Abstract
Translocations in hematologic disease of myeloid or lymphoid origin with breakpoints at chromosome band 12p13 frequently result in rearrangements of the Ets variant gene 6 (ETV6). As a consequence either the ETS DNA-binding domain or the Helix-Loop-Helix (HLH) oligomerization domain of ETV6 is fused to different partner genes. We show here that a t(9; 12)(p24; p13) in a case of early pre-B acute lymphoid leukemia and a t(9; 15; 12)(p24; q15; p13) in atypical chronic myelogenous leukemia in transformation involve the ETV6 gene at 12p13 and the JAK2 gene at 9p24. In each case different fusion mRNAs were found, with only one resulting in an open reading frame for a chimeric protein consisting of the HLH oligomerization domain of ETV6 and the protein tyrosine kinase (PTK) domain of JAK2. The cloning of the complete human JAK2 coding and genomic sequences and of the genomic junction fragments of the translocations allowed a characterization of the different splice events leading to the various mRNAs. JAK2 plays a central role in non–protein tyrosine kinase receptor signaling pathways, which could explain its involvement in malignancies of different hematologic lineages. Besides hop in Drosophila no member of the JAK family has yet been implicated in tumorigenesis.
RECURRING ABNORMALITIES of the short arm of chromosome 9 and 12 have been identified as being among the more common cytogenetic rearrangements in acute lymphoblastic leukemia (ALLs) in children, with reported incidence of between 7% and 15%.1-5 Both regions are frequently involved in structural variations including deletions and unbalanced translocations resulting in the loss of 9p and 12p material, and apparently balanced reciprocal translocations, all involving the regions 9p11-p24 and 12p12-p13.
Cytogenetic alterations of the 12p12-p13 chromosomal bands are detected in about 10% of childhood ALLs.5,6 The translocations involve several partner chromosomes and can be associated with interstitial or terminal deletions that account for up to 50% of the cytogenetically detectable 12p abnormalities.4-7 In fact, recently published results indicate that alterations of the 12p12-p13 region are largely underestimated by standard cytogenetics. ETV6 (also known as TEL) was cloned at the breakpoint of the t(5; 12)(q33; p13) present in myelomonocytic leukemic cells.8 More recently, ETV6 has been shown to be fused to a number of different partners as a result of various leukemia associated translocations.9-15 In ALL, ETV6 was found to be fused to ABL in some rare cases with t(9; 12)(q34; p13),11 and ETV6/AML1 fusion transcripts, resulting from cryptic t(12; 21),12,13 were detected in 22% to 27% of the cases with childhood B-lineage ALL, representing the most common known gene rearrangement in cancer of childhood.16-18 We and others have previously shown that t(12; 21) and deletion of the nontranslocated ETV6 allele are frequently associated.16,17 19
We here report the identification of JAK2 as a fusion partner of ETV6 in t(9; 12)(p24; p13) found in leukemic cells of a child with early B-precursor ALL and in t(9; 15; 12) (p24; q15; p13) found in an adult patient with atypical CML. In both cases this translocation results in the fusion of the HLH domain of ETV6 to the protein tyrosine kinase domain of the receptor associated kinase JAK2.
MATERIALS AND METHODS
Patient material.The first patient, a 19-month-old boy, was admitted to the hospital with hyperthermia, palor, petechiae, splenomegaly, and detectable lymph nodes. Laboratory findings at diagnosis showed 4.4 mmol/L hemoglobine, platelet count 1 × 1010/L, leukocyte count 550 × 109/L, and the presence of 100% blast cells in peripheral blood. The bone marrow (BM) was infiltrated by small blasts of homogeneous size with high nuclear-cytoplasmic ratio, thus establishing a diagnosis of ALL L1 according to the French-American-British (FAB) classification. Immunophenotypic analysis of the blasts showed an early pre-B lineage ALL (CD10+, CD19+, CD20+, CD34+, slg−, Cμ−, HLA-DR+). There were no signs of central nervous system involvement. The patient was enrolled for an intensive chemotherapy regimen according to the European Organisation for Radiotherapy and Chemotherapy (EORTC) protocol 58881. At the end of induction therapy the patient achieved complete hematological remission (CR), which was maintained for 14 months, continuing the treatment according to the protocol. After 7 months of consolidation courses, the patient suffered BM plus overt testicular relapse, with approximately 4% blastic invasion of the BM. Salvage therapy was started and CR was obtained. This second course of this high-dose chemotherapy was interrupted as the patient suddenly became lethargic, hypotonic and mutic, and rapidly developed clinical features of secondary Parkinsonism. A basal ganglia necrosis of toxic origin was diagnosed. Cerebrospinal fluid remained normal. The parents refused further antileukemic therapy. After few months of therapeutic abstention, maintenance therapy was started again. The boy is now in continuous CR of ALL, 31 months after diagnosis. The second patients clinical history was already described.14 Briefly, clinical examination showed a slight splenomegaly. Hematologic data were as follows: hemoglobin level 8.7 g/dL; platelet count 37 × 109; white blood cell count 64 × 109/L with 27% neutrophils, 11% lymphocytes, and 11% lymphoblasts. BM was hypercellular with a proportion of 25% of blast cells with a myeloid morphology, 19% of promyelocytes, 14% of myelocytes, and 14% of eosinophils. The diagnosis of atypical chronic myelogenous leukemia (CML) in transformation was established.
Cytogenetic studies.The initial analysis, at diagnosis, was made on BM cells. Cells were cultured for 16 and 24 hours in vitro without stimulation. Chromosomes were identified using RHG (R-bands by heating using Giemsa) banding techniques and classified according to International System for Cytogenetic Nomenclature (ISCN) (1995).20 The karyotype in the ALL case was abnormal in twelve metaphases, 45,XY, t(9; 12)(p23; p13), t(13; 21). One additional metaphase presented the t(13; 21) as sole rearrangement. The constitutional origin of the t(13; 21) was already known as the mother had undergone a prenatal diagnosis for late in life pregnancy. The same abnormal karyotype was observed at the time of relapse in cells of testicular and medullary biopsies, without additional secondary abnormalities. Cytogenetic analysis performed in the CML case at the time of diagnosis showed clonal chromosomal abnormalities in 100% (6 of 6) of analyzed BM cells described as 46,XY, t(3; 12)(q26; p13), t(9; 15; 12)(p24; q15; p13).
Fluorescence in situ hybridization (FISH) analysis.FISH was performed as previously described.21 22 Biotin-labeled probes were prepared by nick translation (Bio Nick Kit; GIBCO-BRL, Gaithersburg, MD). Whole chromosome painting probes for chromosomes 9 and 12 (Coatasome; Oncor, Gaithersburg, MD) or a chromosome 12 centromere probe were used if necessary to assert the location of the FISH signals. The presence of FISH signals was scored on an average of 23 abnormal metaphases (15 to 52) per probe.
Polymerase chain reaction (PCR) and cloning.Total RNA was isolated from BM using the Trizol reagent (GIBCO-BRL). First-strand cDNA was reverse transcribed from 1 μg of total RNA with MuMLV-reverse transcriptase (GIBCO-BRL) according to standard procedures using the primer R2N6 (5′-CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGC(N)6-3′ ). Nested PCR was performed using primers specific for exon 4 of ETV6 (ALL case), namely ETV6F1 (5′-TTCCACCCTGGAAACTCTATA-3′ ) and ETV6F2 (5′-ATACACACACAGCCGGAGGTC-3′ ) or exon 5 (CML case), ETV6F3 (5′-ATGCACCCTCTGATCCTGAACC-3′ ) and ETV6F4 (5′-GAAGACCTGGCTTACATGAAC-3′ ) in combination with primers R2N R16 (5′-CCAGTGAGCAGAGTGACG-3′ ) and R2N6R2 (5′-GAGGACTCGAGCTCAAGC-3′ ). PCR products were cloned following standard procedures.
The ETV6-JAK2 fusion was confirmed by reverse transcriptase (RT)-PCR on patient RNA using the Titan RT-PCR system (Boehringer Mannheim, Mannheim, Germany), with the following primers: ETV6F1 in combination with JAK2R1 (5′-AAGGTTTGCTAATTCTGCCCACTTTGGTGC-3′ ) specific for exon 17 of JAK2 (ALL case) and primer ETV6F3 in combination with JAK2R2 (5′-TGGTGAGGTTGGTACATCAG-3′ ) specific for exon 12 of JAK2 (CML case).
Long-range PCR was performed using the Expand long template PCR system (Boehringer) with primers ETV6F1 and ETV6F2 in combination with JAK2R1 (ALL case) and primers ETV6F3 and ETV6F4 in combination with JAK2R2 (CML case).
The human JAK2 cDNA was isolated by screening a λDR2 human BM cDNA library (Clontech, Palo Alto, CA) and by 3′RACE on fetal brain cDNA (Clontech). Several overlapping clones were sequenced to obtain the complete sequence from both strands (Genbank accession no. AF005216).
DNA sequencing and analysis.Nucleotide sequence was determined by dideoxynucleotide chain termination with fluorescein isothiocyanate (FITC)-labeled primers and analyzed on ALF sequencer (Pharmacia, Uppsala, Sweden). Database searching was carried out by using the BLASTN algorithm on the NIH Blast server.23
RESULTS
Cytogenetic analysis at diagnosis of a patient with pre-B ALL showed the presence of a t(9; 12)(p24; p13) as the only chromosomal anomaly associated with the disease. FISH using cosmids derived from a contig spanning the ETV6 gene24 located at 12p13 showed the involvement of this gene: a signal on the normal chromosome 12 and the der(9) was observed with cosmid 163E7, containing exons 3 to 5 of ETV6 whereas cosmid 54D5, containing exons 5 to 8, showed a signal on normal and der(12) (Fig 1), suggesting the 12p breakpoint to be located between exon 4 and 5 of ETV6. The second case, a patient diagnosed with CML in blast crisis, was previously studied because of the presence of a t(3; 12) that resulted in a fusion between ETV6 and MDS1-EVI1 on 3q26.14 At that time it was shown that the other ETV6 allele was involved in a complex t(9; 15; 12) (p24; q15; p13). FISH analysis with cosmid 54D5 showed a ‘split’ signal with the probe hybridizing to the der(12) t(3; 12), the der(12)t(9; 15; 12), and the der(9)t(9; 15; 12), suggesting a breakpoint between exon 5 and 6 of ETV6 (Fig 1).
FISH analysis of the ETV6 and JAK2 rearrangements in the ALL case (A and B) and the ALL case (C and D). Hybridization with cosmid 54D5, which spans the 12p13 breakpoint in the CML case, is shown in (A) and (C). Thick arrows indicate the der(12), thin arrows indicate the der(9), and the arrowhead points to the normal 12. (B and D) Hybridization with a mixture of the PACs containing JAK2 and spanning the 9p24 breakpoint. Thin and thick arrows indicate, respectively, the der(9) and the der(12); the arrowhead shows the der(15)t(9; 15; 12); 9N indicates the normal chromosome 9. In (C) and (D) hybridization of a centromere 12 probe is shown in white.
FISH analysis of the ETV6 and JAK2 rearrangements in the ALL case (A and B) and the ALL case (C and D). Hybridization with cosmid 54D5, which spans the 12p13 breakpoint in the CML case, is shown in (A) and (C). Thick arrows indicate the der(12), thin arrows indicate the der(9), and the arrowhead points to the normal 12. (B and D) Hybridization with a mixture of the PACs containing JAK2 and spanning the 9p24 breakpoint. Thin and thick arrows indicate, respectively, the der(9) and the der(12); the arrowhead shows the der(15)t(9; 15; 12); 9N indicates the normal chromosome 9. In (C) and (D) hybridization of a centromere 12 probe is shown in white.
To identify the eventual fusion partner of ETV6, RNA from the patients was reverse transcribed using a random hexanucleotide primer described above. 3′-RACE was performed using nested primers located in exon 4 and 5 of ETV6, respectively. Sequence analysis of the amplification products detected in both cases novel sequences fused to the ETV6 sequence conserving the open reading frame. This novel sequence showed homology to the murine JAK2 kinase25 (90% identity), strongly suggesting that they were derived from the human JAK2.
With these probes, a human JAK2 cDNA was isolated from a human BM cDNA library and completed by 3′-RACE experiments on fetal brain cDNA. In total 4,161 bp of JAK2 cDNA sequence was isolated, including an open reading frame encoding a protein of 1,132 amino acids. This protein is 92% identical (at the amino acid level) to murine JAK2 and 94% identical to the rat JAK2, consistent with its identity as the human JAK2. Using the same probes three P1 artificial chromosome (PAC) clones were isolated from a human genomic PAC library. Analysis of these clones showed that the JAK2 gene consists of 24 exons spanning at least 140 kb (manuscript in preparation). FISH with these genomic clones confirmed the mapping of JAK2 to 9p24 as was previously suggested using a murine probe26 and showed the clones to cross the breakpoint in both leukemias (Fig 1).
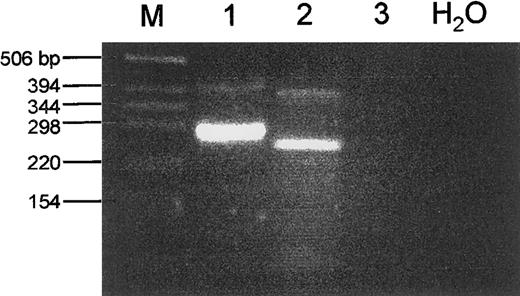
Taken together the data indicated a fusion of exon 4 of ETV6 to exon 17 of JAK2 in the ALL case and a fusion of exon 5 of ETV6 to exon 12 of JAK2 in the CML case. When primers were derived for the respective exons to confirm this directly by RT-PCR, two fragments were unexpectedly detected in the ALL case (Fig 2). Sequence analysis showed the smaller, more abundant, product to result from the splicing of exon 4 of ETV6 to exon 17 of JAK2, while the larger product spliced exon 4 of ETV6 to exon 16 of JAK2. Only the first results in an open reading frame linking the HLH oligomerization domain of ETV6 to part of the JH2 and the complete protein tyrosine kinase domain of JAK2. To confirm this observation at the genomic level, long-range PCR was performed on tumor DNA using primers located in the relevant exons of ETV6 and JAK2. Sequencing then showed that the breakpoint in JAK2 indeed occurred at the genomic level between exons 15 and 16 (Fig 3, top). RT-PCR experiments failed to detect a reciprocal transcript, suggesting that the ETV6/JAK2 fusion has oncogenic properties in this case.
RT-PCR analysis of ETV6/JAK2 fusion products. Total RNA from the ALL case (lane 1), the CML case (lane 2), and normal WBCs (lane 3) was used in a RT-PCR with primers located in the relevant exons of ETV6 and JAK2 (see Materials and Methods section). In each case a major product is observed, together with a minor product of a larger size. The different PCR products are the result of alternative splicing events as shown in Fig 3. Marker size is indicated to the left.
RT-PCR analysis of ETV6/JAK2 fusion products. Total RNA from the ALL case (lane 1), the CML case (lane 2), and normal WBCs (lane 3) was used in a RT-PCR with primers located in the relevant exons of ETV6 and JAK2 (see Materials and Methods section). In each case a major product is observed, together with a minor product of a larger size. The different PCR products are the result of alternative splicing events as shown in Fig 3. Marker size is indicated to the left.
Genomic structure of the different translocations and resulting open reading frames. The upper panel shows the results for the ALL case, the lower panel for the CML case. On top of each panel the genomic structure of the junction fragment is shown. The ETV6 gene is represented by a thin line, the JAK2 gene by a thick line, and a fragment of unknown origin by an open bar. The boxes show the different exons. The sequence of the breakpoints is shown on top of the genomic structure. In the ALL case ETV6 genomic sequences are shown in capitals, JAK2 sequences in lowercase. Two splice variants are observed, the first fusing exon 4 of ETV6 to exon 16 of JAK2. The second splices exon 4 to exon 17 of JAK2. In the CML case exon 5 of ETV6 is spliced either to an exon (U) in the sequence of unknown origin which is then spliced to a cryptic acceptor site within exon 11 of JAK2 or directly to exon 12 of JAK2. ETV6 and JAK2 genomic sequences are indicated in uppercase letters, and a genomic sequence of unknown origin in lowercase letters. Splice donor and acceptor sites for the exon within this sequence are in boldface type. The composition of the fusion proteins encoded by the different splice variants is shown with the sequence of the junction fragments underneath. Arrows indicate the breakpoints. E, EcoRI site; HLH, oligomerization domain of ETV6; JH2, pseudokinase domain of JAK2; PTK, protein tyrosine kinase domain of JAK2.
Genomic structure of the different translocations and resulting open reading frames. The upper panel shows the results for the ALL case, the lower panel for the CML case. On top of each panel the genomic structure of the junction fragment is shown. The ETV6 gene is represented by a thin line, the JAK2 gene by a thick line, and a fragment of unknown origin by an open bar. The boxes show the different exons. The sequence of the breakpoints is shown on top of the genomic structure. In the ALL case ETV6 genomic sequences are shown in capitals, JAK2 sequences in lowercase. Two splice variants are observed, the first fusing exon 4 of ETV6 to exon 16 of JAK2. The second splices exon 4 to exon 17 of JAK2. In the CML case exon 5 of ETV6 is spliced either to an exon (U) in the sequence of unknown origin which is then spliced to a cryptic acceptor site within exon 11 of JAK2 or directly to exon 12 of JAK2. ETV6 and JAK2 genomic sequences are indicated in uppercase letters, and a genomic sequence of unknown origin in lowercase letters. Splice donor and acceptor sites for the exon within this sequence are in boldface type. The composition of the fusion proteins encoded by the different splice variants is shown with the sequence of the junction fragments underneath. Arrows indicate the breakpoints. E, EcoRI site; HLH, oligomerization domain of ETV6; JH2, pseudokinase domain of JAK2; PTK, protein tyrosine kinase domain of JAK2.
A similar RT-PCR analysis of the CML case interestingly also yielded two products, the smaller being more abundant as judged by RT-PCR (Fig 2). The larger amplification product fused exon 5 of ETV6 to a cryptic exon located in the intervening genomic sequence (see below) and a cryptic acceptor site within exon 11 of JAK2. The smaller one fused exon 5 of ETV6 to exon 12 of JAK2, only this resulted in an open reading frame linking the ETV6 HLH oligomerization domain to the JH2 and protein kinase domain of JAK2. Cloning of the genomic breakpoint by long-range PCR indeed detected a 3,600-bp sequence of unknown origin between the ETV6 and JAK2 sequences. This sequence contained the cryptic exon found in the larger RT-PCR product (Fig 3). No reciprocal transcript could be detected by RT-PCR as expected for a three-way translocation. The significance of the complex splice events observed in both cases is not known, but the observation could be relevant for those cases where a fusion of ETV6 apparently does not lead to a fusion protein.
DISCUSSION
The involvement of JAK2 in large number of noncatalytic receptor signaling pathways has been shown extensively over the past few years.27-29 In Drosophila, a dominant gain-of-function mutation of a JAK homologue encoded by the hopsctoch (hop) gene, was reported to result in neoplastic growth. Here a single amino acid change in hop causes a leukemia-like disease in the fly.30 Although constitutive activation of the JAK-STAT pathway was shown in HTLV-1–infected peripheral blood T cells31 and selective growth inhibition of acute lymphoblastic cells has been accomplished, both in vivo and in vitro, by using a specific JAK2 tyrosine kinase blocker,32 no oncogenic activity has been reported for mammalian JAKs.
Reports on an artificial CD16/CD7/Jak2 and an epidermal growth factor receptor/Jak2 fusion protein suggest that Jak2 may become activated by homodimerization.33,34 In these reports it was shown that cross-linking of Jak2 results in autophosphorylation, although the activated pathways seem to differ in both systems. Moreover, in the latter it was shown that activation of Jak2 seems to be sufficient for transducing a growth signal in hematopoietic cells. There is clear evidence that the ETV6 HLH domain provides a self-association motif to the ETV6-PDGFRB oncogenic fusion protein, which results from a t(5; 12) in myelodysplastic disease.35,36 Moreover, this motif was shown to be essential to the chimeric protein to activate the PDGFRB kinase-dependent signaling pathways.35 The same mitogenic properties depending on the HLH domain of ETV6 were shown for the ETV6-ABL oncoprotein associated with t(9; 12).37 In view of these findings a model for the mechanisms of transformation by the ETV6/JAK2 fusion could be proposed where the HLH domain of ETV6 provides a dimerization interface to the kinase domain of JAK2, thus activating JAK2. Interestingly, in the CML case both alleles of ETV6 are affected by translocation. Besides the ETV6/JAK2 fusion, an ETV6/MDS1/EVI1 is also present in the leukemic cells of this patient.14 It is clear that in addition to the effect of the putative fusion proteins that result from the chromosomal rearrangements, the fact that ETV6 can no longer exert its normal function could also contribute to the oncogenic process. However, it should be noted that in both cases a minor mRNA species is formed that potentially codes for an independent HLH domain of ETV6 (Fig 3). The eventual effect of this domain on the ETV6/JAK2 fusion protein remains to be evaluated.
The presence of two fusion genes involving ETV6 in the CML case also raises questions with regard to the involvement of each fusion in the oncogenic process. The involvement of EVI1 in blast crisis in CML has been reported.38 One might speculate that the blast crisis in the present case is related to the ETV6/MDS1/EVI1 fusion, whereas the ETV6/JAK2 fusion would be an earlier event. Unfortunately, no previous samples of the patient are available to test this hypothesis.
The putative involvement of ETV6/JAK2 fusions in both myeloid and lymphoid malignancies is unusual and could be explained by the involvement of the JAK2 kinase in multiple signaling pathways. It should be noted that in the ALL case only part of the JH2 domain is present in the fusion, whereas the complete JH2 domain is present in the fusion found in the myeloid case. Recently it was shown39 that JAK2 activates Raf-1 via p21ras, thus linking JAK2 to the PDGFRB kinase–dependent signaling pathways which appear activated by the t(5; 12) in myeloid malignancies.35 This raises the question of whether the JH2 domain of JAK2, whose specific role is not known, is involved in the determination of the specific pathways activated by the ETV6/JAK2 fusion protein.
In conclusion, we now have the appropriate tools to investigate JAK2 involvement through chromosomal rearrangements in human neoplasia by FISH analysis. Furthermore, we show that dissection of complex chromosomal rearrangements can lead to the identification of different putative actors in leukemiogenesis for each of the observed abnormalities. Also, the occurrence of variant splicing has to be considered during the analysis of other translocations involving ETV6 or JAK2. Finally, to elucidate the potential role of the JH2 domain of JAK2 in the lineage specificity of the putative fusion proteins, a dissection of the pathways activated by the different ETV6/JAK2 fusions will be needed.
ACKNOWLEDGMENT
We thank A. Criel and D. Selleslagh for patient material, B. Brunet for technical support of cytogenetics, and R. Gratery for artwork.
Supported by the CNRS, a grant from the Ligue nationale Française Contre le Cancer, a grant from the Association pour la Recherche sur le Cancer, and Grant No. G 0153.96 of the FWO-Vlaanderen. P.M. is an ‘onderzoeksdirecteur’ and M.B. a “postdoctoraal onderzoeker” of the FWO-Vlaanderen; P.P. is the recipient of a scholarship of the IWT.
Address reprint requests to Peter Marynen, PhD, Center for Human Genetics, University of Leuven, Campus Gasthuisberg, Herestraat 49, B-3000 Leuven, Belgium.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal