Abstract
cDNA isolated from a human B-cell line Raji library was analyzed and shown to encode the full-length cDNA sequence of a novel cell-surface glycoprotein, initially termed HLy9-β. The predicted mature 307-amino acid protein was composed of two extracellular Ig-like domains, a hydrophobic transmembrane region, and an 83-amino acid cytoplasmic domain. The extracellular Ig-like domains presented structural and sequence homology with a group of members of the Ig superfamily that included CD2, CD48, CD58, and Ly9. Northern blot analysis showed that the expression of HLy9-β was predominantly restricted to hematopoietic tissues. Chromosome localization studies mapped the HLy9-β gene to chromosome 1q24, where other members of this Ig superfamily (CD48 and HumLy9) have been mapped. CD84 monoclonal antibodies (MoAbs) were shown to react with cells transfected with the cloned cDNA. These MoAbs were further used to show that CD84 is expressed as a single chain cell-surface glycoprotein of Mr 64,000 to 82,000, which was highly glycosylated. CD84 had a unique pattern of expression, being found predominantly on lymphocytes and monocytes. Thus, the glycoprotein HLy9-β is recognized by MoAbs previously clustered as CD84 and represents a newly identified member of the Ig superfamily that may play a significant role in leukocyte activation.
THE INTERACTION between leukocytes and other cells regulate normal immune function. Along with the polypeptides forming the T-cell and B-cell receptor complexes, a number of surface molecules participate in B-cell and T-cell activation. Some function as cell-cell interaction molecules or as signal-transducing elements, and some can serve both functions.1 A significant number of these molecules are members of the Ig superfamily, which are involved in processes such as adhesion, migration, proliferation, differentiation, and effector functions of leukocytes.2
Extracellular portions of CD2, CD48, and CD58 leukocyte antigens are predicted to have similar molecular structures, and they form a distinct subgroup within the Ig superfamily, named the CD2 family.3 These proteins have similar patterns of conserved disulfide bonds and comprise N-terminal V-set domains, which lack a disulfide bond, and C-terminal C2 set Ig domains. These transmembrane surface glycoproteins are involved in adhesion interactions between T lymphocytes and accessory cells and generate intracellular activation signals.4-6 In vivo studies have shown that the administration of CD2 and CD48 monoclonal antibodies (MoAbs) can inhibit T-cell responses and prolong allograft survival.7,8 Other members of the growing CD2 family are the surface antigens Ly9, 2B4, and the recently described signaling receptor SLAM.9-11
CD84 is a cluster of differentiation that was established during the Fifth International Leukocyte Differentiation Antigen Workshop (Boston, MA, 1994). Three MoAbs, 2G7, 152-1D5, and 153-4D9, defined CD84 as a new surface antigen predominantly expressed by mature B cells and monocytes.12 More recently, biochemical analysis shows that CD84 MoAbs precipitate a single-chain glycoprotein of approximately 72,000 to 86,000 Mr.13
This article describes the molecular cloning, characterization, chromosomal localization, and tissue-specific expression of a new gene of the Ig superfamily that encodes for the cell surface leukocyte antigen CD84.
MATERIALS AND METHODS
Isolation of cDNA clones.A cDNA library from the human Burkitt lymphoma cell line Raji in λgt11 (Clontech Laboratories, Palo Alto, CA) was used to isolate cDNA clones by cross-species hybridization, with a mixture of different polymerase chain reaction (PCR)-generated probes corresponding to sequences reported for murine surface molecule Ly99; the following primers were used to generate the probes: Ly9 #1 (559 bp): sense primer, 5′-AAGAAGCTTCCAAGAATGAGTCAGCAGCAG-3′, and antisense primer, 5′-CATCGTATGTATTTAAATGGGTGTC-3′; Ly9 #2 (470 bp): sense primer, 5′-GACACCCATTTAAATACATACGATG-3′, and antisense primer, 5′-CAGTCTCTTGTAGACAAGCAAGG-3′; and Ly9 #3 (445 bp): sense primer, 5′-CCTTGCTTGTCTACAAGAGACTG-3′, and antisense primer, 5′-CAGAAAATTGTCCATGGCCAGTTG-3′.
cDNA was obtained by reverse transcription polymerase chain reaction (RT-PCR) of total RNA isolated from murine thymus using a First Strand cDNA Synthesis Kit for RT-PCR (Avian Myeloblastosis Virus [AMV], Reverse Transcriptase; Boehringer Mannheim, Mannheim, Germany) and the Taq polymerase Expand High Fidelity (Boehringer Mannheim) following manufacture's instructions. The PCR products were purified, subcloned into pSP64 vector (Promega, Madison, WI), and random priming labeled using [γ-32P] dCTP with the Ready-To-Go DNA labeling kit (Pharmacia Biotech, Uppsala, Sweden). Filter hybridization was performed in ExpressHyb hybridization solution (Clontech Laboratories) at 45°C for 2 hours, followed by low stringency washing in 2× SSC-0.05% sodium dodecyl sulfate (SDS) at room temperature. Plaques that were positive on duplicate filters were picked, and phage insert DNA was isolated. cDNA inserts were removed by EcoRI digestion and subcloned into pSP64 (Promega). Restriction maps were generated and nucleotide sequences were determined by dideoxynucleotide chain termination by using the T7 Sequencing Kit (Pharmacia Biotech).14 A computer search of nucleotide and protein sequences was conducted using the Blast GeneSearch (National Center for Biotechnology Information, National Institutes of Health, Bethesda, MD). Sequence analysis was performed by using PC/Gene Software (IntelliGenetics Inc, Mountain View, CA).
RNA blot analysis.A positively charged nylon membrane, to which poly A+ RNAs from 50 human tissue samples had been immobilized in separated dots (The Human RNA Master Blot), was purchased from Clontech Laboratories. Poly A+ RNA samples on Master Blot have been normalized to the mRNA expression levels of eight different housekeeping genes allowing the determination of the relative expression levels of a target mRNA in different tissues. The nylon membrane includes poly A+ from the following tissues: whole brain, amygdala, caudate nucleus, cerebellum, cerebral cortex, frontal lobe, hippocampus, medulla oblongata, occipital lobe, putamen, substantia nigra, temporal lobe, thalamus, subthalamic nucleus, spinal cord, heart, aorta, skeletal muscle, colon, bladder, uterus, prostate, stomach, testis, ovary, pancreas, pituitary gland, adrenal gland, thyroid gland, salivary gland, mammary gland, kidney, liver, small intestine, spleen, thymus, peripheral leukocytes, lymph node, bone marrow, appendix, lung, trachea, placenta, fetal brain, fetal heart, fetal kidney, fetal liver, fetal spleen, fetal thymus, and fetal lung. Northern blot analysis was performed with nylon membranes purchased from Clontech Laboratories (Multiple Tissue Northern blot) to which 2 μg poly A+ RNA from different tissues (spleen, lymph node, thymus, appendix, lung, and liver) have been blotted after running on a denaturating formaldehyde/1.2% agarose gel. Dot blot and Northern blot membranes were hybridized with a 1,040-bp probe that contained the complete reading frame sequence of the HLy9-β clone and a 2,000-bp human β-actin cDNA as control probe. Hybridization was performed according to the manufacturer's instructions. Membranes were autoradiographed for 72 hours (HLy9-β) or 4 hours (β-actin) at −80°C by using two intensifying screens. Autoradiography was scanned with a Bio-Profile densitometer (Vilber Loumart, Marne la Valleé, France) and the relative dot intensity was determined with Bio-1D Analysis Software (Vilber Loumart). The relative dot intensity value was calculated as the optical density (OD) of the dot minus the OD of the background.
Chromosomal mapping.Fluorescence in situ hybridization (FISH), using a 1,040-bp cDNA probe that contained the complete reading frame sequence of the HLy9-β and a 5.4-kb PCR-generated probe of genomic DNA of the HLy9-β gene (sense oligo: 5′-TCTGCTTGTTCTCATTCTGTCTTCA-3′; antisense oligo: 5′-CAGTAAGAGTTGGGCAGAGAAGAT-3′) were performed as previously described.15 Briefly, 2 μg of each probe was labeled with biotin 16-dUTP (Boehringer Mannheim) by nick translation. A total of 400 ng of labeled probe was precipitated along with 1 μg of Cot-1 DNA (GIBCO/BRL, Gaithersberg, MD) and 1 mg of salmon sperm (Sigma, St Louis, MO), and the pellet was resuspended in a hybridization mix. Ten microliters of the hybridization mix was applied to each slide. After heat denaturation of the probe and the chromosomal spreads, slides were incubated overnight at 37°C. After the posthybridization washes the slides were incubated with avidin-fluorescein isothiocyanate (FITC; Vector Laboratories, Burigame, CA). Slides were mounted with 40 μL of antifade solution (Vector Laboratories) containing 150 ng/mL of 4′,6′-diamidino-2-phenylindole. Slides were studied under an Olympus AH-3 fluorescence microscope (Olympus Optical Co, Ltd, Tokyo, Japan) and images were analyzed with the Cytovision System (Applied Imaging International LTD, Santa Clara, CA).
Cells.Blood was obtained by protocols approved by the Ethics Committee of the Hospital Clinic of Barcelona. Mononuclear cells were isolated by Ficoll-Hypaque density gradient centrifugation. Monocytes were isolated by adherence of blood mononuclear cells to plastic for 60 minutes at 37°C. Cell lines were cultured in complete medium (RPMI 1640 supplemented with 10% fetal calf serum [FCS], antibiotics, and 10 mmol/L glutamine). Cultures of all cell lines were split the day before analysis and were in logarithmic growth.
COS cells were transfected with the HLy9-β cDNA subcloned into the pMT2 expression vector using the diethylaminoethyl-dextran method.16 Cell surface expression was examined after 48 hours by immunohistochemical analysis as described.17 Stable transfectants were produced by cotransfecting the murine pre–B-cell lines 300.19 by using 40 μg of pMT2-HLy9-β and 2 μg of pCI-neo vector containing the neomycin resistance marker (Promega). The 300.19 cells were transfected by electroporation (280 V, 950 μF ) with a Gene Pulser II Apparatus (Bio Rad, Hercules, CA). After electroporation the cells were plated in flat-bottomed 96-well tissue plates by limiting dilution, and selection of stable transfectants was performed by using G418 (1 mg/mL; GIBCO/BRL).18 Twenty clones from the transfection were examined by indirect immunofluorescence screening, and the clone with the highest level of expression was used in this study and maintained in the continuous presence of the selecting drug.
MoAbs.The CD84 MoAbs 2G7 (IgG1), 152-1D5 (IgG1), and 153-4D9 (IgG1); and the MoAbs of the Unknown Panel were obtained from the Fifth International Workshop on Human Differentiation Antigens, Boston MA, 1994.
Immunofluorescence analysis.Cells were kept at 4°C and examined immediately after isolation. Indirect immunofluorescence analysis of viable cells was performed after washing the cells twice. The cells were then incubated for 20 minutes on ice with each MoAb as ascites fluid diluted to the optimal concentration for immunostaining. Isotype-matched murine MoAbs that were unreactive with human leukocytes were used as negative controls. After washing, the cells were treated for 20 minutes at 4°C with FITC-conjugated goat antimouse Ig antibodies (Caltag, South San Francisco, CA). Single- and double-color immunofluorescence analysis was performed on an FACSCalibur (Becton Dickinson, Immnocytometry Systems, San Jose, CA). At least 5,000 cells were analyzed for each sample.
Immunoprecipitation analysis.A total of 40 × 106 cells were washed three times in phosphate buffered saline (PBS), resuspended in 800 μL saline, and surface labeled with 200 μL of Biotin 1 mg/mL (Sigma) for 20 minutes at 4°C. After washing the cells once in RPMI media with 10% FCS and four times in PBS, the cells were lysed in 1 mL of buffer containing 0.5% (vol/vol) NP40 and protease inhibitors as described.19 Immunoprecipitations were carried out using 15 μg of the CD84 MoAbs 2G7 and 152-1D5. Cell lysates were precleared three times for 2 hours using 50 μL (50% vol/vol) of murine Ig-coated beads at 4°C. Cell lysates were precleared again overnight. The precleared lysate was then incubated with 50 μL of CD84 MoAb-coated beads (50% vol/vol) or murine Ig-coated beads with constant rotation at 4°C for 18 hours. Immunoprecipitates were washed and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described.19 The samples were run in the presence of 5% 2-mercaptoethanol (reducing conditions). Mr was determined using prestained standard molecular weight markers (GIBCO/BRL). The proteins were transferred by electroblotting with glycine transfer buffer (96 mmol/L glycine, 12 mmol/L Tris-base in 10% methanol, pH 8.3) to polyvinylidene difluoride (PVDF) membranes (Immobilon; Millipore, Boston, MA). After 2 hours of incubation at room temperature with blocking solution (10% nonfat milk in PBS), avidin peroxidase (100 ng/mL; Sigma) was added for 1 hour. The membranes were washed with PBS with 0.1% of Tween-20, and the blot was developed using Amersham's Enhancer Chemoluminiscence reagent (Amersham International, Little Chalfont, UK). For N-deglycosysation, immunoprecipitates were digested overnight at 37°C with 0.4 U of Peptide-N-glycosidase F (PNGase F; Genzyme Diagnostics, Cambridge, MA) in buffer prepared according to the manufacture's instructions.
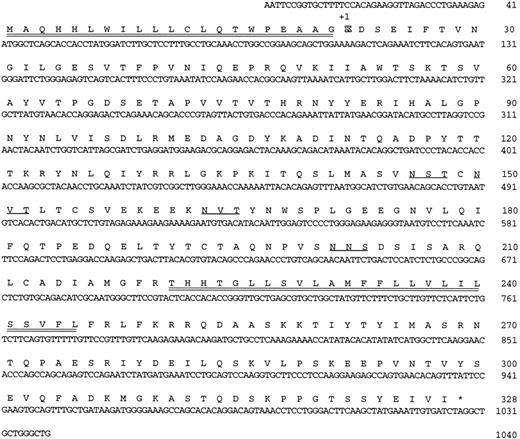
Nucleotide sequence and predicted amino acid sequence of HLy9-β. Potential N-linked glycosylation sites are underlined. Hydrophobic stretches that would serve as a signal peptide region and a transmembrane region are double underlined. * indicates the termination codon.
Nucleotide sequence and predicted amino acid sequence of HLy9-β. Potential N-linked glycosylation sites are underlined. Hydrophobic stretches that would serve as a signal peptide region and a transmembrane region are double underlined. * indicates the termination codon.
RESULTS
Isolation and characterization of HLy9-β cDNA clones and deduced amino acid sequence.Low stringency screening of a human Burkitt lymphoma B-cell line Raji λgt11 library using murine Ly9 cDNAs as probes yielded several positive clones. Four independent cDNA clones, named HLy9-β #1 (∼2.4 kb), HLy9-β #2 (∼1.6 kb), HLy9-β #3 (∼1.3 kb), and HLy9-β #4 (∼1.4 kb), were isolated, and they encoded a cDNA other than that described for HumLy9, the human homologue of murine Ly9 surface molecule.20 After digestion of these clones with EcoRI two bands were observed. All these clones had an overlapping band of approximately 1,100 bp that showed a single open reading frame of 987 bp. The other EcoRI bands corresponded to cDNA encoding 3′ untranslated sequences.
The 1,040-bp nucleotide sequence of clone HLy9-β #1 is shown in Fig 1. The open reading frame encodes a 328-amino acid protein, of which the first 21 amino acids comprise a typical hydrophobic signal peptide that is cleaved from the immature protein. The remaining 307-amino acid protein has a calculated molecular mass of 34,435 Mr before the addition of sugars. There are four potential N-linked glycosylation attachment sites in the mature protein consistent with the observed relative molecular weight on SDS-PAGE of 64,000 to 82,000 Mr. The sequence contains 11 serine and 18 threonine residues, which could be O-glycosylated. There is a single 25-amino acid internal hydrophobic segment that represents the transmembrane region and divides the protein in a 189-amino acid predicted external segment and an 83-amino acid intracytoplasmic region. Analysis of the sequence suggests that the extracellular portion of HLy9-β is composed of two Ig-like domains. The NH2 -terminal domain consisted of a V-like domain, which lacks the usually conserved disulfide bonds between the β sheets. The second domain had the structural features of an Ig-truncated C2 set domain with two putative disulfide bonds. A protein sequence homology search showed that this protein shared significant sequence identity with a group of members of the Ig superfamily, including Ly9, CD48, CD58, and CD2. These data indicate that the HLy9-β gene encodes a protein that belongs to the Ig superfamily.
mRNA analysis for HLy9-β. (A) Dot blot analysis of poly(A)+ RNA from different tissues using labeled HLy9-β and β-actin cDNA as probes. Autoradiography was scanned with a Bio-Profile densitometer (Vilber Loumart, Marne la Valleé, France). (B) Quantification of mRNA in different lymphoid tissues. Dot relative intensity as measured with Bio-1D Analysis Software (Vilber Loumart). (C) Northern blot analysis of HLy9-β.
mRNA analysis for HLy9-β. (A) Dot blot analysis of poly(A)+ RNA from different tissues using labeled HLy9-β and β-actin cDNA as probes. Autoradiography was scanned with a Bio-Profile densitometer (Vilber Loumart, Marne la Valleé, France). (B) Quantification of mRNA in different lymphoid tissues. Dot relative intensity as measured with Bio-1D Analysis Software (Vilber Loumart). (C) Northern blot analysis of HLy9-β.
The predicted cytoplasmic domain of the mature protein contains five tyrosines (Tyr). Interestingly, four of these Tyr (Tyr 241, Tyr 258, Tyr 278, and Tyr 303) contain motifs similar to those previously described as potential SH2 domain binding motifs. These are sequence motifs for kinase molecules such as src-kinase type tyrosine kinases.21 Therefore, sequences in the cytoplasmic tail suggest involvement of this molecule in signaling functions.
Detection of HLy9-β mRNA in hematopoietic tissues.mRNA expression was examined by dot blot analysis using poly A+ RNA isolated from 50 different human tissues, allowing us to determine the relative expression levels of HLy9-β in different tissues and during development. HLy9-β expression appeared to be lymphoid tissue specific: the highest mRNA level was detected in lymph node, spleen, and peripheral leukocytes (Fig 2A and B). Although at lower levels, thymus and fetal spleen also expressed significant levels of mRNA (Fig 2B), whereas it was very low in bone marrow. Significant levels of mRNA for HLy9-β were detected in kidney and, to a lesser extent, in lung (Fig 2A, C, and data not shown). These two were the only nonlymphoid tissues in which detectable levels of mRNA were observed.
Northern blot analysis from different lymphoid tissues and lung showed a band of 4.4 kb (Fig 2C). No smaller RNA species could be detected, even after a prolonged exposure of the membranes, suggesting that the HLy9-β mRNA has a long 3′-untranslated region.
HLy9-β is located at chromosome 1q24.To determine the chromosomal localization of the HLy9-β gene FISH was performed. By using the cDNA probe, pHLy9-β, a consistent but not conclusive hybridization signal on chromosome 1 (band q24) was observed. To locate the gene more precisely, the FISH method was performed using a 5.4-kb genomic fragment of the HLy9-β gene, obtained by PCR. A total of 85% of the metaphases showed signal on 1q24, confirming the localization obtained with the cDNA probe (Fig 3).
Chromosomal localization of HLy9-β gene. Metaphase showing FISH with the HLy9-β 5.4-kb genomic probe. Double spots of hybridization signal on chromosome 1 is indicated by an arrow. The HLy9-β gene was assigned to 1q24.
Chromosomal localization of HLy9-β gene. Metaphase showing FISH with the HLy9-β 5.4-kb genomic probe. Double spots of hybridization signal on chromosome 1 is indicated by an arrow. The HLy9-β gene was assigned to 1q24.
Identification of HLy9-β as CD84.To check whether preexisting MoAbs were able to recognize the gene product of HLy9-β cDNA, the Unknown Panel of MoAbs of the Fifth International Workshop on Differentiation Antigen was screened for MoAbs reactive with pMT2-HLy9-β–transfected cells. Three MoAbs, 2G7, 152-1D5, and 153-4D9, reacted with COS cells transfected with the HLy9-β cDNA, but did not react with cells transfected with the expression vector alone or with an unrelated cDNA (data not shown). These three MoAbs showed an identical and unique pattern of expression and had been clustered as CDw84 during the Fifth International Workshop.12 These MoAbs also specifically stained a murine pre–B-cell line (300.19) stably transfected with the 1,040 bp pHLy9-β cDNA (Fig 4). Expression of CD84 on the transfected cells was determined on the basis of anti-CD84 MoAb reactivity, measured by immunofluorescence staining. The CD84 MoAbs did not react with untransfected parent cells nor with cells transfected with vector alone. In all cases the reactivity pattern of the MoAb was identical. These results show that CD84 MoAbs identify the protein encoded by the HLy9-β cDNA.
CD84 MoAbs reactive with the protein product of the pHLy9-β cDNA. 300.19 cells transfected with HLy9-β cDNA were examined by using the CD84 MoAbs 2G7, 152-1D5, and 153-4D9 in indirect immunofluorescence assays with flow cytometry analysis. Fluorescence histograms generated using 300.19 transfected cells are shown as solid lines. Staining of the 300.19 untransfected cells are shown as dotted lines. A MoAb against CD22 (HB22.7) was unreactive with both transfected and untransfected cell lines. Fluorescence intensity is shown on a three-decade log scale.
CD84 MoAbs reactive with the protein product of the pHLy9-β cDNA. 300.19 cells transfected with HLy9-β cDNA were examined by using the CD84 MoAbs 2G7, 152-1D5, and 153-4D9 in indirect immunofluorescence assays with flow cytometry analysis. Fluorescence histograms generated using 300.19 transfected cells are shown as solid lines. Staining of the 300.19 untransfected cells are shown as dotted lines. A MoAb against CD22 (HB22.7) was unreactive with both transfected and untransfected cell lines. Fluorescence intensity is shown on a three-decade log scale.
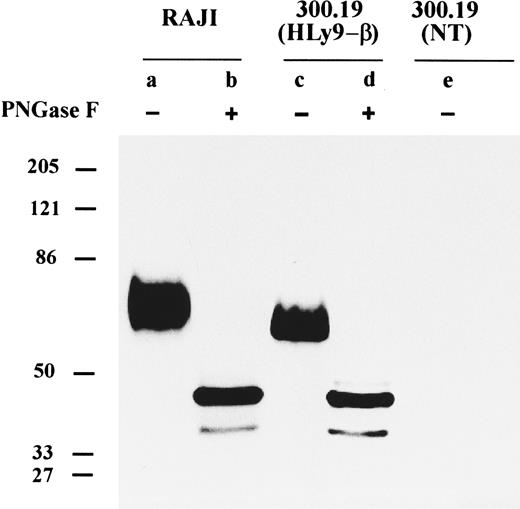
CD84 is highly glycosylated.The CD84 MoAbs were coupled to beads and used to immunoprecipitate the CD84 protein from detergent-solubilized extracts of biotinylated cell lines. Optimum results were obtained using the Raji cell line because the level of CD84 expression was higher than in other cell lines (Fig 5). The CD84 antibodies specifically immunoprecipitated proteins that migrated as a broad single band in the region from ∼64,000 to 82,000 Mr in SDS-polyacrylamide gels (Fig 5).
Immunoprecipitation of cell surface CD84 protein. Detergent lysates of surface-labeled Raji cell line, 300.19 cells transfected with HLy9-β, and 300.19 untransfected cells were immunoprecipitated with the 2G7 plus 152-1D5. The immunoprecipitated materials were treated with or without N-glycosidase (PNGase F ) and analyzed under reducing conditions on a 10% SDS-polyacrylamide gel. Molecular weights (kD) were determined by the migration of a known protein standard.
Immunoprecipitation of cell surface CD84 protein. Detergent lysates of surface-labeled Raji cell line, 300.19 cells transfected with HLy9-β, and 300.19 untransfected cells were immunoprecipitated with the 2G7 plus 152-1D5. The immunoprecipitated materials were treated with or without N-glycosidase (PNGase F ) and analyzed under reducing conditions on a 10% SDS-polyacrylamide gel. Molecular weights (kD) were determined by the migration of a known protein standard.
To determine the molecular mass of the protein in the absence of N-linked sugars, surface-labeled CD84 protein was digested with endoglycosydase F and analyzed by SDS-PAGE. After enzyme digestion, a loss of the ∼64,000 to 82,000 Mr band and the appearance of a major component at ∼45,000 and a fainter band of ∼36,000 Mr was observed (Fig 5). Compatible with this observation CD84 has four potential N-linked glycosylation sites (NXT/S) in the extracellular domain.
Immunoprecipitation with CD84 MoAbs of the 300.19 cell stably transfected with 1,040-bp pHLy9-β cDNA clone showed a broad band similar to the protein that was immunoprecipitated from the Raji cells, except that the Mr was ∼57,000 to 77,000 (Fig 5). After treatment of the immunoprecipitated material with endoglycosydase F, a dramatic reduction in the molecular mass was also observed. A major band of 45,000 and a lighter one of 36,000 Mr identical to those found in immunoprecipitates of the Raji cells line could be detected. These results showed that the differences in molecular mass between the protein in Raji cells and in the transfected cells were because of differences in the degree of glycosylation.
DISCUSSION
In this study, we have isolated cDNA clones encoding for the leukocyte antigen CD84. The structural features of the CD84 protein, predicted from nucleotide sequences derived from the cDNA clone HLy9-β (Fig 1), showed that it encodes a transmembrane type-I glycoprotein containing two Ig-like domains in the extracellular region: a hydrophobic membrane spanning domain, and an 83-amino acid cytoplasmic region (Fig 1). It is likely that the entire coding region of CD84 was identified as transfection of cell lines with the pHLy9-β cDNA-generated cell-surface expression of the protein (Fig 4) and the Mr of the immunoprecipitated protein was similar in both cDNA-transfected cells (∼59,000 to 77,000; Fig 5) and Raji cells (∼64,000 to 82,000; Fig 5). Despite this difference in electrophoretic mobility between the immunoprecipitates from the two cells, the bands observed after treatment with N-glycanase were the same for both cell types (Fig 5). These results show that CD84 undergoes extensive post-translational processing as it was expressed as a single-chain molecule, yet the determined Mr was more than twice the predicted size of the core protein. In fact, differential use of these N-linked sites may explain the heterogeneity in size of the CD84 protein.
Comparison of the CD84 amino acid sequences with other previously identified proteins showed a striking structural homology with the extracellular Ig-like domains of a subgroup of the Ig superfamily surface receptors that includes CD48, CD58, Ly9, and CD2, named the CD2 family. CD84 is most similar to murine and human Ly9, which are predicted to encode a protein with four Ig-like domains in the extracellular region.9 20 In contrast to Ly9, which has four Ig-like domains, CD84 like other members of the CD2 family is composed of only two Ig-like domains. Comparison of Ig-like domains in human Ly9 (HumLy9) and CD84 showed that the first domain of CD84 had 34.6% amino acid sequence identity with domain 1 and 30.3% with domain 3 of HumLy9; and the second domain of CD84 had 38.9% identity with domain 2 and 33.3% with domain 4.
Figure 6 shows sequence homology between CD84, HumLy9, and human CD48. Although the sequences of the domains share relatively little identity (∼25% to 39%), the core structures of the Ig-like domains are remarkably similar. Domain 1 of CD84 lacks a disulfide bond but is otherwise similar to V-like domains found in other proteins of the CD2 family.2,3 The second domain also shares homology with the truncated C2 domain found in this group of members of the Ig superfamily. Four Cys residues, which are conserved in all members of the CD2 family, were found in the putative second extracellular domain of CD84 (Fig 6). These Cys are probably involved in the formation of two disulfide bonds. One pair of these residues at positions 134 and 172 delineates an Ig-like domain creating the characteristic Ig-like loop structure and this domain contained many of the hallmark amino acids that define the C2 set of Ig-like domains.2
Ig-like domains of HLy9-β. HLy9-β Ig-like domains are compared with HumLy9 and human CD48. Solid lines above the sequence predicted β-sheets. Conserved residues appear in stippled boxes. Gaps are indicated with broken line to allow optimal alignment of the proteins. (A) First Ig-Like domain; (B) Second Ig-like domain. * indicates conserved amino acids of the Ig superfamily.
Ig-like domains of HLy9-β. HLy9-β Ig-like domains are compared with HumLy9 and human CD48. Solid lines above the sequence predicted β-sheets. Conserved residues appear in stippled boxes. Gaps are indicated with broken line to allow optimal alignment of the proteins. (A) First Ig-Like domain; (B) Second Ig-like domain. * indicates conserved amino acids of the Ig superfamily.
The 83-amino acid cytoplasmic domain of CD84 contained five tyrosine residues that could serve as potential sites of phosphorylation. Four of these Tyr were contained in two motifs Tyr-X-X-Leu/Ile and Tyr-X-X-Val with a general sequence context that can serve as potential sites for SH2 domain recognition.21 These types of motifs have been shown to be sites of interaction with multiple cytosolic proteins, including tyrosine kinases, and are found in the cytoplasmic domain of different signal transduction molecules, indicating that CD84 may be involved in signal transduction.
The CD84 gene was mapped to chromosome 1q24 (Fig 3), where other members of this Ig superfamily (CD48 and HumLy9) have also been mapped. The genes of the members of this family are located in two pericentric loci on human chromosome 1. Human CD2 and CD58 genes are located on chromosome 1p13, whereas the CD48 and HumLy9 genes are situated on chromosome 1q21-24.20,22,23 These genes are tightly linked with the ATP1A genes on chromosome 1, which have evolved by gene duplication.24 By inference, it is likely that duplication of a primordial gene also gave rise to the structurally related CD2, CD48, CD58, Ly9, and CD84 genes. It has been suggested that Ly9 and mouse CD48 share a common ancestor that duplicated and diverged, to give rise to a precursor of the Ly9 and CD48 genes, with a second duplication event producing the four-domain structure observed in Ly9.9 The sequence similarities between CD84, Ly9, and CD48, together with the mapping of these genes to the same chromosomal region, suggests that these three genes share a common ancestor.
CD84 expression appeared predominately to be lymphoid-tissue restricted as shown by Dot blot and Northern blot analyses (Fig 2A, B, and C) and immunohistological analysis of different tissues (data not shown). The highest levels of CD84 mRNA were detected in lymph node, spleen, and peripheral leukocytes (Fig 2A and B). These results are consistent with the observation that CD84 is expressed by lymphocytes and monocytes.13,25 Staining of lymph node and spleen shows that CD84 is weakly expressed by lymphocytes but strongly in scattered cells in the interfollicular zone, which were most likely macrophages.13 CD84 expression may be an early event in T-lymphocyte development as thymocytes showed low but detectable levels of mRNA (Fig 2) and cell surface expression (data not shown). In contrast very low levels of mRNA were detected in bone marrow, which is consistent with the observation that CD84 is not present in pro-B or pre-B cells.25 26 In only 2 out of the 39 nonlymphoid tissues analyzed, detectable levels of mRNA were observed. Significant levels of mRNA for CD84 could be detected in the kidney and, to a lesser extent, in the lung (Fig 2 and data not shown). The expression of CD84 by these tissues deserves further investigation.
From the structural information obtained in this study, it appears that CD84 is a newly described cell surface molecule that shares homology and chromosomal localization with a group of Ig superfamily members. Although the function of CD84 is still unknown, its structural similarity with other members of the Ig superfamily, as well as the presence of four potential SH2 domain binding motifs found in the cytoplasmic domain, suggest that it may be involved in cellular interactions and signal transduction.
ACKNOWLEDGMENT
We thank Victoria Tovar and Isabel Sánchez for assistance with these experiments.
Supported by Grants No. PB-1562, SAF96-0120, and SAF97-0136 from the Comision Interminsterial de Ciencia y Technologia and Grant No. PIRB-00076 of the Comissió Interdepartamental de Recerca Tecnologia 1995. M.A.d.l.F. is a fellow of the Fondo Investigaciones Sanitarias (97/5246). The GenBank accession number of the cDNA clone is U82988.
Address reprint requests to Pablo Engel, MD, Fundació Clı́nic, Villarroel 170, Barcelona 08036, Spain.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal