Abstract
In contrast to low-grade B-cell lymphomas originating in the gastrointestinal (GI) tract, only few cytogenetic data are available for the large cell, highly malignant variants. We studied 31 large B-cell lymphomas of the GI tract by comparative genomic hybridization (CGH) and fluorescence in situ hybridization using specific DNA probes (FISH). The most frequent aberrations were gains of all or of parts of chromosomes 11 (11 cases), 12 (9 cases), 1q (4 cases), and 3q (4 cases). Losses of parts of chromosome 6q and of parts of the short arm of chromosome 17 (6 cases each) were found most frequently. In four cases a total of seven high-level DNA amplifications was detected. In two of these cases, involvement of specific protooncogenes (RELand MYC) was shown. Some genetic aberrations seemed to be associated with an inferior clinical course: patients with ≥2 aberrations had a significantly shorter median survival. Furthermore, all patients with gains of all or parts of chromosome arm 1q and with high-level DNA amplifications as well as seven of nine patients with gains of all or parts of chromosome 12 died of lymphoma. In conclusion, the pattern of chromosomal gains and losses in large B-cell lymphomas was different from data reported for low-grade (MALT) lymphomas of the stomach and bowel, especially with respect to the high incidence of partial gains of chromosome arm 11q and of all or parts of chromosome 12 and the low frequency of polysomy 3. In addition, our data suggest that chromosomal gains and losses detected by CGH and FISH may predict for the outcome of patients with this tumor entity.
THE MAJORITY OF primary extranodal lymphomas arise in the gastrointestinal (GI) tract. Recently, low-grade lymphoma of the mucosa-associated tissue (MALT) has attracted a lot of attention, and major advances have been made in the understanding of pathogenetic mechanisms as well as in the development of new treatment strategies.1-3 For these low-grade lymphomas, several cytogenetic studies have revealed characteristic chromosomal aberrations. Trisomy 3, which is rarely found in nodal non-Hodgkin's lymphomas (NHL), has been described in about 60% of low-grade GI lymphomas using interphase cytogenetics.4 In marginal zone B-cell lymphomas, which by morphology and by immunophenotype are regarded to be closely related to MALT lymphomas, trisomy 3 has been found with approximately the same incidence by banding techniques.5 In addition, in low-grade B-cell GI NHL the translocation t(11;18)(q21;q21.1) and trisomies of chromosomes 7, 12, and 18 as well as structural aberrations of chromosome 1p have been identified by banding techniques.6-9
In contrast, only few data are available regarding cytogenetic aberrations in large B-cell GI lymphomas (ie, high-grade lymphomas of the GI tract). In two small series complex karyotypes with multiple structural and numerical aberrations were found by G-banding analysis (ref 10, four cases; ref 7, one case). In addition, Du et al11 have described a high incidence of p53 deletions or mutations in primary extranodal high-grade lymphomas, most of which originated in the GI tract. To obtain a comprehensive view of chromosomal gains and losses, we analyzed samples of 31 primary large B-cell lymphomas of the GI tract by fluorescence in situ hybridization using specific DNA probes (FISH) and by comparative genomic hybridization (CGH).12 For FISH studies a DNA probe set allowing the detection of frequent genomic alterations in nodal B-cell lymphomas was used.13 For 30 of the 31 cases, clinical data were available and were correlated with the molecular cytogenetic data.
MATERIALS AND METHODS
Lymphoma classification.
Freshly frozen tumor samples of 31 consecutive patients (14 male, 17 female; age 23 to 89 years; median 71 years) with primary large B-cell lymphomas of the GI tract were analyzed (24 gastric lymphomas, 4 lymphomas of the small intestine, and 3 lymphomas of the ileocoecal region). Examination of routine histology (hematoxylin and eosin [H&E], Giemsa, periodic acid-Schiff [PAS]-stainings) together with CD20-positivity of the tumor cells lead to the classification of these 31 lymphomas as highly malignant B-cell lymphomas with large cell cytology. Subclassification was carried out according to the revised European-American classification of lymphoid neoplasms (REAL-classification).14 The series did not contain cases of Burkitt's or Burkitt-like lymphomas or immunodeficiency-associated large B-cell lymphoma. The majority of tumor samples did not contain any detectable small- or medium-sized (low-grade) component. Seven tumors (marked by an asterisk in Table 1) were secondary large cell lymphomas on the background of marginal zone B-cell lymphoma (of MALT-type). Not all lymphomas were entirely “diffuse”; a few also featured some nodular pattern of growth. Therefore, the tumors were collectively referred to as “large B-cell lymphomas originating in the GI tract.”
Clinical Data of 31 Patients With Primary Large B-Cell Lymphomas of the GI Tract
| Case . | Sex/Age . | Organ . | Initial Stage-150 . | Current Status . | Survival in mo . |
|---|---|---|---|---|---|
| 1 | F/53 | Small intestine | EII1 | CR | 69 |
| 2 | M/54 | Stomach | EIV | CR | 71 |
| 3 | F/75 | Stomach | EIV | Lymphoma-related death | 4 |
| 4 | M/63 | Ileocoecal region | EIII | Lymphoma-related death | 25 |
| 5 | F/83 | Stomach | EIV | CR | 15 |
| 6 | F/89 | Small intestine | EIV | Lymphoma-related death | 0.5 |
| 7 | F/78 | Stomach | EII1 | Lymphoma-related death | 7 |
| 8 | F/62 | Stomach | EII1 | CR | 62 |
| 9 | F/46 | Stomach | EIV | Lymphoma-related death | 12 |
| 10 | M/62 | Stomach | EIV | CR | 46 |
| 11 | F/41 | Stomach | EI2 | CR | 1 |
| 12-151 | M/63 | Stomach | EII1 | Lymphoma-related death | 3 |
| 13-151 | M/46 | Stomach | EII1 | CR | 39 |
| 14-151 | M/68 | Stomach | EII1 | Lymphoma-related death | 0.5 |
| 15-151 | M/79 | Stomach | EII1 | Lymphoma-related death | 3 |
| 16-151 | M/74 | Small intestine | EIV | Lymphoma-related death | 23 |
| 17 | F/43 | Stomach | EI | CR | 86 |
| 18 | F/74 | Ileocoecal region | EII1 | Lymphoma-related death | 101 |
| 19 | M/63 | Stomach | EI | CR | 48 |
| 20 | F/80 | Stomach | EII1 | CR | 40 |
| 21 | F/71 | Stomach | EI2 | CR | 29 |
| 22 | F/74 | Ileocoecal region | EIV | Lymphoma-related death | 13 |
| 23 | M/80 | Stomach | Biopsy-152 | CR | 36 |
| 24 | F/89 | Small intestine | EI1 | CR | 91 |
| 25-151 | F/77 | Stomach | EII1 | Lymphoma-related death | 4 |
| 26-151 | F/41 | Stomach | EII1 | CR | 72 |
| 27 | F/77 | Stomach | EI2 | CR | 47 |
| 28 | M/74 | Stomach | EI2 | CR | 68 |
| 29 | M/81 | Stomach | EI1 | Lymphoma-related death | 3 |
| 30 | M/74 | Stomach | EI1 | CR | 72 |
| 31 | M/23 | Stomach | EI2 | Lost during follow-up | — |
| Case . | Sex/Age . | Organ . | Initial Stage-150 . | Current Status . | Survival in mo . |
|---|---|---|---|---|---|
| 1 | F/53 | Small intestine | EII1 | CR | 69 |
| 2 | M/54 | Stomach | EIV | CR | 71 |
| 3 | F/75 | Stomach | EIV | Lymphoma-related death | 4 |
| 4 | M/63 | Ileocoecal region | EIII | Lymphoma-related death | 25 |
| 5 | F/83 | Stomach | EIV | CR | 15 |
| 6 | F/89 | Small intestine | EIV | Lymphoma-related death | 0.5 |
| 7 | F/78 | Stomach | EII1 | Lymphoma-related death | 7 |
| 8 | F/62 | Stomach | EII1 | CR | 62 |
| 9 | F/46 | Stomach | EIV | Lymphoma-related death | 12 |
| 10 | M/62 | Stomach | EIV | CR | 46 |
| 11 | F/41 | Stomach | EI2 | CR | 1 |
| 12-151 | M/63 | Stomach | EII1 | Lymphoma-related death | 3 |
| 13-151 | M/46 | Stomach | EII1 | CR | 39 |
| 14-151 | M/68 | Stomach | EII1 | Lymphoma-related death | 0.5 |
| 15-151 | M/79 | Stomach | EII1 | Lymphoma-related death | 3 |
| 16-151 | M/74 | Small intestine | EIV | Lymphoma-related death | 23 |
| 17 | F/43 | Stomach | EI | CR | 86 |
| 18 | F/74 | Ileocoecal region | EII1 | Lymphoma-related death | 101 |
| 19 | M/63 | Stomach | EI | CR | 48 |
| 20 | F/80 | Stomach | EII1 | CR | 40 |
| 21 | F/71 | Stomach | EI2 | CR | 29 |
| 22 | F/74 | Ileocoecal region | EIV | Lymphoma-related death | 13 |
| 23 | M/80 | Stomach | Biopsy-152 | CR | 36 |
| 24 | F/89 | Small intestine | EI1 | CR | 91 |
| 25-151 | F/77 | Stomach | EII1 | Lymphoma-related death | 4 |
| 26-151 | F/41 | Stomach | EII1 | CR | 72 |
| 27 | F/77 | Stomach | EI2 | CR | 47 |
| 28 | M/74 | Stomach | EI2 | CR | 68 |
| 29 | M/81 | Stomach | EI1 | Lymphoma-related death | 3 |
| 30 | M/74 | Stomach | EI1 | CR | 72 |
| 31 | M/23 | Stomach | EI2 | Lost during follow-up | — |
Abbreviation: CR, complete remission.
Staging according to Radaszkiewicz et al, 1992.52
Low-grade component present.
Initial stage unknown.
Microscopic dissection of tumor tissue.
To assure a high tumor cell content in the analyzed sample, which is an important prerequisite for CGH studies, in each case 30 serial 10-μm sections from frozen tumor blocks were obtained. For the reliable identification of tumor areas, every fifth section was stained with H&E. To avoid artefacts caused by fixation and staining, only the nonstained serial sections were used for microdissection. In these sections, areas containing high percentages of tumor cells were identified by comparison with the adjacent stained sections. Such areas of interest were marked and dissected under microscopic control. In the first six cases of our series, sections were stained after microdissection to assess the efficiency of this approach. In each of these cases, the dissected area was still surrounded by lymphoma tissue, indicating the high purity of the selected material.
Comparative genomic hybridization.
Genomic DNA was prepared from fresh tumor tissue as described15 using proteinase K digestion and phenol-chloroform extraction. CGH was performed as previously reported.16 Briefly, tumor DNA was labeled with biotin-16-dUTP (Boehringer Mannheim, Mannheim, Germany) and normal human control DNA was labeled with digoxigenin-11-dUTP (Boehringer Mannheim) by a standard nick-translation reaction. One microgram of biotin-labeled tumor DNA, 1 μg of digoxigenin-labeled control DNA, and 70 μg of human Cot-1-DNA (BRL Life Sciences, Gaithersburg, MD) were cohybridized to slides with metaphase cells from blood of a healthy donor. After hybridization for 1 to 2 days and posthybridization washes, control and test DNAs were detected via rhodamine and fluorescein isothiocyanate (FITC), respectively. For identification, chromosomes were counterstained with DAPI (4,6-diamidino-2-phenylindole).
Digital image analysis.
Image analysis was performed using an epifluorescence microscope (Axioplan; Zeiss, Jena, Germany) and the commercially available image analysis systems CYTOVISION (Applied Imaging, Sunderland, UK) or ISIS (MetaSystems, Altluβheim, Germany). For each case, at least 15 metaphase cells were evaluated. Ratio values of 1.25 and 0.75, which have been proven to provide robust criteria for diagnosing overrepresentation and underrepresentation,17 18 were used as upper and lower thresholds for the identification of chromosomal imbalances. Overrepresentations were considered as high-level amplifications when the fluorescence ratio values exceeded 2.0 or when the FITC fluorescence showed strong focal signals and the corresponding ratio profile was diagnostic for overrepresentation. The extension of imbalanced regions was assessed by the comparison of the fluorescence ratio profiles with the corresponding regions in chromosome ideograms. Assignment of highly amplified sequences to chromosomal bands was performed by comparison of signal intensities and DAPI banding on individual chromosomes.
FISH.
For interphase cytogenetic analysis, six probes detecting imbalanced aberrations and probe sets for the diagnosis of the t(11;14)(q13;q32) and the t(14;18)(q32;q21) were used. The probes screening for imbalanced aberrations mapped to chromosomal regions frequently altered in nodal B-NHL and were as follows: YAC clones 866e7 mapping to 3q26 and 963d6 mapping to 6q21 (both obtained from the CEPH YAC library); YAC clone 755b11 mapping to 11q22.3-23.119,20; “cos p16” consisting of a pool of eight overlapping cosmid clones covering approximately 250 kb of the CDKN2 gene on 9p2121; “cos p53” containing four overlapping cosmid clones mapping to 17p13 spanning the p53 tumor-suppressor gene22; and probe “c13S25”consisting of two cosmid clones (ICRFc108I155 and ICRFc108L2145) mapping to the D13S25 locus on 13q14.23 In addition, in one case (no. 16) the cosmid probe cos-myc 7224 containing sequences of theMYC protooncogene was used. For analysis of the t(14;18)(q32;q21), the following probes were used: a YAC clone containing the BCL2 gene (yA153A6) on chromosome 18; for chromosome 14 a pool of cosmid clones cos-Cα1/2, recognizing the Cα1 and Cα2 gene segments proximal to the JH-region,24 and of YAC clone Y6, identifying VH-segments telomeric to the JH-breakpoints in the IgH gene25 was used. For the detection of the t(11;14)(q13;q32) the same probes were used for chromosome 14; for chromosome 11 the differentially labeled 540-kb YAC clone 55g7 spanning the region between the major translocation cluster and the CCND1 gene in the BCL1 locus at 11q13 was applied.20 Hybridization was performed as described previously to nuclei isolated from frozen tissue samples.26In each experiment, two probes coupled to different reporter groups were hybridized simultaneously and served as mutual internal controls. Preparations were only evaluated for a specific DNA probe, if the respective internal control exhibited two hybridization signals in more than 90% of interphase cells. Thus, a high hybridization efficiency was assured. Experiments were evaluated using an epifluorescence microscope (Axioplan; Zeiss) connected to a charged coupled device (CCD) camera. In each case, at least 200 cells were enumerated.
Southern blot analysis.
Southern blot analysis was performed as described.15Briefly, 8 μg of genomic DNA was digested with EcoRI, separated by agarose gel electrophoresis, and transferred to nylon membranes (Boehringer Mannheim). A 777-bp REL-specific probe representing exon 6b was generated by polymerase chain reaction amplification using c-DNA clone Rc/CMV-c-Rel27,28 as template C (kindly provided by P.A. Baeuerle and M.L. Schmitz, Freiburg, Germany). The probe was labeled by random priming using32PdCTP. For control hybridizations, the genomic fragment gMHC-1-D, 4.2 kb in length, from the cardiac β-myosin heavy-chain gene, MYH7, located on 14q12-q13 was used.29 The degree ofREL amplification was determined by densitometric evaluation of the autoradiograph with hybridization signals from probe and control DNAs.
Twenty-one of the 31 cases were analyzed for the presence of the Epstein-Barr virus (EBV) genome. These included 10 of 11 cases with overrepresentations of sequences on 11q. Ten micrograms of cleaved cellular DNA was separated by agarose gel electrophoresis and transferred onto a nylon filter. Hybridization was performed in 50% formamide, 2× sodium chloride/sodium citrate (SSC) buffer at 42°C using the32P-labeled 3.07-kb EBV-Bgl II U fragment as probe. This probe detected the internal repeat I sequence in the EBV genome with a sensitivity better than 0.1 EBV DNA copies per cell.
RESULTS
Comparative genomic hybridization.
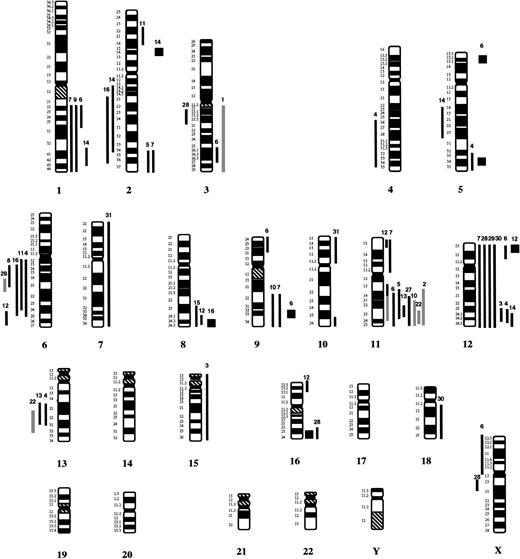
In 21 of the 31 cases, chromosomal imbalances were identified by CGH analysis. Gains of chromosomal material were more frequent than losses (42 gains v 14 losses). The most frequent aberrations were overrepresentations of all or parts of chromosomes 12 (9 cases) and 11 (6 cases) of the long arm of chromosome 1, of parts of chromosome 2 (4 cases each), and of chromosomes 8 and 9 (3 cases each). Gains of parts of chromosomes 5 and 16 were detected in 2 cases each, while a partial gain of chromosome 3 was detected in only one case. The most frequent deletions involved the long arm of chromosome 6 (5 cases) and the long arms of chromosomes 2 and 13 (2 cases each). The gains and losses identified by CGH are summarized in Fig 1. Partial ratio profiles of the cases with gains mapping to chromosome 11 are shown in Fig 2.
Summary of chromosomal imbalances detected by CGH in 31 patients with large B-cell lymphomas of the GI tract. Lines on the left indicate loss of chromosomal material, lines on the right refer to gains of chromosomal material. Squares represent high-level DNA amplifications. Gray lines indicate cases, in which the ratio profiles showed a clear shift toward underrepresentation or overrepresentation; however, the diagnostic thresholds were not reached. In these patients aberrations were confirmed by interphase cytogenetic analysis. The numbers on top of each line refer to the patient analyzed.
Summary of chromosomal imbalances detected by CGH in 31 patients with large B-cell lymphomas of the GI tract. Lines on the left indicate loss of chromosomal material, lines on the right refer to gains of chromosomal material. Squares represent high-level DNA amplifications. Gray lines indicate cases, in which the ratio profiles showed a clear shift toward underrepresentation or overrepresentation; however, the diagnostic thresholds were not reached. In these patients aberrations were confirmed by interphase cytogenetic analysis. The numbers on top of each line refer to the patient analyzed.
Average ratio profiles of chromosomes 11 in cases with overrepresentations of sequences. The percentages below the case numbers indicate the size of the subclone with gain of 11q as determined by interphase cytogenetics. The arrow indicates the chromosomal map position of YAC clone 755b11, which was used for interphase analysis. For cases 2, 6, 10, 12, 13, and 22, images were acquired and evaluated using the Cytovision software package (Applied Imaging). Cases 5, 7, and 27 were evaluated using the ISIS software package (MetaSystems). The ratios of FITC to rhodamine fluorescence are plotted along the chromosomes. The central line (asterisks) indicates a ratio value of 1.0; the lines to the right indicate ratio values of 1.25 (all cases) and 1.5 (all cases except nos. 5 and 7), respectively; and the lines on the left indicate values of 0.75 (all cases) and 0.5 (all cases except nos. 5, 7, and 27), respectively. In case nos. 7 and 27, adequate material was limited and therefore only CGH analysis was performed. #In these cases, the diagnostic thresholds were not reached. However, there is a clear shift of the ratio profiles toward overrepresentation. In these cases, gains of 11q were diagnosed by FISH analysis. For cases 16 and 25, in which gains were diagnosed only in minor subclones, no shifts of the ratio profiles were observed.
Average ratio profiles of chromosomes 11 in cases with overrepresentations of sequences. The percentages below the case numbers indicate the size of the subclone with gain of 11q as determined by interphase cytogenetics. The arrow indicates the chromosomal map position of YAC clone 755b11, which was used for interphase analysis. For cases 2, 6, 10, 12, 13, and 22, images were acquired and evaluated using the Cytovision software package (Applied Imaging). Cases 5, 7, and 27 were evaluated using the ISIS software package (MetaSystems). The ratios of FITC to rhodamine fluorescence are plotted along the chromosomes. The central line (asterisks) indicates a ratio value of 1.0; the lines to the right indicate ratio values of 1.25 (all cases) and 1.5 (all cases except nos. 5 and 7), respectively; and the lines on the left indicate values of 0.75 (all cases) and 0.5 (all cases except nos. 5, 7, and 27), respectively. In case nos. 7 and 27, adequate material was limited and therefore only CGH analysis was performed. #In these cases, the diagnostic thresholds were not reached. However, there is a clear shift of the ratio profiles toward overrepresentation. In these cases, gains of 11q were diagnosed by FISH analysis. For cases 16 and 25, in which gains were diagnosed only in minor subclones, no shifts of the ratio profiles were observed.
In 4 of the 31 patients, a total of 7 high-level DNA amplifications were identified in the tumor samples. These were mapped to the following chromosomal regions: 5p15, 5q33-34 and 9q32-33 (no. 6); 12p12 and 16q23-24 (no. 12); 2p13-15 (no. 14); and 8q24 (no. 16). Partial CGH karyotypes of all chromosomes carrying high-level DNA amplifications are shown in Fig 3.
Partial CGH karyotypes of cases with high-level DNA amplifications. Images were acquired and evaluated using the CYTOVISION software package (Applied Imaging). Numbers at the bottom indicate the respective chromosomes. Hybridization with the tumor DNA is shown as gray level images. The band-like hybridization signals (arrows) indicate highly amplified chromosomal sequences. On the right side of the ideograms, the average ratios of FITC/rhodamine fluorescence are plotted (“ratio profiles”). The central line indicates a ratio value of 1.0; lines to the right indicate ratio values of 1.25 and 1.5, respectively; and lines to the left indicate ratio values of 0.75 and 0.5. n = number of chromosomes analyzed for calculating the respective average ratio profile.
Partial CGH karyotypes of cases with high-level DNA amplifications. Images were acquired and evaluated using the CYTOVISION software package (Applied Imaging). Numbers at the bottom indicate the respective chromosomes. Hybridization with the tumor DNA is shown as gray level images. The band-like hybridization signals (arrows) indicate highly amplified chromosomal sequences. On the right side of the ideograms, the average ratios of FITC/rhodamine fluorescence are plotted (“ratio profiles”). The central line indicates a ratio value of 1.0; lines to the right indicate ratio values of 1.25 and 1.5, respectively; and lines to the left indicate ratio values of 0.75 and 0.5. n = number of chromosomes analyzed for calculating the respective average ratio profile.
Identification of amplified genes.
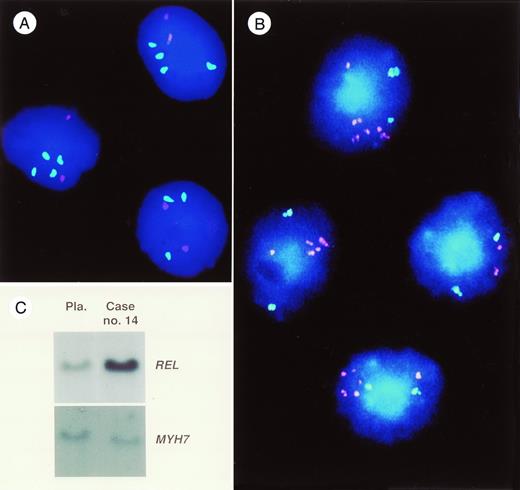
In case 14, a high-level DNA amplification mapping to chromosomal bands 2p13-14 was identified. This is the chromosomal localization of theREL proto-oncogene. Southern blot hybridization showed a sevenfold amplification of this gene (see Fig 4C).
(A) Photomicrograph of a dual-color hybridization obtained with YAC clone 755b11 mapping to 11q22.3-11q23.1 (detected via FITC, green) and YAC clone 866e7 mapping to 3q26 (detected via rhodamine, red) to cells of patient no. 6. In three cells, three or four green hybridization signals are seen indicating an overrepresentation of sequences on band 11q22.3-11q23.1 in these cells. In contrast, only two red hybridization signals are present indicating a normal copy number of sequences on band 3q26. (B) Photomicrograph of a dual-color FISH experiment with cosmid probe cos-myc 72, containing sequences of the MYC proto-oncogene detected via rhodamine (red) and the YAC clone 963d6 mapping to band 6q21 detected via FITC (green). In this case (no. 16), CGH analysis showed a strong bandlike hybridization signal at chromosomal band 8q23-24.3. In three of the cells a tight cluster of red hybridization signals is visible indicating an amplification of these sequences. In contrast, two green signals are seen in all cells. (C) Southern blot analysis of DNA of case no. 14 (right) and DNA from a placenta (left) serving as an internal control. Probes for the REL proto-oncogene andMYH7 (control) are marked on the right side. Note the high intensity of the REL signal in this case. Densitometric evaluation showed a sevenfold amplification of this gene.
(A) Photomicrograph of a dual-color hybridization obtained with YAC clone 755b11 mapping to 11q22.3-11q23.1 (detected via FITC, green) and YAC clone 866e7 mapping to 3q26 (detected via rhodamine, red) to cells of patient no. 6. In three cells, three or four green hybridization signals are seen indicating an overrepresentation of sequences on band 11q22.3-11q23.1 in these cells. In contrast, only two red hybridization signals are present indicating a normal copy number of sequences on band 3q26. (B) Photomicrograph of a dual-color FISH experiment with cosmid probe cos-myc 72, containing sequences of the MYC proto-oncogene detected via rhodamine (red) and the YAC clone 963d6 mapping to band 6q21 detected via FITC (green). In this case (no. 16), CGH analysis showed a strong bandlike hybridization signal at chromosomal band 8q23-24.3. In three of the cells a tight cluster of red hybridization signals is visible indicating an amplification of these sequences. In contrast, two green signals are seen in all cells. (C) Southern blot analysis of DNA of case no. 14 (right) and DNA from a placenta (left) serving as an internal control. Probes for the REL proto-oncogene andMYH7 (control) are marked on the right side. Note the high intensity of the REL signal in this case. Densitometric evaluation showed a sevenfold amplification of this gene.
In case 16, a strong bandlike signal was observed on chromosomal bands 8q23-24.3. This is the region where the MYC proto-oncogene is localized. FISH analysis using an MYC-specific probe showed a tight cluster of signals indicating the presence of an amplification of this gene. In contrast, simultaneous hybridization with the probe YAC963d6 resulted in two distinct hybridization signals in more than 90% of cells (see Fig 4B).
Interphase cytogenetics.
To define the cut-off levels for the various DNA probes, hybridization experiments to cells obtained from frozen samples of normal tonsillar tissue (n = 5) were performed. By analogy to previous interphase studies (see eg, ref 30), the cut-off levels were defined by the mean + 3 SD of the respective results in the experiments with the five tonsils. Thus, a deletion was diagnosed whenever the percentage of cells exhibiting ≤1 hybridization signal exceeded the mean + 3 SD of the percentage of cells exhibiting ≤1 hybridization in the control experiments. For the various probes, the cut-off values for diagnosing a deletion are given in parentheses: YAC 963d6 (3.4%); p16 (9.75%); D13S25 (9.6%); p53 (12.4%). Overrepresentations were diagnosed if cells exhibited ≥3 hybridization signals in percentages exceeding the respective cut-off values: YAC 866e7 (8%); p16 (6.6%); YAC 755b11 (3.1%); D13S25 (12.6%). A t(11;14) was diagnosed if more than 3.5% of interphase cells exhibited two chromosome 11, three chromosome 14 signals, and one cohybridization of a chromosome 11 and a chromosome 14 signal (mean + 3 SD of the respective result in the experiments with the five tonsils). A t(14;18) was diagnosed if more than 4.5% of interphase cells exhibited two chromosome 18 signals, three chromosome 14 signals, and one cohybridization of a chromosome 18 and a chromosome 14 signal.
In 29 of the 31 large B-cell lymphomas, sufficient material for interphase cytogenetics was available. For detection of the t(11;14) and the t(14;18), a subset of 18 cases was investigated. The most frequent aberration was a gain of sequences on the long arm of chromosome 11, which was detected in 9 of 29 cases (percentage of cells with ≥3 hybridization signals: 13% to 85%; median 41%). In 6 of 29 cases, a deletion of the p53 tumor-suppressor gene was identified (13.5% to 80% of cells; median 30%). Only 4 of 29 cases exhibited a gain of sequences on the long arm of chromosome 3 (12%, 26%, 13.5%, and 75% of cells, respectively). In three cases, a gain of sequences on the short arm of chromosome 9 was detected (16%, 29%, and 78% of cells). In an additional case (no. 4) a hyperploidy was diagnosed. In regions with normal genomic content based on the CGH profiles (ie, 6q21 and 3q26), interphase analysis showed more than two hybridization signals in the majority of cells (see Table2). By contrast, interphase cytogenetic analysis using the “cos p16” probe resulted in two hybridization signals in more than 60% of nuclei, demonstrating the presence of a p16 deletion in this case. Deletions within chromosomal band 13q14 were identified in four cases (89%, 21%, two cases 13.5%). One case exhibited a gain on chromosome band 13q14. On chromosomal band 6q21 four cases showed only one hybridization signal in high percentages of cells (92%, 67.5%, 65%, and 46.5%).
FISH Data and Corresponding CGH Karyotypes
| Case . | . | FISH Data* . | (%) . | CGH Karyotype† . |
|---|---|---|---|---|
| 1 | 3q26: | ≥3 signals | (26) | rev ish enh(3q12q29)‡ |
| 2 | 11q22.3-23.1: | ≥3 signals | (21) | rev ish enh(11q13q25)‡ |
| 4 | hyperploidy (mean): | rev ish enh(5q33q35,12q23q24.2), dim(4q25q35,6q12q24,13q14q31) | ||
| (33% dipl.; 20% tripl.; 44.5% tetrapl.) | ||||
| 6q21: | 2 signals | (92) | ||
| 9p21 (p16): | 2 signals | (60.5) | ||
| 13q14: | 2 signals | (89) | ||
| 17p13 (p53): | 2 signals | (56.5) | ||
| 1 signal | (38) | |||
| 5 | 11q22.3-23.1: | ≥3 signals | (61) | rev ish enh(2q35q37,11q13q23) |
| 17p;13 (p53): | ≤1 signal | (58) | ||
| 6 | 3q26: | ≥3 signals | (77) | rev ish enh(1q21q25,3q25q27,9p24p22,11q13q25), dim(Xp22.3p11.2), amp(5p15,9q33) |
| 11q22.3-23.1: | ≥3 signals | (80) | ||
| 9p21 (p16): | ≥3 signals | (78) | ||
| 17p13 (p53): | ≤1 signal | (88.5) | ||
| 8 | 6q21: | ≤1 signal | (46.5) | rev ish dim(6q13q21) |
| 10 | 11q22.3-23.1: | ≥3 signals | (55) | rev ish enh(9q22q34,11q21q25‡) |
| 11 | 6q21: | ≤1 signal | (65) | rev ish enh(2p23p16),dim(6q12q24) |
| 17p13 (p53): | ≤1 signal | (15.5) | ||
| 12 | 3q26: | ≥3 signals | (12) | rev ish enh(8q24.1q24.2,11p15,11q12q24,16p13.3p13.2) |
| 11q22.3-23.1: | ≥3 signals | (41.5) | dim(6q25q26),amp(12p12p13,16q23q24) | |
| 13q14: | ≥3 signals | (40) | ||
| 13 | 11q22.3-23.1: | ≥3 signals | (37.5) | rev ish enh(11q23q24),dim(13q14q31) |
| 13q14: | ≤1 signal | (21) | ||
| t(14;18) | (6) | |||
| 14 | 17p13 (p53): | ≤1 signal | (13.5) | rev ish enh(1p32p43,12q24.1q24.3),dim(2q14.1q34,5q14q23),amp(2p14) |
| 16 | 3q26: | ≥3 signals | (13.5) | rev ish dim(2q21q36,6q13q25),amp(8q23q24.3) |
| p16: | ≥3 signal | (16) | ||
| 11q22.3-23.1: | ≥3 signals | (13.5) | ||
| 21 | 13q14: | ≤1 signal | (13.5) | — |
| 22 | 11q22.3-23.1: | ≥3 signals | (34.5) | rev ish enh(11q23q24)‡ |
| 13q14: | ≤1 signal | (13.5) | rev ish dim(13q22q32)‡ | |
| 23 | 17p13 (p53): | ≤1 signal | (24.5) | — |
| 24 | 9p21 (p16): | ≥3 signals | (42.5) | |
| 25 | 11q22.3-23.1: | ≥3 signals | (13.5) | — |
| 28 | t(11;14) | (9) | rev ish enh(12,16q23q24),dim(Xq13q21) | |
| 29 | 6q21: | ≤1 signal | (67.5) | rev ish enh(12),dim(6q21q23) )‡ |
| Case . | . | FISH Data* . | (%) . | CGH Karyotype† . |
|---|---|---|---|---|
| 1 | 3q26: | ≥3 signals | (26) | rev ish enh(3q12q29)‡ |
| 2 | 11q22.3-23.1: | ≥3 signals | (21) | rev ish enh(11q13q25)‡ |
| 4 | hyperploidy (mean): | rev ish enh(5q33q35,12q23q24.2), dim(4q25q35,6q12q24,13q14q31) | ||
| (33% dipl.; 20% tripl.; 44.5% tetrapl.) | ||||
| 6q21: | 2 signals | (92) | ||
| 9p21 (p16): | 2 signals | (60.5) | ||
| 13q14: | 2 signals | (89) | ||
| 17p13 (p53): | 2 signals | (56.5) | ||
| 1 signal | (38) | |||
| 5 | 11q22.3-23.1: | ≥3 signals | (61) | rev ish enh(2q35q37,11q13q23) |
| 17p;13 (p53): | ≤1 signal | (58) | ||
| 6 | 3q26: | ≥3 signals | (77) | rev ish enh(1q21q25,3q25q27,9p24p22,11q13q25), dim(Xp22.3p11.2), amp(5p15,9q33) |
| 11q22.3-23.1: | ≥3 signals | (80) | ||
| 9p21 (p16): | ≥3 signals | (78) | ||
| 17p13 (p53): | ≤1 signal | (88.5) | ||
| 8 | 6q21: | ≤1 signal | (46.5) | rev ish dim(6q13q21) |
| 10 | 11q22.3-23.1: | ≥3 signals | (55) | rev ish enh(9q22q34,11q21q25‡) |
| 11 | 6q21: | ≤1 signal | (65) | rev ish enh(2p23p16),dim(6q12q24) |
| 17p13 (p53): | ≤1 signal | (15.5) | ||
| 12 | 3q26: | ≥3 signals | (12) | rev ish enh(8q24.1q24.2,11p15,11q12q24,16p13.3p13.2) |
| 11q22.3-23.1: | ≥3 signals | (41.5) | dim(6q25q26),amp(12p12p13,16q23q24) | |
| 13q14: | ≥3 signals | (40) | ||
| 13 | 11q22.3-23.1: | ≥3 signals | (37.5) | rev ish enh(11q23q24),dim(13q14q31) |
| 13q14: | ≤1 signal | (21) | ||
| t(14;18) | (6) | |||
| 14 | 17p13 (p53): | ≤1 signal | (13.5) | rev ish enh(1p32p43,12q24.1q24.3),dim(2q14.1q34,5q14q23),amp(2p14) |
| 16 | 3q26: | ≥3 signals | (13.5) | rev ish dim(2q21q36,6q13q25),amp(8q23q24.3) |
| p16: | ≥3 signal | (16) | ||
| 11q22.3-23.1: | ≥3 signals | (13.5) | ||
| 21 | 13q14: | ≤1 signal | (13.5) | — |
| 22 | 11q22.3-23.1: | ≥3 signals | (34.5) | rev ish enh(11q23q24)‡ |
| 13q14: | ≤1 signal | (13.5) | rev ish dim(13q22q32)‡ | |
| 23 | 17p13 (p53): | ≤1 signal | (24.5) | — |
| 24 | 9p21 (p16): | ≥3 signals | (42.5) | |
| 25 | 11q22.3-23.1: | ≥3 signals | (13.5) | — |
| 28 | t(11;14) | (9) | rev ish enh(12,16q23q24),dim(Xq13q21) | |
| 29 | 6q21: | ≤1 signal | (67.5) | rev ish enh(12),dim(6q21q23) )‡ |
*FISH data: The chromosomal map position of the DNA probes and the percentage of cells with the respective signal number are listed.
CGH-karyotype: CGH results are described according to the ISCN nomenclature; rev ish refers to reverse in situ hybridization, enh (enhanced) and dim (diminished) indicate gains or losses of chromosomal material in the indicated region and amp refers to amplification. Bold karyotypes indicate correspondence of FISH and CGH results. Double daggers (‡) mark CGH karyotypes, which showed a clear shift toward overrepresentation or underrepresentation; however the diagnostic thresholds were not reached (see also Fig 2).
Both the t(11;14) and the t(14;18) were detected in only 1 of 18 cases each (nos. 28 and 13, respectively). The percentages of cells harboring the translocation were below 10%. In Fig 4, representative examples of FISH experiments are shown. The data of the interphase analyses are listed in Table 2.
Comparison of FISH and CGH.
In 11 instances, aberrations were detected both by FISH and CGH. In 20 instances, gains or losses were identified by interphase analyses; however, the respective CGH profiles were not in the diagnostic range. In the majority of these cases (n = 13) this was caused by the presence of aberrations in subclones of the analyzed samples. In other cases, the size of the imbalanced segments most likely was below the spatial resolution of CGH (see Fig 2). For a reliable detection, gains or losses of at least 10 Mbp are required (see refs 31 and 32). In 44 instances, aberrations identified by CGH were mapped to chromosomal regions, for which no DNA probes were used in our series. These data support a combined approach exploiting the advantages of both CGH (comprehensive screening of the genome) and FISH (detection of small aberrations, even in subclones of the tumor).
EBV status.
Of the 21 cases studied, only 1 case (no. 1) showed the presence of EBV sequences by Southern blot analysis.
Correlation of molecular cytogenetic data with the clinical course.
Of 30 patients, 17 are still alive and in complete remission 1 to 91 months (median, 47 months) after diagnosis. One patient (no. 31) was lost during follow-up. The remaining 13 patients died from lymphoma 0.5 to 101 months (median, 4 months) after diagnosis. The patients with less complex molecular cytogenetic karyotypes (0 or 1 aberration, n = 15) had a significantly higher probability of survival than patients with two or more aberrations (median survival 48 monthsv 13 months; P = .022, Wilcoxon test). Furthermore, although the patient number is limited, there seemed to be a correlation between specific genetic findings and prognosis. Seven of nine patients with gains of all or part of chromosome 12 died 0.5 to 25 months (median, 3 months) after diagnosis. Similarly, the four patients carrying a gain of the long arm of chromosome 1 and four patients with high-level DNA amplifications died from lymphoma 0.5 to 12 months (median, 0.5 months) and 0.5 to 23 months (median, 0.5 months) after diagnosis, respectively. Survival data for the most frequent genetic aberrations are listed in Table 3.
Clinical Course in Different Molecular Cytogenetic Subgroups
| . | Total No. of Patients . | Patients Alive (all in complete remission) . | Median Survival (all patients; mos) . | . |
|---|---|---|---|---|
| 30 | 17 | 29 (0.5-101) | ||
| 0 and 1 aberration | 15 | 11 | 48 (3-101) | P < .022 |
| ≥2 aberrations | 15 | 6 | 13 (0.5-72) | |
| +11q | 11 | 5 | ||
| p53 deletion | 6 | 3 | ||
| Gains on chromosome 12 | 9 | 2 | ||
| +1q | 4 | 0 | ||
| Amplifications | 4 | 0 |
| . | Total No. of Patients . | Patients Alive (all in complete remission) . | Median Survival (all patients; mos) . | . |
|---|---|---|---|---|
| 30 | 17 | 29 (0.5-101) | ||
| 0 and 1 aberration | 15 | 11 | 48 (3-101) | P < .022 |
| ≥2 aberrations | 15 | 6 | 13 (0.5-72) | |
| +11q | 11 | 5 | ||
| p53 deletion | 6 | 3 | ||
| Gains on chromosome 12 | 9 | 2 | ||
| +1q | 4 | 0 | ||
| Amplifications | 4 | 0 |
DISCUSSION
Because of the limited availability of fresh tumor tissue for chromosomal banding analyses, only few cytogenetic data of large B-cell lymphomas originating in the GI tract have been reported. In this study, different molecular cytogenetic techniques were used to achieve a comprehensive analysis of genetic aberrations in these lymphomas. Comparative genomic hybridization was combined with interphase cytogenetic analysis using a set of DNA probes allowing the detection of frequent chromosomal anomalies in B-cell neoplasias. With this approach, aberrations were detected in 26 of the 31 tumors.
The most frequent alteration in our study was a gain of material on chromosome 11 identified in 11 of the 31 cases. Based on the CGH data, the smallest commonly overrepresented region spanned chromosomal bands 11q22-q23. In this region, proto-oncogenes are located, for which a possible pathogenetic role in hematological malignancies has been demonstrated (eg MLL/ALL1,33,34RCK,35LPC,36BOB1,37 and PLZF38). Gains of chromosome 11 have been described rarely in nodal lymphomas of patients without immunosuppression or in low-grade gastrointestinal lymphomas. In contrast, it is frequent in secondary lymphomas occurring in immunocompromised hosts, such as organ transplant recipients or human immunodeficiency virus–infected subjects.39 Virtually all of these secondary lymphomas are associated with EBV infection, and frequently involvement of the GI has been observed.40 In our series, the EBV genome was identified in tumor cells of only 1 of 21 patients. This finding is in line with the clinical data: none of the patients had any evidence of an underlying disorder causing immunosuppression. Thus, a direct link between EBV infection and GI lymphomas with specific aberrations on 11q was not substantiated, and this aberration may be a characteristic feature of large B-cell lymphomas of the GI tract.
In contrast to low-grade MALT lymphomas including GI lymphomas, in which trisomy 3 was found in more than 50% of cases,4,7 we identified gains of this chromosome only in 4 of our 31 cases with large B-cell lymphomas: gains of parts of chromosome 3 were found in 2 of 7 tumors with low-grade component and only 2 of 24 cases without low-grade component. Thus, aberrations involving chromosome 3 may be less important in putatively primary high-grade GI lymphomas. This is in support of a previously published interphase cytogenetic study using a chromosome 3–specific repetitive DNA probe. In this series, trisomy 3 was less frequent in high-grade than in low-grade MALT lymphomas.11
In addition to a comprehensive study of chromosomal imbalances, a subset of our series (n = 18) was also investigated for the presence of the t(11;14) and the t(14;18), which are among the most common balanced aberrations in NHL. These aberrations were found in only one case each. Percentages of cells carrying the respective aberration were low (<10%), and in both cases other aberrations were present in higher percentages of cells. These findings indicate that neither the t(11;14) nor the t(14;18) are among the primary genetic events in large B-cell lymphoma of the GI tract. A similarly low incidence of the t(14;18) has been reported before.7 41
In four cases, a total of seven high-level DNA amplifications were identified. All of these cases had multiple molecular cytogenetic aberrations. The incidence of such DNA amplifications was similar to other lymphomas.42-45 Several of the amplified regions coincide with the chromosomal localizations of proto-oncogenes, which may be of pathogenetic relevance in the respective cases. Using a candidate gene approach, in two instances the involvement of such proto-oncogenes could be demonstrated: in one case with a high-level DNA amplification mapping to chromosomal bands 2p13-15, an amplification of the REL proto-oncogene was shown using Southern blot hybridization. Such REL amplifications have been described before in extranodal lymphomas.43,46 In another case with a high-level DNA amplification mapping to chromosomal band 8q24, amplification of the MYC proto-oncogene was shown. This gene is known to be rearranged much more frequently in GI lymphomas than in nodal lymphomas.47 However, MYCamplification has not been described in GI lymphomas before.
Despite the limited number of patients, some molecular cytogenetic findings seemed to be associated with the clinical outcome. One important factor was the complexity of the genetic alterations. Patients with two or more aberrations had a significantly shorter median survival (13 months) than patients with one aberration or a normal molecular cytogenetic karyotype (48 months). In addition, all five patients, in whom no chromosomal rearrangements were found, had a favorable clinical course with long-lasting complete remissions. These data are in line with results from two large studies investigating various types of nodal NHL.48 49 Furthermore, some specific chromosomal imbalances, ie, gains on 1q and on chromosome 12, and the presence of high-level DNA amplifications were associated with a particularly unfavorable course of disease.
All patients with gains of all or parts of 1q and/or high-level DNA amplifications as well as 7 of 9 patients with gains of all or parts of chromosome 12 died of lymphoma (median survival: 7 months for 1q, 3 months for high-level DNA amplifications, and 3 months for gains on chromosome 12). In a large study of nodal diffuse large cell NHL, aberrations on 1q have been shown to be associated with poor prognosis.48 With respect to gains on the long arm of chromosome 12, no prognostic impact has been described in high-grade B-cell NHL. For chromosome arm 12q, in our series the commonly overrepresented region extended from bands 12q24.1 to 12q24.3. This is the chromosomal localization of a recently described gene,BCL7A, which has been demonstrated to be of pathogenetic relevance in a Burkitt lymphoma cell line.50 In a previous study based on nine cases of NHL the presence of homogeneously staining regions, the banding hallmark of gene amplifications, was associated with poorer prognosis.51
In conclusion, the combined molecular cytogenetic approach resulted in a comprehensive analysis of chromosomal gains and losses in large B-cell lymphomas of the GI tract. The pattern of aberrations was different from low-grade (MALT) lymphomas of the stomach and bowel, especially with respect to the high incidence of partial gains of chromosome 11q and of all or parts of chromosome 12 and the low frequency of polysomy 3. In addition, our data suggest that the gains and losses detected by CGH and FISH may predict for the outcome of these lymphomas.
ACKNOWLEDGMENT
We gratefully acknowledge Profs Christian Herfarth, Michael Wannenmacher, and Armin Quentmaier (all of Heidelberg, Germany), as well as Drs Werner Schaupp (Weinheim, Germany) and Rolf Sippel (Mosbach, Germany) for providing clinical data of some of the patients. We thank Dr Richard Schlenk (Heidelberg, Germany) for support in the analysis of our clinical data and Sabine Gantner for skillful technical assistance.
Supported by the Deutsche Forschungsgemeinschaft (Grant No. Be 1454/5-1) and the Tumorzentrum Heidelberg/Mannheim.
Address reprint requests to Martin Bentz, MD, Medizinische Klinik und Poliklinik V, Hospitalstr.3, 69115 Heidelberg, Germany.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal