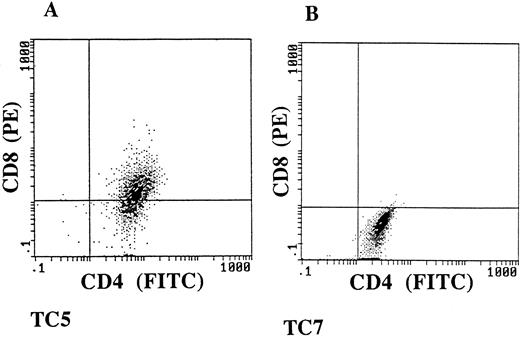
Isolation of CD4+ and CD4+CD8dim+ cytotoxic T-lymphocyte clones from Cou TIL-cultures.
To further study the specificity of the cultured tumor-infiltrating lymphocytes, we developed cloned T-lymphocyte populations from tumor fragment cocultures obtained from patient Cou. The T-lymphocyte clones were obtained by limiting dilution, using as feeder cells irradiated allogeneic PBL stimulated with PHA. Ten CD3+ TCR α/β+ MHC class I+ and MHC class II+ long-term growing T-cell clones were studied for their phenotype and cytotoxic activity (data not shown). Two of them were selected for their ability to kill autologous fresh tumor cells isolated from the skin tumor (Table 2). The phenotypic analysis of these two autologous tumor-specific T-cell clones, named TC5 and TC7, showed that TC5 was double-positive CD4+CD8dim+, and TC7 single-positive CD4+CD8− (Fig6). These two clones have maintained stable reactivity and phenotype in culture for more than 1 year. We next showed that TC5 and TC7 T-cell clones could also have a similar lytic activity against the two cultured Vβ13+ tumor cell lines Cou-LS and Cou-LB established, respectively, from the skin and from the blood, previously mentioned. Both tumor cell lines were lysed at levels ranging from 15% to 35%, and the cytotoxicity levels were increased for higher effector-to-target ratios (Table 2). It is interesting to postulate that enhancement of the killing observed in the presence of an anti-CD3 MoAb could be caused by the fact that both the effector cells and the target tumor cells were T lymphocytes. The cytotoxic activity of these two T-cell clones was highly specific, as they were able to lyse neither allogeneic tumor T cells nor the natural killer–sensitive target cells K562 (Table 2).
Cytotoxic Activity of TC5 and TC7 Clones Against Autologous Tumor Cells and the Tumor Cell Lines Cou LS and Cou LB
| T-cell Clones . | Autologous Tumor T Cells E/T Ratio* . | Cou-LS E/T Ratio . | Cou-LB E/T Ratio . | Allogeneic Tumor T Cells E/T Ratio . | K562 E/T Ratio . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 50/1 . | 10/1 . | 50/1 . | 25/1 . | 50/1 . | 10/1 . | 50/1 . | 10/1 . | 50/1 . | 10/1 . | |
| TC5 | 42† | 13 | 33 | 16 | 24 | 10 | 3 | 3 | 2 | 2 |
| TC5 + CD3 MoAb | ND | 47 | 36 | 49 | 26 | ND | ND | |||
| TC7 | 27 | 12 | 17 | 10 | 23 | 11 | 2 | 2 | 3 | 0 |
| TC7 + CD3 MoAb | ND | 48 | 31 | 33 | 19 | ND | ND | |||
| T-cell Clones . | Autologous Tumor T Cells E/T Ratio* . | Cou-LS E/T Ratio . | Cou-LB E/T Ratio . | Allogeneic Tumor T Cells E/T Ratio . | K562 E/T Ratio . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 50/1 . | 10/1 . | 50/1 . | 25/1 . | 50/1 . | 10/1 . | 50/1 . | 10/1 . | 50/1 . | 10/1 . | |
| TC5 | 42† | 13 | 33 | 16 | 24 | 10 | 3 | 3 | 2 | 2 |
| TC5 + CD3 MoAb | ND | 47 | 36 | 49 | 26 | ND | ND | |||
| TC7 | 27 | 12 | 17 | 10 | 23 | 11 | 2 | 2 | 3 | 0 |
| TC7 + CD3 MoAb | ND | 48 | 31 | 33 | 19 | ND | ND | |||
Abbreviation: ND, not done.
Cytotoxicity assays were performed according to a standard51Cr-release method. Assays at various effector to target cell (E/T) ratios with 5 × 10351Cr-labeled target cells/well were performed in triplicate, using 96-well V-bottomed microtiter plates.
Results are expressed as percentages of specific lysis.
TC5 and TC7 have a CD4+CD8dim+ and CD4+CD8− phenotype. Double immunostaining flow-cytometric analysis was performed, using an FITC-conjugated anti-CD4 MoAb, and a PE-conjugated anti-CD8 MoAb.
TC5 and TC7 have a CD4+CD8dim+ and CD4+CD8− phenotype. Double immunostaining flow-cytometric analysis was performed, using an FITC-conjugated anti-CD4 MoAb, and a PE-conjugated anti-CD8 MoAb.
The cytotoxic activity of TC5 and TC7 T cell clones was restricted by MHC class I molecules.
To further define the restriction element of the cytotoxic activity of TC5 and TC7, we performed blocking experiments, using a panel of MoAbs. Results from a representative experiment are shown in Fig 7. The monomorphic anti-class I MoAb W6/32 induced a significant inhibition of the specific antitumoral cytotoxic activity of TC5 and TC7, whereas, as expected from previous phenotypic analyses indicating that the tumor cell lines were MHC class II–negative, the anti–MHC class II MoAb L243 had no effect. At the effector cell level, neither CD4 nor CD8 molecules were involved in the effector cell cytotoxic activity of both T-cell clones. Further, blocking experiments performed with the B1.23.2 MoAb, which reacts preferentially with HLA-B and HLA-C molecules, indicated that only TC5 T-cell clone cytotoxic activity was strongly inhibited by this MoAb, whereas TC7 was not blocked. These results suggest that TC5 reacts with a peptide presented by HLA-B or HLA-C molecules, whereas TC7 recognizes a peptide presented by HLA-A molecules.
The cytotoxic activity of the clones TC5 (Fig 4A and C) and TC7 (Fig 4B and D) on the autologous tumor cell line Cou-LS is blocked by anti–MHC class I MoAbs. Anti-CD4 and anti-CD8 MoAbs were added to effector cells for 30 minutes, and cells were washed before use. W6/32, B1.23.2, and L243 were added to the51Cr-labeled target cells for 30 minutes and cells were washed before use.
The cytotoxic activity of the clones TC5 (Fig 4A and C) and TC7 (Fig 4B and D) on the autologous tumor cell line Cou-LS is blocked by anti–MHC class I MoAbs. Anti-CD4 and anti-CD8 MoAbs were added to effector cells for 30 minutes, and cells were washed before use. W6/32, B1.23.2, and L243 were added to the51Cr-labeled target cells for 30 minutes and cells were washed before use.
CDR3 size analysis of Vβ/Cβ PCR products in skin tumor and PBL.
We next examined CDR3 size Vβ distribution in the skin tumor and in PBL by runoff analysis as described previously.30 The RNA was reverse transcribed and amplified by PCR with the use of 24 Vβ subfamily primers and 1 fluorescent Cβ-specific primer. The labeled PCR products were analyzed on an automated DNA sequencer to determine the VDJ (CDR3) size distribution and the intensity of the signals. The results from tumor sample and PBL of patient Cou are shown in Fig 8. The profiles, which reflected the CDR3 size diversity in a given Vβ subfamily, could be divided into two categories: some displayed five to eight peaks spaced by three nucleotides each in a nearly Gaussian distribution (such as Vβ1 or Vβ6 in tumor, or Vβ3 in PBL). Others contained one or several dominant peaks highly suggestive of clonal expansions (such as Vβ7, Vβ8, Vβ13, Vβ15, Vβ22, and Vβ23). It must be noted that these dominant peaks were detected both in the tumor and in PBL, which is consistent with the fact that PBL harbored 30% of circulating tumor cells, which express Vβ7/Jβ2.3, Vβ13/Jβ2.5, and Vβ22/Jβ2.5.
Vβ-Cβ VDJ junction CDR3-size distribution profiles in skin tumor (A) and PBL (B) from patient Cou. cDNA made from total RNA extracted from the tumor biopsy sample or the PBL was subjected to PCR amplification with Vβ-specific primers and a fluorescent Cβ primer. The labeled PCR products were subjected to electrophoresis and analysis on an automated DNA sequencer. The fluorescence profiles (x-axis, Vβ-Cβ size in nucleotides; y-axis, fluorescence intensity) of the 24 Vβ subfamilies are shown.
Vβ-Cβ VDJ junction CDR3-size distribution profiles in skin tumor (A) and PBL (B) from patient Cou. cDNA made from total RNA extracted from the tumor biopsy sample or the PBL was subjected to PCR amplification with Vβ-specific primers and a fluorescent Cβ primer. The labeled PCR products were subjected to electrophoresis and analysis on an automated DNA sequencer. The fluorescence profiles (x-axis, Vβ-Cβ size in nucleotides; y-axis, fluorescence intensity) of the 24 Vβ subfamilies are shown.
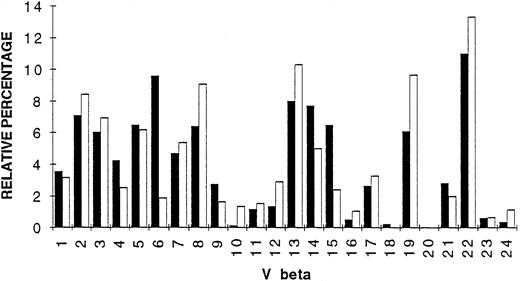
Comparative analysis of TCR Vβ gene segment expression.
To evaluate Vβ gene segment expression in tumor and PBL of patient Cou, Vβ-Cβ PCR amplification (30 cycles) was performed, using a fluorescent Cβ primer as described previously.30 Labeled PCR products were directly analyzed on the automated sequencer and the intensities of the peaks were obtained at the end of the electrophoresis run. The intensities of the different peaks present in all Vβ subfamilies were calculated and represented as histograms. Figure 9 summarizes the results of TCR Vβ gene segment usage in the tumor and PBL of patient Cou. As shown in Fig9, the TCR repertoire is diverse with the presence of most Vβ genes. Comparisons between tumor and PBL repertoires show some differences, namely that Vβ6 gene segment was overexpressed in the tumor (five times more expressed in the tumor than in PBL). Vβ15 and Vβ14 genes were, respectively, 2.7 and 1.7 times more expressed in the tumor than in PBL.
Vβ gene segment usage in skin tumor (▪) and PBL (□) of patient Cou. The Vβ subfamily gene usage was determined by quantitative PCR, and expressed as percentage (histogram bars) of the sum of the fluorescence intensities present in all detected peaks obtained after PCR amplification with a fluorescent Cβ primer.
Vβ gene segment usage in skin tumor (▪) and PBL (□) of patient Cou. The Vβ subfamily gene usage was determined by quantitative PCR, and expressed as percentage (histogram bars) of the sum of the fluorescence intensities present in all detected peaks obtained after PCR amplification with a fluorescent Cβ primer.
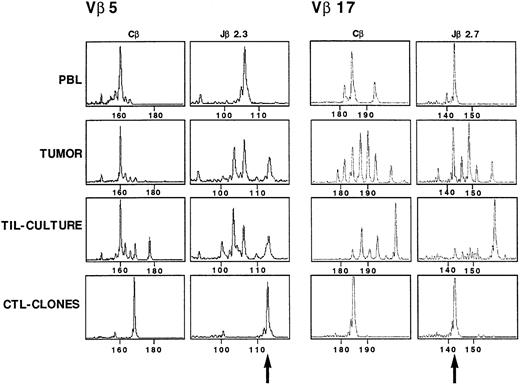
Characterization and in situ detection of the TCR transcripts in TC5 and TC7 CTL clones.
The TCRβ gene segment usage by TC5 and TC7 CTL clones was determined using an RT-PCR approach and a panel of Vβ and Jβ primers as previously described for the tumor cell line. TC5 expressed a Vβ5/Jβ2.3 rearrangement, and TC7 a Vβ17/Jβ2.7 rearrangement (Fig 10). The junctional region of TC5 and TC7 TCRβ chain was then analyzed by cloning and sequencing amplified Vβ5/Jβ2.3 and Vβ17/Jβ2.7 transcripts, respectively. Table 3 shows the nucleic acid sequences of both TC5 and TC7 junctional regions. Amplified Vβ5/Cβ and Vβ17/Cβ PCR products from PBL, tumor, and TIL-culture RNA samples were successively copied with Cβ and Jβ primers. TC5 clone gave a monoclonal peak of 113 nucleotides when copied with Jβ2.3 primer, which was also detected in the initial tumor, and in the TIL-culture, but not detected in PBL (Fig 10). TC7 clone gave a monoclonal peak of 142 nucleotides when copied with Jβ2.7 primer, which was detected in uncultured tumor, and was dominant in PBL. It is noteworthy that it was less expanded in the TIL-culture.
CDR3 size profiles of TCR Vβ5 and TCR Vβ 17 transcripts in samples from patient Cou analyzed with fluorescent Cβ, Jβ2.3, and Jβ2.7. cDNA made from total RNA extracted from TC5 (Vβ5) and TC7 (Vβ17) CTL clones, TIL-culture, tumor suspensions, and PBL from the patient were amplified in a PCR reaction primed by Vβ5/Vβ17– and Cβ–specific primers. The unlabeled amplification products were elongated with nested fluorescent Cβ, Jβ2.3, and Jβ2.7. Aliquots were subjected to electrophoresis and analysis on an automated sequencer. Arrows indicate the signal corresponding to the two CTL clones TCR transcripts.
CDR3 size profiles of TCR Vβ5 and TCR Vβ 17 transcripts in samples from patient Cou analyzed with fluorescent Cβ, Jβ2.3, and Jβ2.7. cDNA made from total RNA extracted from TC5 (Vβ5) and TC7 (Vβ17) CTL clones, TIL-culture, tumor suspensions, and PBL from the patient were amplified in a PCR reaction primed by Vβ5/Vβ17– and Cβ–specific primers. The unlabeled amplification products were elongated with nested fluorescent Cβ, Jβ2.3, and Jβ2.7. Aliquots were subjected to electrophoresis and analysis on an automated sequencer. Arrows indicate the signal corresponding to the two CTL clones TCR transcripts.
VDJ Sequences of TC5 and TC7
| Clone . | Vβ . | Framework . | CDR3 . | Framework . | Jβ . |
|---|---|---|---|---|---|
| TC5 | 5 | TCG TCG | AGC TCC CGC CTT GGA | TCG TGT | 2.3 |
| TC7 | 17 | GCC AGC TC | A CCT GGG CGA CAG TT | C GAG CAG TAC | 2.7 |
| Clone . | Vβ . | Framework . | CDR3 . | Framework . | Jβ . |
|---|---|---|---|---|---|
| TC5 | 5 | TCG TCG | AGC TCC CGC CTT GGA | TCG TGT | 2.3 |
| TC7 | 17 | GCC AGC TC | A CCT GGG CGA CAG TT | C GAG CAG TAC | 2.7 |
DISCUSSION
The skin lesions in CTCL contain an heterogeneous cell infiltrate including malignant cells which are usually CD3+CD4+ lymphocytes that phenotypically resemble mature T-helper cells. Although these tumor cells comprise the majority of cell population, there is usually an admixture of nonneoplastic tumor-infiltrating T lymphocytes, macrophages, and other inflammatory cells. Staining of the clonal population with anti-Vβ MoAbs has shown that in early mycosis fungoides the clonal proliferation can be restricted to the epidermal part of the infiltrate, suggesting that the dermal part of the infiltrate is mainly composed of reactive lymphocytes.20 Other studies have shown that CTCL skin lesions contain TIL expressing an activated cytotoxic phenotype,24 and that these TIL could influence the long-term survival of patients with CTCL.32 Thus, CTCL represents a unique tumor model where both the tumoral and the nonmalignant infiltrating lymphocytes are T lymphocytes. In the present study, we examined whether CTCL TIL exhibit reactivity against the autologous tumor. We show that T lymphocytes obtained from CTCL skin fragments mechanically dispersed into single cell suspension or from Sézary syndrome PBL displayed cytotoxicity against autologous tumor cells after several days in culture with rIL-2 and rIL-7. This observation is in agreement with results of previous studies, which have shown that IL-7 can increase the generation and propagate the long-term growth of anti-tumor CTL.33
IL-7 has also been shown to be a potent growth factor for CTCL tumor cells.34 Exposure of Sézary cells to IL-7 increases IL-2 receptor and IL-7 receptor expression, and IL-7 and IL-2 have synergistic effects on the growth of CTCL tumor cells.34,35IL-7 is produced by keratinocytes,36 and keratinocyte-derived IL-7 is a growth factor for CTCL-derived cell lines.34 IL-7 transgenic mice develop a progressive cutaneous disorder with a cutaneous lymphoid infiltrate.37IL-7 mRNA is found in disease-involved skin and blood CTCL cells,35 and most CTCL express IL-7 receptor.38These findings suggest that IL-7 may represent an important cytokine for the pathophysiology of CTCL, with possible autocrine or paracrine growth-stimulating properties. In addition, the use of IL-7 may contribute to maintain in vitro the immunogenicity of CTCL tumor cells. We established long-term tumor T-cell lines from the skin and from the blood of patient Cou by culturing them with IL-7 and IL-2. The same Vβ transcripts were found to be expressed by the two long-term T-cell lines and by fresh tumor cells, showing that both T-cell lines were derived from the same clone of tumor cells.
To further study the phenotype and the function of tumor-infiltrating lymphocytes, T-cell clones were isolated from CTCL skin fragments of patient Cou. Two cytotoxic T-cell clones were selected initially for their ability to kill the tumor cells obtained from the previously frozen tumor fragment. These two T-cell clones, namely TC5 and TC7, had, respectively, a CD3+α/β+CD4+CD8dim+and CD3+α/β+CD4+CD8−phenotype. TC5 and TC7 also had a specific cytotoxic activity directed against the skin- and the blood-derived tumor cell lines Cou-LS and Cou-LB. These results suggest that, in contrast to recent results presented with melanoma-specific CTL clones,39 fresh tumor cells and cultured tumor cell lines express the same tumor antigens. In addition, these results show that identical tumor peptides are presented by the tumor cells both in the skin and in the blood. We performed blocking experiments to define the restriction element of these two T-cell clones. We found that the cytotoxic activities of TC5 and TC7 were inhibited by the monomorphic anti–MHC class I MoAb W6/32. In contrast, TC5 and TC7 were differentially affected in their cytotoxic activity by the MoAb B1.23.2, which reacts with HLA-B and HLA-C molecules. These results suggest that the TC5 T-cell clone detects a peptide presented by HLA-B or -C molecules, whereas TC7 recognizes a distinct peptide presented by HLA-A. The majority of CD4+ clones are MHC class II–restricted, whereas CD8+ clones are mainly MHC class I–restricted. However, CD4+ MHC class I–restricted mature T-cell clones have already been reported, indicating that the generally accepted model for thymic selection may not be absolute.40-42 In addition, the lysis of tumor cells expressing class I but not class II molecules by cytotoxic CD4+43,44 and CD4+CD8+45T-cell clones has already been reported. In these models, the cytotoxic activity was restricted by the class I antigens, and CD4 and class II interactions were not essential for the lytic interactions.44 45
There is only one report on CD8+ class I–restricted tumor-specific cytotoxic T-cell clones in CTCL,25 showing several major differences with our study: (1) the cytotoxic T cells were isolated from the blood of patients with Sézary syndrome; (2) they were isolated from the CD8-enriched fraction of PBL; (3) cloned CD8 cells did not lyse unmodified autologous tumor targets, and the cytotoxic activity of these clones required the preculture of the tumor targets with autologous gamma-irradiated tumor cultured in medium containing 15% fetal bovine serum; (4) cultured tumor cells expressed variable amounts of MHC class I molecules. In our study, the tumor cell lines expressed constant amounts of MHC class I histocompatibility molecules and were MHC class II−. It must be noted that in contrast to cutaneous lymphomas of B-cell origin which are usually MHC class II+, CTCL do not always express MHC class II gene products. Thus, the present study is the first demonstration of the presence inside the cutaneous infiltrate of CD4+ and CD4+CD8dim+ cytotoxic tumor-specific T-cell clones.
Finally, it was important to assess whether the tumor-specific T-cell clones were preferentially expanded in vivo. The TCR Vβ gene segment usage of the tumor cell lines and the cytotoxic T-cell clones was determined, using RT-PCR and a panel of Vβ and Jβ primers. TC5 and TC7 were, respectively, found to express Vβ5/Jβ2.3 and Vβ17/Jβ2.7 rearrangements. Three TCRβ rearrangements corresponding to Vβ7/Jβ2.3, Vβ13/Jβ2.5, Vβ22/Jβ2.5 were found to be expressed by the skin- and the blood-derived tumor cell lines. These three rearranged gene segments were related to a trisomy in the tumor cells. Various chromosomal abnormalities, including chromosome 7, have been reported in CTCL.46However, this is the first report of three TCR Vβ transcripts associated to a trisomy 7 in CTCL tumor cells. Using anti-TCR Vβ MoAbs, only Vβ13 was clonally detected on the cell membrane. As expected from CDR3-sequencing, Vβ22 and Vβ7 were not detected by the specific MoAbs on tumor cell surface. This result excluded the possibility that another TCR Vβ could be present on tumor cell membrane, since the expression of two TCR transcripts has already been reported.47 CDR3 analysis of Vβ-Cβ PCR products showed several dominant peaks, both in tumor and PBL. Vβ-specific amplification revealed comparable representations of the Vβ7, Vβ13, and Vβ22 segments, corresponding to the tumor cells that were present both in the skin tumor and in the blood, indicating that the trisomy 7 was already present in the tumor cells and was not a consequence of the in vitro culture. Furthermore, PCR analysis on genomic DNA of the tumor indicated the presence of the three rearrangements detected at the cDNA level. Comparison of the CDR3-size distribution of TCR-Vβ transcripts of both T-cell clones showed that TC5 and TC7 were present in the lesional tumor site and expanded at a various extent in TIL-culture. In addition, TC7 was also found in PBL, whereas TC5 was totally absent from patient PBL. These results show the presence within the tumor infiltrate of CTCL of tumor-specific, class I–restricted cytotoxic T lymphocytes, some of which can be present in the blood. These findings suggest that these antitumoral effector T cells participate to the antitumor reactivity of the host immune system. However, the CTCL tumor cells could be able to avoid immune destruction by several mechanisms, such as abnormal T-lymphocyte signal transduction48 or the secretion of soluble inhibitory factors, such as transforming growth factor-β or IL-10. Indeed, overexpression of IL-10 has recently been shown in advanced CTCL that could contribute to the downregulation of this specific antitumor reactivity.49 Fas-L expression could be another strategy used by tumor cells to escape immune rejection.50
In conclusion, these results constitute a unique model for the study of human T-T lymphocyte interactions. In addition, they represent a fundamental step for further isolation and characterization of the class I peptides that are important for the priming of antitumor responses in CTCL, including the TCR idiotype expressed by the tumor.51 Identification of peptide antigens will enable the development of peptide vaccine therapies in CTCL.
M.B. and H.E. contributed equally to this work.
Supported by grants from INSERM, Paris XII University, and Association pour la Recherche sur le Cancer.
Address reprint requests to Martine Bagot, MD, PhD, INSERM U448, Faculté de Médecine, 8 Avenue du Général Sarrail, 94010, Créteil, France.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal