Abstract
Numerous transcription factors allow hematopoietic cells to respond to lineage- and stage-specific cytokines and/or to act as their effectors. The transcription factors PU.1 and c-Myb are essential for hematopoiesis, most likely acting at distinct stages of differentiation, but sharing a common set of target genes. To determine whether PU.1 and c-Myb are functionally interrelated, murine bone marrow (BM) cells and 32Dcl3 murine myeloid precursor cells were infected with a retrovirus carrying a PU.1 cDNA and assessed for myeloid colony formation and for granulocytic differentiation, respectively. Compared with noninfected normal BM cells or to cells infected with an empty virus, hematopoietic precursor cells expressing PU.1 formed an increased number of interleukin-3 (IL-3) and granulocyte colony-stimulating factor (G-CSF )–stimulated colonies. Moreover, granulocytic differentiation of 32Dcl3 cells constitutively expressing PU.1 was accelerated, as indicated by morphology and by expression of differentiation markers. Downregulation of c-Myb protein levels by expression of an antisense c-myb construct was also associated with a faster kinetics of 32Dcl3 granulocytic differentiation. Sequence analysis of the 5′ flanking region of the c-myb gene revealed a consensus PU box at position +16 to +21 able to specifically interact in electrophoretic mobility shift assays with either bacterially synthesized PU.1 protein or whole cell extracts from differentiated 32Dcl3 cells. Transient expression of PU.1 in cotransfection assays in different cell lines resulted in inhibition of chloramphenicol acetyl transferase activity driven by different segments of the c-myb promoter. Moreover, such an effect was dependent on an intact PU box. Thus, the ability of PU.1 to potentiate terminal myeloid differentiation appears to involve downregulation of c-myb expression, an essential step during differentiation of hematopoietic precursor cells.
HEMATOPOIETIC CELL differentiation involves differential expression of specific genes essential for the acquisition of the mature blood cell phenotype. The molecular mechanisms associated with these events are not fully understood, but transcription factors are thought to play a major role, and gene targeting experiments have shown that some of these factors, such as GATA-1,1 GATA-2,2 tal-1/SCL,3 Ikaros,4 c-myb,5 PU.1,6-8 and AML-1,9 are required for normal in vivo hematopoiesis.
The proto-oncogene c-myb is the cellular homologue of the v-myb gene, which was first identified in the avian myeloblastosis virus (AMV) and encodes a DNA binding protein that acts as a transcription activator. The DNA binding domain, which consists of three highly conserved imperfect repeats of 51 to 52 amino acids, each containing three regularly spaced tryptophan residues, resides in the N-terminal region of the protein. This DNA binding domain is evolutionarily well conserved and has been found in yeast, maize, and Drosophila c-myb–related proteins. Several findings support the important role of c-Myb in hematopoiesis: (1) it is preferentially expressed in hematopoietic progenitor cells and its levels decrease as cells differentiate10; (2) exposure of progenitor cells to c-myb antisense oligonucleotides inhibits formation of hematopoietic colonies in vitro11; (3) constitutive expression of c-Myb blocks both murine erythroleukemia cell differentiation, induced by treatment with erythropoietin, dimethylsulfoxide, or N,N′-hexamethylene bisacetamide,12-15 and granulocyte colony-stimulating factor (G-CSF )–induced differentiation of murine 32Dcl3 myeloid precursors16,17; and (4) homozygous null mutation of the c-myb gene in mice is associated with loss of hematopoietic precursors and embryonic lethality.5 An important mechanism for regulation of c-myb expression involves transcriptional pausing within the first intron,18 but there is also evidence for regulation at the level of transcription initiation,19-21 and c-myb itself has been shown to regulate its own promoter.22 Only a few targets of c-myb have been identified, including the T-cell receptor δ chain enhancer,23 mim-1,24 cdc-2,25 CD4,26 CD34,27 human c-myb,22,28 heat shock protein (HSP)-70,29 and c-fms promoters.30,31 It is not yet understood how the function of c-Myb as transcription factor is related to the regulation of the early stages of hematopoiesis; a possible mechanism involves enhanced expression of receptors for early-acting hematopoietic growth factors that may allow expansion of the progenitor cell pool.32
PU.1 is a member of the ets family of transcription factors originally described in B lymphocytes and macrophages33 but also found in all hematopoietic lineages, except T-cell lines and mature T lymphocytes.34 The PU.1 protein is the product of the protooncogene Spi-1, which is the site of insertional mutagenesis in Friend virus–induced murine erythroleukemia.35 PU.1 was defined based on its interaction with a purine-rich sequence in the SV40 enhancer called the PU box.36 Functional PU.1 binding sites have been identified in the mouse β–major globin intervening sequence 2 (IVS2),37 in the promoters of the murine major histocompatibility complex (MHC) class II gene,34 the murine Spi-1 gene,38 the macrophage scavenger receptor,39 FcγRI,40 FcγRIII,41 CD11b,42 CD18,43,44 immunoglobulin J-chain,45,46 the G-CSF receptor,47 the enhancers of Igκ,48 λ,49 and μ chains.50 Like c-Myb, PU.1 plays an important role in hematopoiesis because blocking of PU.1 function in CD34+ progenitor cells impairs in vitro hematopoiesis,51 and PU.1 overexpression in mouse bone marrow (BM)-derived macrophages stimulates proliferation of these cells.52 Moreover, mice in which the PU.1 gene has been inactivated by homologous recombination6-8 exhibit multilineage defects, as indicated by an impairment of erythroblast maturation and the inability to generate B and T lymphocytes, monocytes and granulocytes, and osteoclasts.
During blood formation, c-myb and PU.1 genes are sequentially expressed in BM cells. Although highly expressed in undifferentiated precursors, c-myb is absent in mature cells,53 whereas PU.1 is barely detectable in CD34+ progenitor/precursor cells but expressed at increasing levels as these cells differentiate.51
Recent work suggests that c-Myb and PU.1 regulate a common set of target genes. The activity of the macrophage colony-stimulating factor (M-CSF ) receptor promoter, for example, is upregulated by PU.1 and downregulated by c-Myb,31 and the activity of the neutrophil elastase gene promoter is positively and cooperatively regulated by PU.1, c-Myb, and C/EBP.54 Accordingly, mechanisms of cross-regulation between PU.1 and c-Myb might exist that are important in finely regulating the kinetics of hematopoietic differentiation. To investigate these possible functional interrelationships, we studied the ability of PU.1-infected normal murine BM cells to form myeloid colonies in vitro and of PU.1-infected 32Dcl3 myeloid precursors to differentiate in response to G-CSF stimulation. We also assessed the ability of PU.1 to regulate c-myb expression by examining the interaction of PU.1 with the c-myb promoter.
MATERIALS AND METHODS
Plasmid Constructs
pSV-PU.1.The murine PU.1 cDNA33 was kindly provided by Dr R. Maki. A 1.1-kb cDNA fragment containing the full-length murine PU.1 coding sequence was subcloned into the EcoRI site of the pSV40 polylinker vector19 in the sense orientation.
LXSN-PU.1.1.1-Kb EcoRI PU.1 cDNA fragment was subcloned in the sense orientation into the EcoRI site of the pLXSN vector.55
LXSN-mybTAS.The murine c-myb transactivation domain was synthesized by reverse transcription polymerase chain reaction (RT-PCR) amplification of total RNA from 32Dcl3 cells, and primers were used that corresponded to nucleotides 701 to 720 and the reverse complementary nucleotides 1099 to 1118 of the murine c-myb cDNA sequence.56 The 417-bp amplified fragment was subcloned into the pCRII vector (Invitrogen Corp, San Diego, CA), sequenced, and then subcloned in the antisense orientation at the EcoRI site of the pLXSN retroviral vector. pGEX-2TK-PU.1, which directs the expression of the GST-PU.1 fusion protein, was prepared as follows: pGEX-2TK vector (Pharmacia Biotech, Uppsala, Sweden) was linearized with BamHI, filled-in with Klenow enzyme, and then digested with EcoRI restriction endonuclease. The pSV-PU.1 plasmid was digested with PvuII and EcoRI, and the released 1.0-kb fragment corresponding to the PU.1 cDNA was ligated into the vector. Sequencing of the resulting plasmid and Western blot analysis of the bacterially produced fusion protein with an anti-PU.1 polyclonal antibody (Santa Cruz Biotechnology, Inc, Santa Cruz, CA) confirmed the correct frame of the construct and the production of a protein with the expected size.
B1-CAT, S1-CAT, and P1-CAT constructs have been described.22
P1s-CAT.A 112-bp fragment corresponding to nucleotides −78 to +34 of the human c-myb gene 5′ flanking region was PCR-amplified using a 5′ primer (AACAAGCTTCTGCAGGGGCGCCAGA) and a 3′ primer (GGGTCTAGAGGAGGAGGAAACAGGT). The resulting fragment was digested with HindIII and Xba I, subcloned in pCAT-Basic vector (Promega, Madison, WI), and sequenced.
P1m-CAT.A 112-bp fragment corresponding to nucleotides −78 to +34 of the human c-myb gene 5′ flanking region, containing a mutated PU box was PCR-amplified, subcloned in pCAT-Basic vector, and sequenced. The forward 5′ primer used was the same as above, whereas the reverse primer sequence was 5′ GGGTCTAGAGGAGGACCAAACAGGT 3′.
Cell Lines and Preparation of Viral Stocks
The murine interleukin-3 (IL-3)–dependent 32Dcl3 cell line has been described.57 Cells were maintained in Iscove's modified Dulbecco medium (IMDM), supplemented with 10% heat-inactivated fetal calf serum (FCS), 2 mmol/L L-glutamine, and 10% conditioned medium from WEHI-3B murine myelomonocytic cells as a source of crude IL-3.58 For induction of differentiation, 32Dcl3 cells were washed twice with phosphate-buffered saline (PBS) and resuspended (2 × 105 cells/mL) in IMDM containing 10% heat-inactivated FCS and 10% conditioned medium from U87MG human glioblastoma cells as a source of crude G-CSF.59 For immunofluorescence studies, exponentially growing and G-CSF–treated 32Dcl3 cells were collected at different times and incubated for 30 minutes at 4°C in PBS containing 0.1% gelatin, 0.01% sodium azide, and 5% human serum in the presence of fluorescein isothiocyanate (FITC)-conjugated Mo1 monoclonal antibody (PharMingen, San Diego, CA). Cells were washed twice and specific fluorescence was analyzed by flow cytometry in an EPICS Profile Analyzer (Coulter Corp, Hialeah, FL).
The murine ecotropic GP+E86 packaging line60 was maintained with Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS and 2 mmol/L L-glutamine. Viral stocks were generated by electroporation of pLXSN, LXSN-PU.1, and LXSN-mybTAS in GP+E86 cells, and virus-producing cell transfectants were obtained after a 10- to 14-day selection in G418-containing medium (1 mg/mL). Virus-containing supernatants were collected 3 days after monolayer formation of G418-resistant GP+E86 colonies. Viral titers (colony-forming units per milliliter) were determined by infection of Rat-2 fibroblasts and scoring the number of G418-resistant colonies.
Colony Formation Assays of Virus-Infected BM Cells
Murine BM cells were obtained from 4- to 6-week-old mice treated with 5-fluorouracil (150 mg/kg of body weight) 6 days before harvest. Cells were cultured at 106 cells/mL in IMDM supplemented with 10% fetal bovine serum (FBS), L-glutamine, penicillin/streptomycin, 50 μmol/L 2-mercaptoethanol, and 5 U/mL of murine IL-3 and granulocyte-macrophage colony-stimulating factor (GM-CSF ). After 24 hours, cells were resuspended in helper-free retroviral stocks supplemented with the same growth factors and Polybrene (4 μg/mL). Retrovirus-containing fresh media were added every 6 hours up to 48 hours. After an additional 24 hours, cells were washed three times and used immediately for clonogenic assays. Cells were plated in MethoCult H4230 semisolid medium (Stem Cell Technologies, Vancouver, BC) in the presence of the indicated concentrations of growth factors (mIL-3, 5 U/mL; mG-CSF, 10 U/mL; mKit ligand, 10 ng/mL), and colonies were scored 9 to 12 days later.
Northern Blot Analysis
Total RNA was prepared as described,61 and 5 μg from each sample was fractionated in a 1.2% agarose denaturing gel and transferred onto nitrocellulose membranes. 32P-labeled probes were murine PU.1 cDNA, murine c-myb, and murine G-CSF receptor cDNA. The same filter was sequentially hybridized with the c-myb, the G-CSF receptor, and β-actin probes at 42°C in 50% formamide, 5 × saline sodium citrate buffer (SSC), 0.5% sodium dodecyl sulfate (SDS), 1% Denhardt's solution, 0.2% Dextrane sulfate, 100 μg/mL sonicated, and denatured salmon sperm DNA and washed at high stringency (0.2× SSC, 0.1% SDS at 65°C).
Western Blot Analysis
Cells were lysed in HEPES buffer (10 mmol/L HEPES, pH 7.5, 150 mmol/L NaCl, 10% glycerol, 1 mmol/L ethylenediaminetetraacetic acid [EDTA], 1 mmol/L dithiothreitol [DTT]) containing 0.5% (vol/vol) Nonidet P-40, in the presence of protease inhibitors (1 mmol/L phenylmethylsulfonyl fluoride (PMSF ), 10 μg/mL leupeptin, 25 μg/mL aprotinin, 100 μg/mL pepstatin). Lysates were clarified and equal amounts of protein from each sample were fractionated in 8.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose. Membranes were blocked with 5% nonfat dry milk in PBS containing 0.05% Tween 20. PU.1 protein was detected with an anti-PU.1 polyclonal antiserum (Santa Cruz Biotechnology, Inc) followed by incubation with an anti-rabbit immunoglobulin antiserum conjugated with horseradish peroxidase (Amersham Life Science Inc, Buckinghamshire, UK) and chemiluminescence analysis. c-Myb protein was detected through the use of a commercial anti–c-Myb monoclonal antibody (type I; UBI, Lake Placid, NY) followed by incubation with horseradish peroxidase-conjugated sheep anti-mouse immunoglobulin antiserum (Amersham) and chemiluminescence analysis. Anti–β-actin monoclonal antibody (Oncogene Science, Inc, Cambridge MA) was used as a control.
Electrophoretic Mobility Shift Assay
GST-PU.1 recombinant protein was produced in BL-21 (DH3) cells transformed with pGEX-2TK-PU.1 as described.17 Whole cell extracts from 32Dcl3, U937, and Jurkat cells were prepared as described.62 Five microgram of rPU.1 protein or 15 μg of total extracts was used in each electrophoretic mobility shift assay (EMSA) reaction. Briefly, lysates were incubated on ice for 15 minutes with 0.12 μg/μL of poly di:dc in 20 mmol/L HEPES (pH 7.9), 50 mmol/L NaCl, 1.5 mmol/L MgCl2 , 0.5 mmol/L DTT, 0.2 mmol/L EDTA, 1% Nonidet P-40, and 10% glycerol. Unlabeled double-stranded oligodeoxynucleotides (50 ng = 100-fold molar excess) were used as specific and nonspecific competitor and included in the reactions when needed. Either 2 μg of a purified anti-PU.1 rabbit polyclonal antibody (Santa Cruz) or 2 μg of an unrelated rabbit polyclonal antibody (Santa Cruz) were used to determine the specificity of the binding. 32P–end-labeled double-stranded oligonucleotide probe (0.5 ng) corresponding to wild-type or mutated nucleotides (+6 to +40) of the c-myb gene were added to the binding reaction. Reaction mixtures were electrophoresed at 10 V/cm in native 5% PAGE in 0.25× Tris-borate-EDTA at 4°C.
Transfection and Chloramphenicol Acetyl Transferase (CAT) Assays
Jurkat cells (ATCC) were maintained in RPMI 1640 medium supplemented with 10% heat-inactivated FCS and 2 mmol/L L-glutamine. 32Dcl3 myeloid precursor cells were maintained in IMDM supplemented with 2 mmol/L L-glutamine and 10% WEHI-3B conditioned medium as source of crude IL-3. Transient transfection of hematopoietic cells were performed as described63; in particular, cells were washed twice in serum-free medium, resuspended in the same medium at 107 cells per 0.5 mL, and electroporated (Gene Pulser [BioRad, Richmond, VA; 960 μF], 280 V [Jurkat], or 250 V [32Dcl3]) with 15 μg of reporter plasmids and 30 μg of effector plasmid. After electroporation, cells were incubated for 5 minutes on ice and resuspended in 10 mL of complete medium. Plasmid (3 μg) containing the β-galactosidase cDNA driven by the SV40 promoter was included as an internal control for transfection efficiency.
Tk−ts13 hamster fibroblasts64 were maintained in DMEM medium supplemented with 10% heat-inactivated FCS and 2 mmol/L L-glutamine. Cells were transfected by the calcium phosphate precipitation method with 2 μg of reporter plasmids and 5 μg of effector plasmid as described.65 Plasmid (1 μg) containing β-galactosidase cDNA was included as internal control for transfection efficiency.
Cells were collected 48 hours after transfection and proteins were extracted in hypotonic buffer by 3 cycles of freezing and thawing and normalized for transfection efficiency by the β-galactosidase assay as recommended by the manufacturer (Promega). Cell lysates were incubated with acetyl coenzyme A and 14C-chloramphenicol for 90 minutes at 37°C. CAT levels were assayed by thin-layer chromatography followed by autoradiography and by scintillation counting of acetylated chloramphenicol.
RESULTS
Ectopic Expression of PU.1 in Murine BM Cells Increases Granulocytic Colony Formation
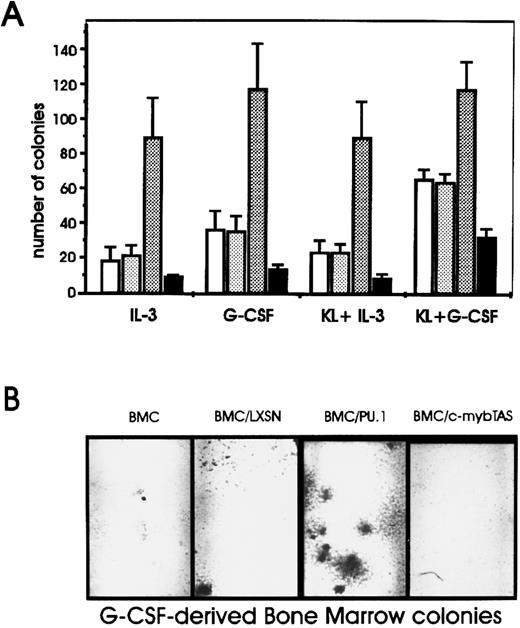
A retroviral vector carrying the PU.1 cDNA (LXSN-PU.1) was used to transduce PU.1 expression into murine BM cells, and the effects of PU.1 expression on normal hematopoietic progenitor cells were assessed based on the ability of LXSN-PU.1–infected cells to form myeloid colonies in methylcellulose in the presence of various cytokines used individually or in combination. The insert-less virus had no effect on growth factor–stimulated colony formation, whereas infection with the PU.1-encoding virus induced a marked increase in the size of the colonies (not shown) and a fourfold stimulation in the number of IL-3– and G-CSF–derived colonies (Fig 1). Similar effects were observed upon stimulation of colony formation with GM-CSF (not shown). Colony-forming unit–macrophage (CFU-M) colony formation was not assessed. Because blocking of c-myb expression has been shown to inhibit colony formation of BM cells,11 a virus carrying the transactivation domain of murine c-myb in the antisense orientation (LSXN-mybTAS) was included as a control. Infection of BM cells with this virus diminished the number of colonies by 50% in all conditions (Fig 1A), consistent with previous studies using antisense oligodeoxynucleotides.11
Effect of ectopic PU.1 expression on the number of colonies derived from BM cells. (A) Noninfected murine BM cells (□) or cells infected with PU.1 (LXSN-PU.1) (▧), the c-myb transactivation domain in antisense orientation (LXSN-mybTAS) (▪), or the insert-less (LXSN) () retroviral constructs. After 48 hours, 2.5 × 104 cells were plated in methylcellulose in the presence of the indicated growth factors. Colonies were counted 10 days later. Error bars indicate ± SD for three independent experiments. (B) Photomicrographs of G-CSF–derived BM colonies.
Effect of ectopic PU.1 expression on the number of colonies derived from BM cells. (A) Noninfected murine BM cells (□) or cells infected with PU.1 (LXSN-PU.1) (▧), the c-myb transactivation domain in antisense orientation (LXSN-mybTAS) (▪), or the insert-less (LXSN) () retroviral constructs. After 48 hours, 2.5 × 104 cells were plated in methylcellulose in the presence of the indicated growth factors. Colonies were counted 10 days later. Error bars indicate ± SD for three independent experiments. (B) Photomicrographs of G-CSF–derived BM colonies.
Effect of Ectopic PU.1 Expression on G-CSF–Induced Granulocyte Differentiation of 32Dcl3 Cells
Because PU.1 expression in murine marrow progenitors potentiated the ability of these cells to respond to G-CSF (Fig 1), we assessed the effects of constitutive PU.1 expression in the myeloid precursor 32Dcl3 cell line, which depends on the presence of IL-3 for cell growth and readily differentiates into mature granulocytes upon removal of IL-3 and addition of G-CSF.53
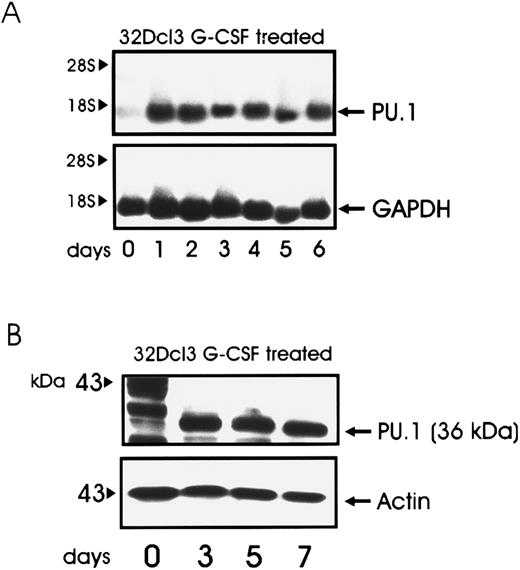
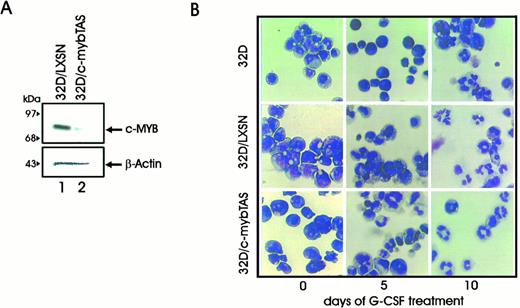
32Dcl3 cells expressed low levels of PU.1 when growing in IL-3; however, such levels increased when cells were deprived of IL-3 and the medium was supplemented with G-CSF (Fig 2, panel A and B). To study the effect of PU.1 overexpression in the process of granulocytic differentiation, 32Dcl3 cells were infected with a retrovirus carrying the murine PU.1 cDNA under the control of the MoMLV long terminal repeat (LXSN-PU.1). Mixed populations were chosen to avoid clonal variability. After selection in the presence of G418, expression of the transgene was assessed by Northern (Fig 3A) and Western blotting (Fig 3B). Growth kinetics of cells infected with LXSN or the PU.1-containing virus were identical to that of parental 32D cells in the presence of IL-3, and PU.1 expression did not abrogate IL-3 requirements for cell growth (data not shown). When cells were induced to differentiate by treatment with G-CSF, parental and LXSN-infected cells continued to grow for several days before terminal differentiation, as described.57 In contrast, 32D/PU.1 cells showed limited proliferation potential in the presence of G-CSF (Fig 4A, see page 1833). After 3 days of G-CSF stimulation, May-Grünwald-Giemsa staining of cytospin smears revealed evident features of differentiation in 32D/PU.1 cell cultures (Fig 4B). In contrast, parental and 32D/LXSN cells remained blastic during the initial days of G-CSF–induced differentiation, showing only terminal differentiation after 8 to 10 days of culture in the presence of G-CSF (Fig 4B).
Kinetics of PU.1 expression in G-CSF–treated 32Dcl3 cells. Total RNA (A) was isolated at the indicated times, and 5 μg of each sample was electrophoresed, blotted, and hybridized to a 32P-labeled PU.1 cDNA probe. Total lysate (30 μg) was used for immunoblotting (B) with the anti-PU.1 antibody.
Kinetics of PU.1 expression in G-CSF–treated 32Dcl3 cells. Total RNA (A) was isolated at the indicated times, and 5 μg of each sample was electrophoresed, blotted, and hybridized to a 32P-labeled PU.1 cDNA probe. Total lysate (30 μg) was used for immunoblotting (B) with the anti-PU.1 antibody.
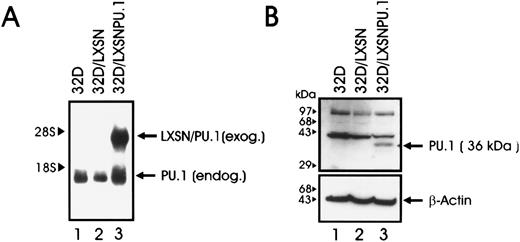
(A) PU.1 mRNA expression in parental and retrovirus-infected 32Dcl3 cells. Total RNA was isolated from infected 32D/LXSN cells (lane 2) or 32D PU.1 cells (lane 3) after selection in G418, and 5 μg from each sample was electrophoresed, blotted, and hybridized to a 32P-labeled PU.1 cDNA probe. Parental 32D cells (lane 1) were used as control. Arrows indicate the position of endogenous and exogenous PU.1 mRNAs. (B) Western blot analysis of PU.1 expression in 32D/PU.1 cells. Total lysates of parental 32Dcl3 cells (lane 1), and cells infected with the LXSN retrovirus (32D/LXSN; lane 2) or with the LXSN retrovirus carrying the PU.1 cDNA (32D/PU.1, lane 3) were subjected to SDS-PAGE, transferred onto nitrocellulose, and incubated with an anti-PU.1 polyclonal antibody. The membrane was then stripped and incubated with an anti–β-actin antibody as a control for equal loading.
(A) PU.1 mRNA expression in parental and retrovirus-infected 32Dcl3 cells. Total RNA was isolated from infected 32D/LXSN cells (lane 2) or 32D PU.1 cells (lane 3) after selection in G418, and 5 μg from each sample was electrophoresed, blotted, and hybridized to a 32P-labeled PU.1 cDNA probe. Parental 32D cells (lane 1) were used as control. Arrows indicate the position of endogenous and exogenous PU.1 mRNAs. (B) Western blot analysis of PU.1 expression in 32D/PU.1 cells. Total lysates of parental 32Dcl3 cells (lane 1), and cells infected with the LXSN retrovirus (32D/LXSN; lane 2) or with the LXSN retrovirus carrying the PU.1 cDNA (32D/PU.1, lane 3) were subjected to SDS-PAGE, transferred onto nitrocellulose, and incubated with an anti-PU.1 polyclonal antibody. The membrane was then stripped and incubated with an anti–β-actin antibody as a control for equal loading.
(A) Growth kinetics of parental and retrovirus-infected 32D cells in G-CSF–supplemented culture medium. Cells were seeded at 2.5 × 105 cells/mL; viable cells were determined by trypan blue dye exclusion test. (B) Morphology of parental, 32D/LXSN, and 32D/PU.1 cells in the presence of IL-3 or after 3, 5, and 10 days of stimulation with G-CSF. Cells were removed from the cultures, cytocentrifuged onto slides, air-dried, and stained with May-Grünwald-Giemsa.
(A) Growth kinetics of parental and retrovirus-infected 32D cells in G-CSF–supplemented culture medium. Cells were seeded at 2.5 × 105 cells/mL; viable cells were determined by trypan blue dye exclusion test. (B) Morphology of parental, 32D/LXSN, and 32D/PU.1 cells in the presence of IL-3 or after 3, 5, and 10 days of stimulation with G-CSF. Cells were removed from the cultures, cytocentrifuged onto slides, air-dried, and stained with May-Grünwald-Giemsa.
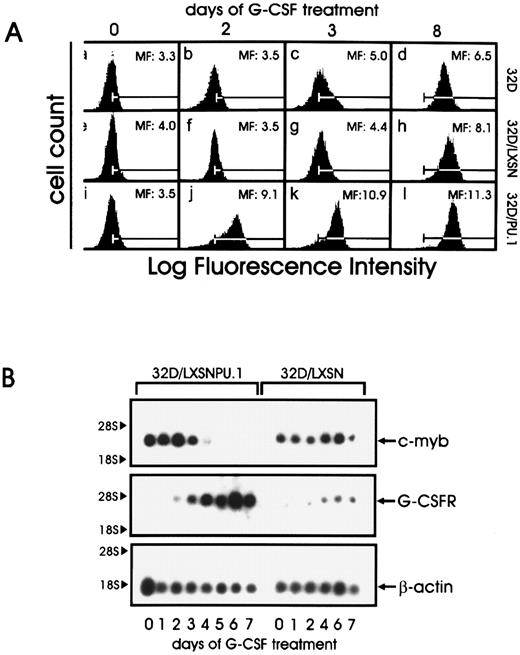
(A) CD11b expression in parental and retrovirus-infected 32D cells. Cells were cultured in the presence of G-CSF for the indicated times, stained with fluoresceinated Mo1 monoclonal antibody, and analyzed by flow cytometry. Mean fluorescence intensity is shown in each panel; the bars represent the background staining of an isotype control antibody. (B) G-CSF receptor and c-myb expression during differentiation of 32D cells constitutively expressing PU.1 32D/PU.1 and 32D/LXSN cells were cultured in the presence of G-CSF. Total RNA was isolated at the indicated times, and 5 μg of each sample was electrophoresed, blotted, and hybridized to a c-myb cDNA probe. The filter was then stripped and hybridized to a G-CSF receptor cDNA and to a β-actin probe.
(A) CD11b expression in parental and retrovirus-infected 32D cells. Cells were cultured in the presence of G-CSF for the indicated times, stained with fluoresceinated Mo1 monoclonal antibody, and analyzed by flow cytometry. Mean fluorescence intensity is shown in each panel; the bars represent the background staining of an isotype control antibody. (B) G-CSF receptor and c-myb expression during differentiation of 32D cells constitutively expressing PU.1 32D/PU.1 and 32D/LXSN cells were cultured in the presence of G-CSF. Total RNA was isolated at the indicated times, and 5 μg of each sample was electrophoresed, blotted, and hybridized to a c-myb cDNA probe. The filter was then stripped and hybridized to a G-CSF receptor cDNA and to a β-actin probe.
Mac-1 (CD11b) is a leukocyte integrin characteristic of mature granulocytes. Flow cytometry (Fig 5A) revealed upregulated Mac-1 expression in 32D/PU.1 cells as early as 2 days after G-CSF treatment (compare mean fluorescence in panels b, f, and j in Fig 5A), whereas Mac-1 upregulation was evident in 32D and 32D/LSXN cells only after 8 days of induction with G-CSF (Fig 5A).
Another marker of granulocytic differentiation is the G-CSF receptor itself, whose mRNA levels increase as cells differentiate. Compared with 32D/LXSN control cells, G-CSF receptor mRNA levels were upregulated earlier and at higher levels in 32D/PU.1 cells (Fig 5B). By contrast, c-myb is highly expressed in blastic cells and is absent in differentiated cells. Northern blot analysis indicated c-myb downregulation by day 5 in 32D/PU.1 cells, whereas c-myb levels remained constant at least until day 7 of G-CSF treatment in control LXSN-infected cells (Fig 5B). β-Actin mRNA levels were used as internal control for RNA loading.
Effect of c-myb Antisense Transcripts on Granulocytic Differentiation
Constitutive expression of c-myb blocks the final stages of erythromyeloid differentiation without altering the proliferative status of the cells.15 However, loss of c-Myb function in hematopoietic progenitors impairs hematopoiesis, most likely as a consequence of blocking the proliferative phases of this process. Thus, downregulation of c-myb expression in lineage-committed cells able to respond to a late-acting growth factor such as G-CSF might potentiate terminal cell differentiation. To test this hypothesis, 32Dcl3 cells were infected with a retroviral vector carrying the c-myb transactivation domain in the antisense orientation (LXSN-mybTAS) and selected in the presence of G418.
Western blot analysis confirmed that c-Myb protein levels were markedly decreased in LXSN-myb TAS–infected cells (Fig 6A). After 5 days in the presence of G-CSF, the cell cultures infected with the virus carrying the c-myb antisense construct (LXSN-myb TAS cells) showed an increase in the number of intermediate and segmented cells and a clear decrease in the number of blast cells (Fig 6B and Table 1), indicating that inhibition of c-Myb expression directs 32Dcl3 precursor cells toward a more mature phenotype.
(A) Western blot analysis of c-myb levels in 32D cells carrying a c-myb construct in the antisense orientation. (B) G-CSF–induced differentiation of 32Dcl3 cells carrying a c-myb antisense construct (32D/myb TAS cells). Photomicrographs of 32Dcl3, 32D/LXSN, and 32D/mybTAS cells in the presence of IL-3 or after 5 and 10 days of stimulation with G-CSF. Representative microphotographs of May-Grünwald-Giemsa–stained cytospin smears are shown.
(A) Western blot analysis of c-myb levels in 32D cells carrying a c-myb construct in the antisense orientation. (B) G-CSF–induced differentiation of 32Dcl3 cells carrying a c-myb antisense construct (32D/myb TAS cells). Photomicrographs of 32Dcl3, 32D/LXSN, and 32D/mybTAS cells in the presence of IL-3 or after 5 and 10 days of stimulation with G-CSF. Representative microphotographs of May-Grünwald-Giemsa–stained cytospin smears are shown.
Response of 32D/mybTAS Cells to G-CSF
| . | Percentage of Cell Types . | ||
|---|---|---|---|
| . | Blasts . | Intermediate . | Segmented . |
| 32D | 21 | 52 | 27 |
| 32D/LXSN | 14 | 55 | 31 |
| 32D/mybTAS | 2 | 37 | 61 |
| . | Percentage of Cell Types . | ||
|---|---|---|---|
| . | Blasts . | Intermediate . | Segmented . |
| 32D | 21 | 52 | 27 |
| 32D/LXSN | 14 | 55 | 31 |
| 32D/mybTAS | 2 | 37 | 61 |
32D cells, insert-less infected cells (32D/LXSN), and cells infected with the c-myb transactivation domain in antisense orientation (32D/mybTAS) were grown for 5 days in the presence of G-CSF (10% of U87MG conditioned medium). Cytospin smears were stained by May Grünwald-Giemsa and 100 cells were counted. Representative of three different experiments with similar results.
PU.1 Binds to the c-myb Promoter
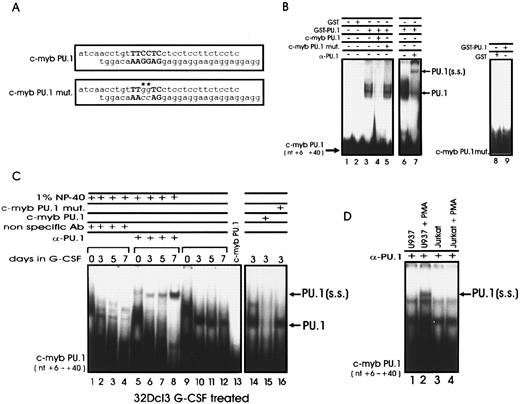
The temporal pattern of c-myb and PU.1 expression during differentiation of CD34+ hematopoietic progenitor cells and the requirements of both genes for normal hematopoiesis5,6,9,47 raise the possibility of cross-regulation in PU.1 and c-myb expression involving transcription mechanisms of gene activation. A search of PU.1 binding sites in the 5′ flanking region of the c-myb gene revealed a consensus PU box corresponding to nucleotides +16 to +21, relative to the transcription initiation site.22 To determine whether PU.1 binds to this site, a recombinant PU.1-glutathione-S-transferase (GST) fusion protein was allowed to interact in EMSA with a 32P-labeled double-stranded oligonucleotide corresponding to nucleotides +6 to +40 of the c-myb gene (Fig 7A). A retarded complex was detected with recombinant GST-PU.1 protein, but not with GST protein alone (Fig 7B, lanes 2 and 3). Binding was specifically competed by a 100-fold molar excess of unlabeled wild-type probe, but not by the same amount of an oligonucleotide in which guanines 18 and 19 were mutated to cytosines (Fig 7A and B, lanes 4 and 5). In addition, the retarded complex was specifically supershifted by an anti-PU.1 but not by a nonrelated rabbit polyclonal antibody (Fig 7B, lanes 6 and 7). No retarded complex was observed when the rPU.1 protein was incubated with mutated double-stranded oligodeoxynucleotides used as probe (Fig 7B, lane 6). The mutation in the two guanine residues of the PU box has been shown to prevent PU.1 binding,33 an additional indication that the binding of PU.1 to the c-myb promoter occurs specifically through this site.
PU.1 interacts with a PU box in the c-myb promoter. (A) Wild-type and mutated sequences of the PU box–containing oligonucleotides corresponding to nucleotides +6 to +40 of the human c-myb promoter. The mutated nucleotides are indicated by asterisks. (B) EMSA of PU.1 binding to the c-myb promoter. The double-stranded c-myb wild-type oligonucleotide was 32P-labeled and incubated with GST protein (lane 2), GST-PU.1 recombinant fusion protein (lanes 3 and 6), GST-PU.1 in the presence of a 100-fold molar excess of wild-type (lane 4) or mutated (lane 5) unlabeled probe, and GST-PU.1 in the presence of the anti-PU.1 antibody (lane 7). (B) EMSA were also performed with GST or GST-PU.1 and the PU box–mutated c-myb promoter fragment as probe (lanes 8 and 9). (C) EMSA performed with whole cell extracts from G-CSF–treated 32Dcl3 cells and PU box–containing c-myb double-stranded oligonucleotide as probe. Binding was performed in the presence of an anti-PU.1 antibody (lanes 5 to 8) or in the presence of a nonrelated rabbit polyclonal antibody (lanes 1 to 5). Binding was also performed in the presence of a 100-fold molar excess of either unlabeled wild-type or mutated PU box–containing c-myb double-stranded oligonucleotide used as specific or nonspecific competitor (lanes 15 and 16), respectively. (D) EMSA with whole cell extracts from U937 and Jurkat cells were performed as controls (lanes 1 to 4). PU.1 indicates the DNA/PU.1 protein complex; PU.1(ss) indicates the supershifted DNA-PU.1/anti-PU.1 Ab complex.
PU.1 interacts with a PU box in the c-myb promoter. (A) Wild-type and mutated sequences of the PU box–containing oligonucleotides corresponding to nucleotides +6 to +40 of the human c-myb promoter. The mutated nucleotides are indicated by asterisks. (B) EMSA of PU.1 binding to the c-myb promoter. The double-stranded c-myb wild-type oligonucleotide was 32P-labeled and incubated with GST protein (lane 2), GST-PU.1 recombinant fusion protein (lanes 3 and 6), GST-PU.1 in the presence of a 100-fold molar excess of wild-type (lane 4) or mutated (lane 5) unlabeled probe, and GST-PU.1 in the presence of the anti-PU.1 antibody (lane 7). (B) EMSA were also performed with GST or GST-PU.1 and the PU box–mutated c-myb promoter fragment as probe (lanes 8 and 9). (C) EMSA performed with whole cell extracts from G-CSF–treated 32Dcl3 cells and PU box–containing c-myb double-stranded oligonucleotide as probe. Binding was performed in the presence of an anti-PU.1 antibody (lanes 5 to 8) or in the presence of a nonrelated rabbit polyclonal antibody (lanes 1 to 5). Binding was also performed in the presence of a 100-fold molar excess of either unlabeled wild-type or mutated PU box–containing c-myb double-stranded oligonucleotide used as specific or nonspecific competitor (lanes 15 and 16), respectively. (D) EMSA with whole cell extracts from U937 and Jurkat cells were performed as controls (lanes 1 to 4). PU.1 indicates the DNA/PU.1 protein complex; PU.1(ss) indicates the supershifted DNA-PU.1/anti-PU.1 Ab complex.
Experiments with whole cell extracts from G-CSF–treated 32Dcl3 cells gave similar results (Fig 7C). A retarded complex was observed after incubation of the PU-box–containing c-myb promoter fragment with extracts from 32Dcl3 cells cultured for 3, 5, and 7 days with 10% U87MG conditioned medium as source of crude G-CSF (Fig 7C, lanes 10 to 12). The specificity of the binding was assessed using as competitors both the mutated double-stranded oligonucleotide and the wild-type c-myb fragment (nucleotides 6 to 40), containing the PU box (Fig 7C, lanes 15 and 16). The presence of PU.1 in the complex was determined by including in the binding reactions a specific anti-PU.1 rabbit polyclonal antibody. As expected, a supershifted complex was observed when the extracts from G-CSF–treated 32Dcl3 cells were incubated with the anti-PU.1 Ab (Fig 7, lanes 5 to 8) but not with a nonrelated rabbit polyclonal antibody (Fig 7, lanes 1 to 4). The binding reactions were performed in the presence of 1% NP-40 to prevent the possible association of PU.1 with other proteins that might hinder the binding of the anti-PU.1 antibody to the formed DNA-protein complex. As control, EMSA with the anti-PU.1 Ab were also performed with the PU box–containing c-myb double-stranded oligonucleotide and extracts from U937 cells in which PU.1 expression is enhanced by treatment with phorbol esters or from Jurkat cells that do not express PU.1 at detectable levels (Fig 7D).
PU.1 Transrepresses c-myb Promoter Activity
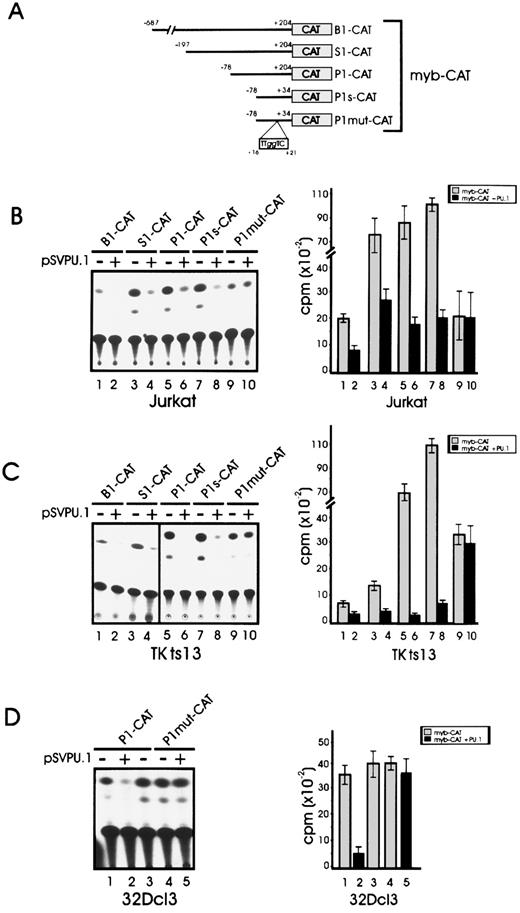
To determine whether the binding of PU.1 is functionally relevant for the ability of c-myb promoter to regulate the expression of a linked reporter gene, cotransfection experiments were performed in cell lines not expressing PU.1 (TK−ts13 and Jurkat cells) or expressing PU.1 at low levels (32Dcl3 cells). Reporter constructs containing serial deletions of c-myb promoter driving the CAT gene (B1-CAT, S1-CAT, P1-CAT, and P1s-CAT; Fig 8A) were transiently transfected in Jurkat cells (B1−, S1−, and P1-CAT) and in 32Dcl3 myeloid precursor cells (P1-CAT only), together with an effector plasmid carrying the PU.1 cDNA driven by the SV40 promoter. As shown in Fig 8B and D, expression driven by the c-myb promoter constructs was repressed in the presence of PU.1 (Fig 8B, lanes 2, 4, and 6; and D, lane 2) as compared with CAT activity in the presence of a cotransfected empty vector (Fig 8B, lanes 1, 3, 5, and 7; D, lane 3). However, the effect was more apparent (approximately 70% inhibition of promoter activity) when the shorter P1 construct was used. Similar results were obtained when CAT activity was assayed in the nonhematopoietic Tk−ts13 hamster fibroblasts transiently cotransfected with the c-myb reporter plasmids and the PU.1 expression vector (Fig 8C, lanes 2, 4, 6, and 8). Moreover, the results of CAT assays performed in Jurkat, Tk−ts13 and 32Dcl3 cells using a c-myb reporter construct specifically mutated in the PU box (Fig 8B and C, lanes 9 and 10; D, lanes 4 and 5) indicated that the negative effect of PU.1 on the activity of the c-myb promoter is dependent on an intact PU box.
PU.1 represses transcription driven by the c-myb promoter. (A) Schematic representation of c-myb promoter-CAT constructs used for transient transfection CAT assays. Numbering is relative to nucleotide +1.19 Location of the mutated PU.1 binding site is shown. (B) CAT activity in Jurkat cells transiently cotransfected with the c-myb-CAT constructs (wild-type and mutated) and a PU.1 expression vector (lanes 2, 4, 6, 8, and 10) or the empty vector (lanes 1, 3, 5, 7, and 9). (C) CAT activity in Tk−ts13 hamster fibroblasts transiently cotransfected with the c-myb–CAT constructs (wild-type and mutated) and a PU.1 expression vector (lanes 2, 4, 6, 8, and 10) or the empty vector (lanes 1, 3, 5, 7, and 9). Error bars indicate ±SD for three independent experiments. (D) CAT activity in 32Dcl3 cells transiently transfected with wild-type P1-CAT (lane 1) or mutant P1mut-CAT (lane 3) c-myb promoter in the presence of pSVPU.1 (lanes 2 and 5) or of pSV vector (lane 3).
PU.1 represses transcription driven by the c-myb promoter. (A) Schematic representation of c-myb promoter-CAT constructs used for transient transfection CAT assays. Numbering is relative to nucleotide +1.19 Location of the mutated PU.1 binding site is shown. (B) CAT activity in Jurkat cells transiently cotransfected with the c-myb-CAT constructs (wild-type and mutated) and a PU.1 expression vector (lanes 2, 4, 6, 8, and 10) or the empty vector (lanes 1, 3, 5, 7, and 9). (C) CAT activity in Tk−ts13 hamster fibroblasts transiently cotransfected with the c-myb–CAT constructs (wild-type and mutated) and a PU.1 expression vector (lanes 2, 4, 6, 8, and 10) or the empty vector (lanes 1, 3, 5, 7, and 9). Error bars indicate ±SD for three independent experiments. (D) CAT activity in 32Dcl3 cells transiently transfected with wild-type P1-CAT (lane 1) or mutant P1mut-CAT (lane 3) c-myb promoter in the presence of pSVPU.1 (lanes 2 and 5) or of pSV vector (lane 3).
DISCUSSION
Cellular differentiation is the process by which precursor cells acquire the characteristics of mature cell types through transcription factor–induced activation and repression of specific programs of gene expression. Such lineage-specific patterns of gene expression are likely controlled by more than one transcription factor acting as positive or negative regulators of stage-specific processes. Among such factors, PU.1 and Myb have a well-defined role in hematopoiesis. Thus, we investigated the effects of perturbed PU.1 or c-Myb expression in hematopoietic precursor cells and asked whether PU.1 and c-Myb expression are functionally and mechanistically interrelated.
Colony formation assay in semisolid medium to assess in vitro differentiation of BM cells has revealed that downregulation of PU.1 expression impairs in vitro hematopoiesis.51 Consistent with this observation, overexpression of PU.1 in BM cells potentiates the ability of these cells to form myeloid colonies in vitro in response to G-CSF stimulation (Fig 1). To further investigate this function, the effects of PU.1 overexpression were assessed in 32Dcl3 cells. When cultured in the presence of G-CSF, these cells readily differentiated into granulocytes, concomitant with a rapid increase in PU.1 expression. 32Dcl3 cells constitutively expressing high levels of PU.1 did not differentiate spontaneously into granulocytes; however, upon treatment with G-CSF, the kinetics of granulocytic differentiation of PU.1-infected 32D cells was much faster than that of parental cells, with the PU.1-infected cells fully differentiated as early as 2 to 3 days after G-CSF stimulation.
Inhibition of c-myb expression also impairs in vitro hematopoiesis.11 It has been proposed that c-Myb is essential for the proliferative status of progenitor cells and that down-regulation of c-myb expression might be crucial for the occurrence of terminal differentiation.12,14 16
PU.1 overexpression in BM cells potentiated the ability of these cells to form myeloid colonies in vitro, and overexpression of PU.1 in 32Dcl3 cells enhanced granulocytic differentiation in response to G-CSF (Figs 1 and 4). Thus, we reasoned that these effects might reflect not only the induction of genes necessary for terminal differentiation, but also the downregulation of the expression of genes involved in maintaining the cells in an undifferentiated state. In 32Dcl3 cells constitutively expressing PU.1, treatment with G-CSF was associated with a decrease of c-myb mRNA levels with a kinetics faster of that observed in parental cells. However, c-myb mRNA was expressed at high levels during the early stages of G-CSF–induced differentiation of PU.1-expressing 32D cells, suggesting that the effects of PU.1 on c-myb expression might involve direct and differentiation-related mechanisms of gene regulation. Downregulation of c-Myb protein levels in 32Dcl3 cells infected with a retrovirus carrying a c-myb antisense construct was also associated with enhanced granulocytic differentiation in response to G-CSF, but the effect was not as dramatic as that induced by PU.1 overexpression. This is probably caused by c-Myb–independent effects exerted by PU.1 overexpression. Together, these results are consistent with the possibility that c-myb expression is negatively regulated by PU.1, directly or in association with PU.1-dependent differentiation-related mechanisms.
Sequence analysis of the c-myb gene 5′-flanking region revealed a consensus PU box at position +16 to +21 relative to the transcription initiation site. We investigated whether this element plays a role in the regulation of c-myb expression by (1) assessing PU.1 binding in EMSA; (2) determining whether PU.1 transrepresses c-myb promoter activity in cotransfection assays in hematopoietic and nonhematopoietic cells; and (3) investigating whether an intact PU.1 binding site is required for such effects. Although a primary mode of regulation of c-myb expression involves transcriptional pausing in the first intron,18 substantial evidence indicates that c-myb regulation can also occur at the promoter level.21,22,28 We used the human c-myb promoter for our studies; however, the PU box is also 100% conserved in the murine promoter,66 consistent with the importance of this segment of the 5′ flanking region in the regulation of c-myb expression. Moreover, like most genes regulated by PU.1, c-myb lacks a consensus TATA box, and its PU box is near the major transcription initiation site. The ability of an ets family member like PU.1 to act as a negative regulator of transcription is not unprecedented; it has been recently reported that π1, a member of the ets family, negatively regulates the bcl-2 promoter,67 and that PU.1 itself transrepresses the activity of the immunoglobulin H enhancer68 and of the I-Aβ gene in a DNA binding–dependent manner.69 In addition to its role as regulator of promoter activity in a DNA binding–dependent manner, PU.1 has also been shown to bind RNA and interfere with the process of gene splicing, at least in vitro.70 Because the PU.1 binding site in the c-myb gene lies in the 5′ flanking region immediately downstream of the major site of transcription initiation, we cannot exclude an RNA-dependent mechanism of regulation of gene expression.
Our findings of negative regulation of the c-myb promoter by PU.1 are consistent with the temporal pattern of expression of these two genes. c-Myb is expressed in hematopoietic progenitor/precursor cells, and its levels decrease as cells differentiate. By contrast, differentiated B cells, monocytes, and granulocytes express high levels of PU.1 mRNA. In CD34+ BM cells, c-myb levels are high, whereas PU.1 is barely detectable. When these cells differentiate in vitro along the myeloid or the erythroid lineage, PU.1 expression is upregulated.51 In the erythroid lineage, the expression declines when cells terminally differentiate, but mature myeloid cells express high levels of PU.1. On the contrary, c-Myb expression is no longer detectable in both lineages when cells acquire a mature phenotype.
Our data suggest that PU.1 plays a role in the downregulation of c-myb expression during normal hematopoiesis. Consistent with this hypothesis, a null mutation of the c-myb gene in mice results in the failure to generate definitive fetal hematopoiesis with consequent death of the mice at day 14.5 of development. Homozygous deletion of the PU.1 gene has been shown to cause embryonic lethality occurring at about day 17.5 in one study,6 whereas in another study7 death of the mice occured immediately after birth. Regardless of the reasons of these apparent discrepancies, it is clear that, compared with c-Myb, PU.1 is required at a later stage of development.
In summary, PU.1 is a transcription factor with an important role in allowing progenitor/precursor cells to complete their differentiation program. Such a process might depend, in part, on the induction of genes specific for differentiated cells, but also on the repression of genes such as c-myb, whose expression, if maintained at high levels, interferes with terminal differentiation.
ACKNOWLEDGMENT
We thank Dr Susan Weil for independent evaluation of May-Grünwald-Giemsa slides and Marina Hoffman for critical reading of the manuscript.
T.B. and D.P. are equal contributors.
Supported in part by grant no. CA46782 from the National Institutes of Health to B.C. T.B. was supported by a Doctores y Tecnologos fellowship of the Ministerio de Educacion and Ciencia of Spain.
Address reprint requests to Dr Bruno Calabretta, Department of Microbiology and Immunology, Jefferson Cancer Institute, Bluemle Life Sciences Bldg, 233 S 10th St, Room 630, Philadelphia, PA 19107.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal