FANCONI ANEMIA (FA) is a rare autosomal recessive disease characterized by multiple congenital abnormalities, bone marrow (BM) failure, and cancer susceptibility. The mean age of onset of anemia is 8 years, and the mean survival is 16 years. Death usually results from complications of BM failure. Considerable progress in the field of FA research has resulted from the recent cloning of two of the five known FA genes. The purpose of the current review is to describe the structure and function of the FA genes and to outline novel approaches to FA diagnosis and therapy, based on the availability of these genes.
CLINICAL COURSE OF FA
The congenital abnormalities and clinical course of FA have been extensively reviewed.1-9 Patients with FA have growth retardation and abnormalities of the skin (generalized hyperpigmentation and/or café au lait spots), upper extremities (frequently with defects in the thumbs or forearms), kidneys, and gastrointestinal system. Male FA patients have underdeveloped gonads and defective spermatogenesis. The large range of organ systems affected in FA implicates the FA genes in a general developmental process required during normal human embryogenesis.
The hematologic complications of FA have also been extensively reviewed.1,4 10 FA patients develop macrocytosis and pancytopenia, typically during the first decade of life. Deficiencies in platelets or red blood cells usually precede white blood cell abnormalities. The patients have “fetal-like” erythropoiesis, with increased i antigen and hemoglobin F, and generally have high serum erythropoietin levels. The progression to pancytopenia is highly varied among FA patients.
At least 20% of patients with FA develop cancers.1 Because many FA patients die of other causes before they might have developed cancer, the actuarial risk of cancer is even higher.11 FA patients primarily develop acute myeloblastic leukemia; however, cancers of several organ systems including the skin, gastrointestinal, and gynecological systems have been described. The skin and gastrointestinal tumors are usually squamous cell carcinomas.12-14 In addition, FA patients receiving androgen therapy for BM failure are prone to liver tumors. Cancer tends to be a disease of older FA patients. The average age of patients who develop cancers is 15 years for leukemia, 16 years for liver tumors, and 23 years for other tumors. There is a slightly higher risk of cancer for female FA patients, irrespective of the increased risk of gynecological cancers.15
DIAGNOSIS OF FA
The diagnosis of FA exploits the sensitivity of FA cells to bifunctional alkylating agents. FA cells have increased spontaneous chromosomal breakage that is amplified by the addition of the cross-linking agents, diepoxybutane (DEB) or mitomycin C (MMC). Similar spontaneous, but not DEB-induced, chromosomal changes are observed in Bloom's syndrome and ataxia telangiectasia.16,17 The DEB test is a highly sensitive and specific test for FA.18 Although useful in the diagnosis of FA patients, the DEB test does not distinguish FA carriers from the general population. Also, the DEB test can give false-negative results, particularly in patients with cellular mosacism.19 The DEB test has been used successfully in prenatal diagnosis of FA. Prenatal diagnosis has been performed by chorionic villus sampling (CVS) at 9 to 12 weeks of gestation or by amniocentesis at 16 weeks in many cases of suspected FA.20 21
The diagnosis of FA is complicated by the wide variability in FA patient phenotype.2 The differential diagnosis of FA includes other genetic syndromes including neurofibromatosis, VACTERL association, and TAR (thrombocytopenia with absence of radii). Although the DEB test is highly effective in discriminating FA from these syndromes, the test remains underutilized.18 Further confounding diagnosis, approximately 33% of patients with FA have no obvious congenital abnormalities.6 These FA patients may first present with BM failure or cancer.22 Despite this range in phenotypic variation, there exists no obvious correlation between the severity of the disease and the extent of cellular sensitivity to DEB.
TREATMENT OF FA
The treatment of FA is similar, but not identical, to the treatment of other forms of acquired aplastic anemia. Patients are treated with supportive care (ie, blood transfusions) for their BM failure. FA patients respond transiently to therapy with androgens and the cytokines, granulocyte-macrophage colony-stimulating factor (GM-CSF )23 and G-CSF.24 The treatment of choice for FA is allogenic BM transplant with a histocompatible sibling donor. In one recent study,25 18 patients with FA had allogeneic BM transplants from matched sibling donors (MSD). Seventeen patients had sustained engraftment and transfusion independence. Still, FA patients have severe toxicity from graft-versus-host disease (GVHD), perhaps resulting from an increased cellular sensitivity to endogenous cytokines released during GVHD. T-cell depletion has reduced GVHD for BM transplantation from unrelated donors.26 Umbilical cord blood also offers a potential source of hematopoietic stem cells for FA patients without sibling matches.27-31
LEUKEMIA IN FA
Leukemia in FA patients differs significantly from leukemia in the general pediatric and adolescent populations.1,22,32 Treatment of leukemia in FA patients is largely ineffectual, with many patients dying within 1 to 3 months of diagnosis. Many of the FA patients who develop leukemia have pre-existing myelodysplastic syndrome, or preleukemia. Many of these patients have cytogenetic abnormalities in chromosomes 1 and 733,34 resulting from complete or partial chromosomal deletion or translocation. Leukemic cells from FA patients usually do not have abnormalities in the p53 gene.35 In one case, when a leukemic FA patient did achieve a remission, the marrow remained myelodysplastic.22
THE CELLULAR PHENOTYPE OF FA CELLS
Because of the cellular sensitivity to cross-linking agents, FA is often compared with other syndromes of drug sensitivity and genomic instability, including ataxia telangiectasia (AT), Xeroderma pigmentosum (XP), Cockayne syndrome (CS),36,37 Bloom's syndrome, and HNPCC38 (Table 1). The eight genes for XP encode proteins that comprise a DNA excision repair complex.39,40 ATM, the gene mutated in the inherited human disease ataxia-telangiectasia (AT), encodes a protein with homology to PI-3 kinase,41 suggesting that the ATM protein plays a role in sensing DNA damage and signaling the induction of p53.42 More recent evidence shows that the ATM protein product associates with meiotically pairing chromosomes.43,44 The gene for Bloom's syndrome encodes a protein related to a known yeast protein that regulates the cell cycle.45 46 Therefore, the Bloom's syndrome protein may regulate a G2/M checkpoint in the normal mammalian cell cycle. The genomic instability of these syndromes may result from a cellular defect in one of several processes, including DNA repair, cell-cycle regulation, or DNA replication.
Diseases of Genomic Instability
| Disease . | Damaging Agent . | Neoplasm . | Function . |
|---|---|---|---|
| FA | Cross-linking agents | Acute myeloblastic leukemia, hepatic, gastrointestinal, and gynecological tumors | Unknown |
| XP | UV light | Squamous cell carcinomas | Excision repair |
| AT | Ionizing radiation | Lymphoma | Afferent pathway to p53 |
| Bloom's syndrome | Alkylating agents | Acute lymphoblastic leukemia | Cell-cycle regulation |
| Cockayne's syndrome | UV light | Basal cell carcinoma | Transcription coupled repair |
| Hereditary nonpolyposis colon cancer (HNPCC) | Unknown | Adenocarcinoma of colon, ovarian cancer | DNA mismatch repair |
| Disease . | Damaging Agent . | Neoplasm . | Function . |
|---|---|---|---|
| FA | Cross-linking agents | Acute myeloblastic leukemia, hepatic, gastrointestinal, and gynecological tumors | Unknown |
| XP | UV light | Squamous cell carcinomas | Excision repair |
| AT | Ionizing radiation | Lymphoma | Afferent pathway to p53 |
| Bloom's syndrome | Alkylating agents | Acute lymphoblastic leukemia | Cell-cycle regulation |
| Cockayne's syndrome | UV light | Basal cell carcinoma | Transcription coupled repair |
| Hereditary nonpolyposis colon cancer (HNPCC) | Unknown | Adenocarcinoma of colon, ovarian cancer | DNA mismatch repair |
Cellular Abnormalities of FA
| . | References . |
|---|---|
| Sensitivity to cross-linking agents | Schroeder, 1964 (47) |
| German, 1966 (48) | |
| Prolongation of G2 phase of cell cycle | Kaiser, 1982 (49) |
| Kubbies, 1985 (51) | |
| Sensitivity to oxygen | |
| Grow poorly at ambient O2 | Schlindler, 1988 (52) |
| Overproduction of O2 radicals | Korkina, 1992 (50) |
| Deficient O2 radical defense | Gille, 1987 (53) |
| Deficiency in superoxide dismutase | Joenje, 1979 (67); Mavelli, 1982 (68) |
| Sensitivity to ionizing radiation (G2 specific) | Bigelow, 1979 (54) |
| Overproduction of tumor necrosis factor-α | Rosselli, 1992 (55) |
| Direct defects in DNA repair | |
| Accumulation of DNA adducts | Takeuchi, 1993 (56) |
| Defective in repair of DNA cross-links | Fujiwara, 1977 (57) |
| Genomic instability | |
| Spontaneous chromosome breakage | Auerbach, 1989 (3) |
| Hypermutable (by deletion mechanism) | Papadopoulo, 1990 (58) |
| Increased apoptosis | Willingale-Theune, 1989 (59) |
| Kupfer, 1996 (60) | |
| Wang, 1996 (64) | |
| Defective p53 induction | Rosselli, 1995 (61) |
| Kupfer, 1996 (60) | |
| Intrinsic stem cell defect | |
| Decreased colony growth in vitro | Daneshbod-Skibba, 1980 (62) |
| Alter, 1991 (10) | |
| Decreased gonadal stem cell survival | Whitney, 1996 (63) |
| . | References . |
|---|---|
| Sensitivity to cross-linking agents | Schroeder, 1964 (47) |
| German, 1966 (48) | |
| Prolongation of G2 phase of cell cycle | Kaiser, 1982 (49) |
| Kubbies, 1985 (51) | |
| Sensitivity to oxygen | |
| Grow poorly at ambient O2 | Schlindler, 1988 (52) |
| Overproduction of O2 radicals | Korkina, 1992 (50) |
| Deficient O2 radical defense | Gille, 1987 (53) |
| Deficiency in superoxide dismutase | Joenje, 1979 (67); Mavelli, 1982 (68) |
| Sensitivity to ionizing radiation (G2 specific) | Bigelow, 1979 (54) |
| Overproduction of tumor necrosis factor-α | Rosselli, 1992 (55) |
| Direct defects in DNA repair | |
| Accumulation of DNA adducts | Takeuchi, 1993 (56) |
| Defective in repair of DNA cross-links | Fujiwara, 1977 (57) |
| Genomic instability | |
| Spontaneous chromosome breakage | Auerbach, 1989 (3) |
| Hypermutable (by deletion mechanism) | Papadopoulo, 1990 (58) |
| Increased apoptosis | Willingale-Theune, 1989 (59) |
| Kupfer, 1996 (60) | |
| Wang, 1996 (64) | |
| Defective p53 induction | Rosselli, 1995 (61) |
| Kupfer, 1996 (60) | |
| Intrinsic stem cell defect | |
| Decreased colony growth in vitro | Daneshbod-Skibba, 1980 (62) |
| Alter, 1991 (10) | |
| Decreased gonadal stem cell survival | Whitney, 1996 (63) |
In addition to cross-linking agent sensitivity, FA cells have several other phenotypic abnormalities3,10 47-68 (outlined in Table 2). A detailed description of these studies is beyond the scope of this review. Many of these cellular assays have been performed on FA cells from multiple complementation groups. Accordingly, it remains unclear whether these cellular abnormalities correspond to all FA complementation groups or to only a subset. Most of the abnormalities described for FA cells are probably epiphenomena and do not relate directly to the primary cellular defect in each FA complementation group. Therefore, a true understanding of the primary defect in FA, such as DNA repair, cell-cycle regulation, or prevention of apoptosis, can only come from a molecular understanding of the newly cloned FA proteins.
CLONING OF FA GENES
The complementation analysis of FA cells, using somatic cell fusion studies, has allowed the identification of at least five complementation groups.69-73 At least three of the five groups (A, C, and D) map to discrete chromosomal loci72,74-76 (Table 3). Therefore, FA is a genetically heterogenous disorder, unlike the syndromes, ataxia-telangiectasia (AT)41 and Bloom's syndrome,45 which arise from mutations in single genes.
Complementation Groups of FA
| Subtype . | Estimated Percentage of FA Patients . | Chromosome Location . | Protein Product . |
|---|---|---|---|
| A | 66.0 | 16q24.3 | 163 kD (predicted nuclear localization) |
| B | 4.3 | ? | |
| C | 12.7 | 9q22.3 | 63 kD (cytoplasmic localization) |
| D | 4.3 | 3p22-26 | |
| E | 12.7 | ? | |
| Subtype . | Estimated Percentage of FA Patients . | Chromosome Location . | Protein Product . |
|---|---|---|---|
| A | 66.0 | 16q24.3 | 163 kD (predicted nuclear localization) |
| B | 4.3 | ? | |
| C | 12.7 | 9q22.3 | 63 kD (cytoplasmic localization) |
| D | 4.3 | 3p22-26 | |
| E | 12.7 | ? | |
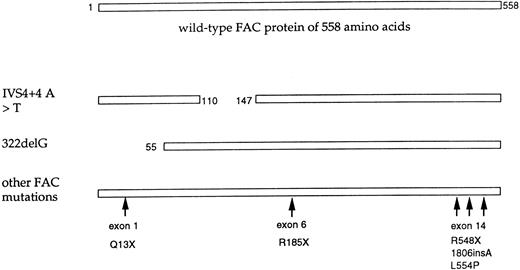
Mutations of the FAC polypeptide found in patients with FA. The wild-type FAC protein (558 amino acids) is shown schematically at the top. The indicated FAC alleles are also shown. The IVS4+4 A < T mutant allele encodes a protein with an in-frame deletion of 37 amino acids.80,91 The 322delG has a frame shift mutation in exon 1.77 This allele encodes an amino terminal truncated FAC protein with translation reinitiation from methionine 55.81 The IVS4+4 A < T mutation and 322delG account for approximately 75% of FAC mutations. Less common mutant alleles of FAC are also shown. At least three of these mutations fall in exon 14, encoding the carboxy terminus of the FAC protein. An allele with a C to T nucleotide substitution at base 808 (exon 6) is predicted to encode a protein with premature termination at amino acid 185.83 The 1806insA mutation is predicted to encode a truncated protein of 526 amino acids.86 A nucleotide substitution of T to C, at base 1913, encodes a protein with an (L554P) mutation.77 Recent studies suggest that this mutant protein is expressed in FAC cell lines91 and has dominant negative activity when overexpressed.94 This allele is the only known missense mutant and suggests that the carboxy terminus of FAC has functional relevance (see text). The relative frequency of these various FAC alleles has recently been described.82
Mutations of the FAC polypeptide found in patients with FA. The wild-type FAC protein (558 amino acids) is shown schematically at the top. The indicated FAC alleles are also shown. The IVS4+4 A < T mutant allele encodes a protein with an in-frame deletion of 37 amino acids.80,91 The 322delG has a frame shift mutation in exon 1.77 This allele encodes an amino terminal truncated FAC protein with translation reinitiation from methionine 55.81 The IVS4+4 A < T mutation and 322delG account for approximately 75% of FAC mutations. Less common mutant alleles of FAC are also shown. At least three of these mutations fall in exon 14, encoding the carboxy terminus of the FAC protein. An allele with a C to T nucleotide substitution at base 808 (exon 6) is predicted to encode a protein with premature termination at amino acid 185.83 The 1806insA mutation is predicted to encode a truncated protein of 526 amino acids.86 A nucleotide substitution of T to C, at base 1913, encodes a protein with an (L554P) mutation.77 Recent studies suggest that this mutant protein is expressed in FAC cell lines91 and has dominant negative activity when overexpressed.94 This allele is the only known missense mutant and suggests that the carboxy terminus of FAC has functional relevance (see text). The relative frequency of these various FAC alleles has recently been described.82
BIOCHEMISTRY AND MOLECULAR BIOLOGY OF FAC
FAC was cloned by functional complementation of an Epstein-Barr virus (EBV)-immortalized type C FA cell line, HSC536.77 As predicted by the complementation test, the FAC cDNA corrects the MMC sensitivity and DEB sensitivity of FA(C) cell lines, but does not correct the MMC sensitivity of FA cells derived from type A, B, D, or E patients. Cells derived from patients with FA(C) have mutations in both alleles of the FAC gene, consistent with the autosomal recessive inheritance of the FA syndrome. The FAC cDNA encodes a 558–amino acid polypeptide (63 kD) with no homology to other proteins in GenBank. Moreover, the FAC gene is composed of 14 exons,78 spans approximately 80 kb, and maps to human chromosome 9q22.3.78 The murine FAC gene has been isolated, but no FAC homologues from other species have been reported. Murine FAC is only 66% identical to the human protein, but is able to functionally complement human cells from FA(C) patients.79
Mutational analysis of the FAC gene has shown a relatively small number of characteristic mutations, represented in specific ancestral backgrounds (Fig 1). The IVS4+4 A > T mutation is found in patients of Ashkenazi-Jewish ancestry and accounts for greater than 80% of FA in this population.80 Patients homozygous for this mutation have more severe FA, with multiple congenital abnormalities and early onset of hematological disease.81,82 The 322delG mutation is found in patients of Northern European ancestry, particularly from Holland. Patients homozygous for this mutation have a comparatively mild FA, with fewer congenital abnormalities and later onset of hematological disease.82 Additional mutant alleles of FAC have been identified with mutations in exon 1,82 exon 6,83 and exon 14.77 84-86 A total of seven pathogenic mutations have now been identified, of which three are located in exon 14. Most of these mutations result in either truncation or internal deletion of the FAC protein. Only one pathogenic missense mutation (L554P) has been described. An additional change (D195V) is a polymorphism82; the FAC polypeptide containing this amino acid substitution has normal activity when expressed in an FA(C) indicator cell line (A. D'Andrea, unpublished observation, January 1997).
Analysis of the FAC mRNA and protein have provided some insight into the cellular function of the FAC gene. The FAC mRNA is expressed in multiple cell types and organ systems, consistent with a general function of FAC in organismal development. Increased expression of the FAC mRNA has been observed in the skeletal system, suggesting a more specialized function of FAC in bone development.87 The FAC mRNA exists as three different sizes of 2.3, 3.2, and 4.6 kb.77 The existence of multiple mRNA transcripts appears to result, at least in part, from variable lengths of the 3′ untranslated region. In addition, there is evidence that the FAC mRNA undergoes alternative splicing and exon skipping.83 For instance, a naturally occurring mRNA splice variant, lacking exon 13 sequences, has been detected by reverse transcriptase-polymerase chain reaction (RT-PCR) in normal and FA cell lines. The functional significance of these mRNA splice variants and encoded proteins remains unknown.
The FAC gene contains at least two alternative transcription initiation sites, suggesting the possibility of alternative mechanisms of transcriptional regulation.88 Still, there is little evidence for variable expression of the FAC mRNA under different physiological conditions. The FAC mRNA is not induced by MMC and is not differentially expressed during cell-cycle progression. Recent evidence shows some cell variation of FAC protein expression,89 suggesting possible posttranslational mechanisms of FAC regulation.
The FAC protein is a soluble cytoplasmic protein,90,91 suggesting that it does not play a direct role in DNA repair. The protein remains in the cytoplasm, irrespective of cell-cycle stage or cellular treatment with cross-linking agents. If the FAC protein does interact with DNA directly, it may do so during a narrow window of time in the cell cycle, when the nuclear membrane is degraded. Recent evidence suggests that FAC binds to a complex of cytoplasmic proteins of either 60, 50, or 34 kD.92 The level of FAC protein varies slightly during cell-cycle progression, reaching peak expression during the G2/M transition of the cell cycle. Other evidence shows that the FAC protein binds to the mitotic cyclin-dependent kinase cdc2, suggesting that it has a cellular function that is regulated during the G2/M transition.89 The interaction of FAC with cdc2 suggests a molecular mechanism for the prolonged G2 phase of FA cells.49,51 For instance, the FAC protein may be required for normal cdc2 activity during G2/M progression. Alternatively, the FAC protein may be part of a DNA repair pathway downstream of the cdc2 kinase. In addition, the FAC protein appears to bind to GRP94.93 GRP94 is a 94-kD member of the molecular chaperone family and is expressed in the endoplasmic reticulum where it plays a role in protein transport. The precise cellular function of the FAC protein or the importance of these molecular interactions remains unclear.
STRUCTURE/FUNCTION ANALYSIS OF THE FAC PROTEIN
The mutant FAC proteins expressed by FA(C) patients have allowed the identification of critical functional domains of FAC. All FAC cell lines analyzed to date express at least some isoform of the FAC protein.81 No patient-derived cell lines with true null mutations in the FAC gene have been identified, suggesting that at least partial FAC protein function may be required for viability. Patients homozygous for the IVS4+4 A > T mutation [severe FA(C)] express a truncated 55-kD isoform of the FAC protein. This protein is an “in-frame” deletion of the FAC protein, missing 37 amino acids encoded by exon 4. In contrast, patients homozygous for the 322delG mutation (mild FAC) express a truncated 50-kD isoform of the FAC protein. This protein results from translation reinitiation at an internal methionine residue, downstream of the exon 1 mutation. This 50-kD form of FAC is also expressed in normal cells, suggesting that the internal reinitiation product may have a normal cellular function.81,91 The 55-kD and 50-kD isoforms of FAC, although expressed at normal levels in FA cells, fail to confer MMC resistance. These results suggest that these N-terminal–deleted domains of the FAC protein are required for normal FAC function in vivo. In addition, the HSC536 cells (described above) express the abnormal FAC(L554P) protein. This mutant protein does not confer normal MMC resistance85,91 and may have dominant inhibitory activity when overexpressed with the normal FAC polypeptide.94 This mutation, and several other patient-derived mutations in exon 14 (Fig 1), suggest that the C-terminus of the FAC protein is critical for its in vivo function. Further deletion analysis of the C-terminus of the FAC protein confirms the functional importance of the C-terminus. How these critical functional domains of the FAC protein relate to its intracellular function remains unknown. For instance, these regions of the FAC protein may be required for its ability to bind to other critical regulatory proteins in the cell. Recent evidence shows that the C-terminus of FAC binds to cdc289 while a central region binds to GRP94.93
POSSIBLE CELLULAR FUNCTIONS OF THE FAC POLYPEPTIDE
The cellular function of the FAC polypeptide has been analyzed by comparing isogenic cell lines, differing only by the expression of the normal FAC protein.60,77 95 Expression of the wild-type FAC protein in FA(C) cells corrects several phenotypic abnormalities of the parental cell line, including its MMC sensitivity, its G2 accumulation, and its chromosomal instability. Although overexpression of FAC corrects the MMC sensitivity of FA(C) cells to a normal (wild-type) level, it does not render cells superresistant to MMC. Accordingly, it is unlikely that the FAC protein performs an enzymatic role in the cell, such as efflux of MMC or scavenging of MMC or oxygen radical byproducts.
Interestingly, MMC-sensitive cells acquire resistance to MMC by multiple mechanisms, any one of which may be related to FAC. First, overexpression of the P-glycoprotein, p120, confers cellular resistance to MMC.96 Second, mitomycin C requires bioreduction to exert its cytoxic effect.97 Several cellular enzymes, including NAD(P)H:quinone oxidoreductase, are required for the activation of MMC to its ultimate DNA damaging state. Cellular inactivation of NAD(P)H:quinone oxidoreductase results in decreased MMC-generated DNA damage and increased cellular resistance to MMC.98 Third, several investigators have found that overexpression of ribosomal protein S3 can partially rescue the phenotype of FA cells99 (M. Kelley, personal communication, November 1996). Whether these mechanisms of cellular resistance to MMC bear any relevance to the FA gene pathway remains unknown.
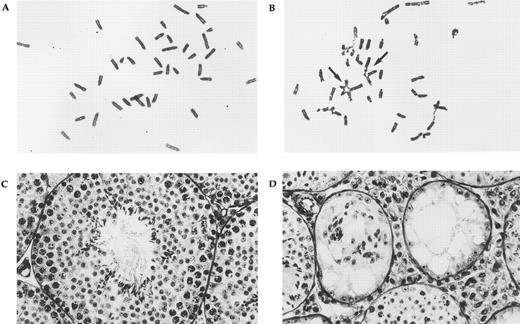
Phenotypes of FAC knockout mice. Panels (A) (wild-type control) and (B) (FAC knockout mouse) show the results of chromosome breakage analysis. Cells were exposed to 20 ng/mL MMC for 48 hours. Multiple radial formation (arrows), gaps, and breaks can be seen in the mutant, but not control cells. Testicular histology of age-matched wild-type control (C) and FAC knockout mouse (D) at the same magnification. The normal control shows the cross-section of a seminiferous tubule containing all stages of spermatogenesis. In the mutant animal, two small abnormal seminiferous tubules can be seen. Spermatogenesis is absent and only Sertoli cells remain. Hypertrophy of interstitial tissue is also evident.
Phenotypes of FAC knockout mice. Panels (A) (wild-type control) and (B) (FAC knockout mouse) show the results of chromosome breakage analysis. Cells were exposed to 20 ng/mL MMC for 48 hours. Multiple radial formation (arrows), gaps, and breaks can be seen in the mutant, but not control cells. Testicular histology of age-matched wild-type control (C) and FAC knockout mouse (D) at the same magnification. The normal control shows the cross-section of a seminiferous tubule containing all stages of spermatogenesis. In the mutant animal, two small abnormal seminiferous tubules can be seen. Spermatogenesis is absent and only Sertoli cells remain. Hypertrophy of interstitial tissue is also evident.
Conflicting studies have analyzed the possible interaction of FAC with the p53 pathway. p53 is a major cellular determinant of genomic stability.100 Some studies61 suggest that FA cells, like AT cells,42 are defective in the induction of p53. Other studies suggest that FA cells are functional for p53 induction.60,95 In more recent studies, overexpression of normal FAC protein in FA(C) cells corrected a cellular defect in apoptosis.64,65 These results implicate the FAC polypeptide in a general function in the prevention of cellular apoptosis. Consistent with these studies, anti-sense oligonucleotides to FAC inhibit hematopoietic cell growth, suggesting that FAC plays a role in maintaining blood cell viability.66 The molecular mechanism of enhanced cellular survival by FAC expression remains unknown.
MOUSE MODELS
Two murine models of FA(C) were developed using targeted recombination in embryonic stem cells.63,101 In one knockout mouse, exon 8 of the FAC gene was deleted101 and replaced by a neomycin resistance gene; in the other mouse model, exon 9 was deleted.63 These mutations are likely to represent null alleles. The phenotype of homozygous FAC mutant animals was similar in the two strains. FAC mutants are viable and show no obvious birth defects of the skeletal system or urinary system. Cells derived from these animals show the classic hypersensitivity to bifunctional cross-linking agents. Similar chromosomal abnormalities and G2 cell-cycle abnormalities were also observed (Fig 2A). Nonetheless, pancytopenia did not develop during the first year of life. Similarly, no leukemia or increased cancer susceptibility was observed during this time span. Although no peripheral blood abnormalities were detectable, mice with deleted exon 9 had an age-dependent decrease in burst-forming units-erythroid (BFU-E) and colony-forming unit GM (CFU-GM) progenitor assays. In addition, hematopoietic progenitor cells showed a distinct hypersensitivity to interferon-γ (IFN-γ). Other mitotic inhibitors had no differential effect. Increased cell susceptibility to IFN-γ is mediated by fas-induced apoptosis.63 102 Cells derived from the FAC knockout mouse showed increased levels of fas expression at lower levels of IFN-γ. It is possible that IFN-γ hypersensitivity is the major pathogenetic mechanism in the development of progressive anemia in FA patients. The relationship between this phenotype and the cellular response to DNA crosslinks is currently unclear.
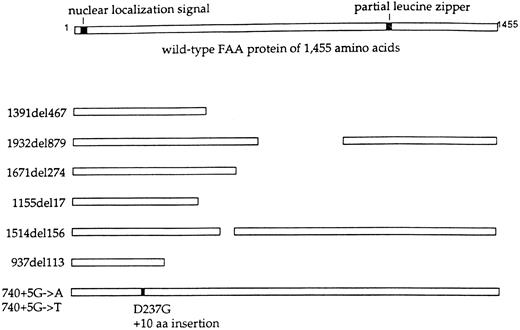
Mutations of the FAA polypeptide. The wild-type FAA protein (1455 amino acids), predicted from the cloned FAA cDNA,108 109 is shown schematically at the top. The protein contains a putative bipartite nuclear localization signal at the N-terminus and an incomplete leucine zipper. The indicated FAA alleles are shown at the left. The mutant FAA polypeptides, predicted by the mutant alleles, are shown schematically. A mutant protein with an in-frame deletion is predicted by the 1514(del)156 allele. Detection of these predicted mutant proteins in FA(A) cells awaits the generation of an FAA specific antiserum.
Mutations of the FAA polypeptide. The wild-type FAA protein (1455 amino acids), predicted from the cloned FAA cDNA,108 109 is shown schematically at the top. The protein contains a putative bipartite nuclear localization signal at the N-terminus and an incomplete leucine zipper. The indicated FAA alleles are shown at the left. The mutant FAA polypeptides, predicted by the mutant alleles, are shown schematically. A mutant protein with an in-frame deletion is predicted by the 1514(del)156 allele. Detection of these predicted mutant proteins in FA(A) cells awaits the generation of an FAA specific antiserum.
The major phenotypic abnormality in both mutant mouse strains was a reduction in the number of male and female germ cells (Fig 2B). No stage-specific defect of spermatogenesis was observed, making a meiotic defect very unlikely. This is in contrast to the aberrations in meiosis that have been observed in AT and mismatch repair mice.103-105 The reduction in germ cells in FAC knockout mice was present at birth, indicating a developmental defect. With respect to this germ cell phenotype, FAC mice are similar to w and steel mice, which have defects in the c-kit receptor106 and its ligand, stem cell factor,107 respectively.
The unexpected germ-cell and IFN-γ hypersensitivity phenotypes observed in FAC knockout mice have added another level of complexity to the understanding of FA. Models for the function of the FA pathway must ultimately explain these observations, along with several other cellular abnormalities (Table 2). The availability of the two murine knockouts will aid in the development and refinement of these models and will provide an experimental system for studying FA therapies.
BIOCHEMISTRY AND MOLECULAR BIOLOGY OF FAA
The cloning of FAA was reported only very recently108,109 and therefore much less is known about the FAA protein. The major FAA mRNA transcript is 5.5 kb long and found in low abundance in all adult tissues tested. In addition to this major mRNA species, multiple transcripts, both larger and smaller, can be seen on Northern blot, suggesting a complex pattern of mRNA splicing of the gene. The 5.5-kb mRNA contains an open reading frame of 4,368 nucleotides (nt), predicting a 1,455–amino acid protein with a relative molecular mass of 163 kD. Functional complementation has been achieved with this protein, but the 5′ end of the mRNA has not yet been definitively mapped. Because no STOP codons have been found in the cDNA, 5′ to the putative translation START site, an additional 5′ protein coding sequence may exist. Similar to FAC, the predicted FAA protein has no significant homologies to other proteins in sequence databases. Also, no common motifs are shared between the two FA proteins. The predicted FAA polypeptide has two overlapping bipartite nuclear localization signals, located at amino acids 18-34 and 19-35. In addition, a partial leucine zipper consensus sequence is found at position 1069-1090. These homologies are consistent with a function of FAA in the nucleus, perhaps as a DNA binding protein. The chromosomal location of FAA at 16q23.4, between markers D16S3121 and D16S303,75 was confirmed by direct methods. The gene spans approximately 80 kb and consists of at least 43 exons.109a A locus involved in loss of heterozygosity in breast cancer has been mapped to the same region110 111 but is unlikely to be identical to the FAA gene.
Several FAA gene mutations have already been described, but no predominant alleles are apparent. However, patients of South African descent were not examined in the two cloning reports.108,109 Because of the known linkage dysequilibrium in this population75 which has the highest known incidence of FA,112 it appears likely that an Afrikaner founder mutation will be identified. In addition, there exists a high prevalence of complementation group A among patients from Italy,113 suggesting a possible predominant allele in these patients. Interestingly, six of the first eight reported FAA mutations caused deletions of the FAA cDNA. The cDNA deletions may result from splice site mutations or genomic deletions. Two additional alleles were splice mutations, while no missense alleles were reported. The predicted effects of these changes on the FAA protein are depicted in Fig 3. Four of the eight mutations are predicted to result in severely truncated proteins. One interesting allele (1514del156) predicts an in-frame deletion of 52 amino acids, indicating the potential functional importance of these residues. The high frequency of independent deletions is unusual for an autosomal recessive disorder and suggests the possibility of repeat sequences which predispose to genomic rearrangements. If additional analysis confirms the high incidence of mutations leading to amino acid deletions, the protein truncation test114 may prove to be useful in the study of FAA patients.
PROSPECTS FOR THE CLONING OF OTHER FA GENES
At least three and possibly more FA genes remain to be cloned.69,72 The two FA genes which have been isolated to date were originally identified by functional complementation of MMC hypersensitivity,77,109 underscoring the power of this experimental approach. However, positional cloning techniques have also been successful.108 Functional complementation by cDNA libraries is a technically difficult endeavor, which tends to yield many false-positive cDNA clones. Prior knowledge of the chromosomal location of an FA gene can greatly aid in rapidly narrowing down the number of candidate cDNAs. Because FA is a genetically heterogeneous disorder, mapping by linkage analysis requires that a large number of families first be categorized into a complementation group by somatic cell hybrid experiments. This was accomplished for the FAA locus, because the vast majority of FA patients belong to this group.75 Other complementation groups are more rare, making linkage analysis difficult for other FA genes. This limitation can be overcome by the use of microcell-mediated chromosome transfer to map FA genes. The feasibility of this approach has been shown for FAD, a rare complementation group for which only three families have been identified.76
As more FA genes are cloned, it becomes likely that novel members of the pathway will be identified as proteins that interact with already known FA proteins. Intense efforts to isolate additional FA genes are ongoing in several laboratories and are likely to be successful in the near future.
MODELS OF THE FA GENE PATHWAY
Based on the striking clinical similarities among FA patients from all complementation groups, the proteins encoded by the FA genes probably interact within the cell. The FA proteins may interact by a direct physical association or by a functional interaction. Several models are possible. First, the FA proteins may physically associate in a protein complex. Such a complex may serve as a defense mechanism, protecting the cell from DNA or protein cross-link damage. Second, the proteins may function sequentially in an enzymatic cascade, resulting in the repair or removal of cross-linked cellular macromolecules. Third, the FA proteins may interact in a signal transduction cascade. Such a pathway may include several components, including a “sensor” of crosslinking damage in the cytoplasm, a “transducer” for transmitting signals to the nucleus, and an “inducer” of new gene transcription, new DNA repair processes, or cell cycle arrest. Consistent with this latter model, the FAC protein may perform the sensor function in the cytoplasm and the FAA protein may translocate to the nucleus upon cellular exposure to cross-linking agents. Analysis of the FAC and FAA proteins with specific antisera, following cellular exposure to cross-linking agents, may help to distinguish among these possible models. Recent evidence suggests that FAA and FAC proteins bind and form a protein complex (G. Kupfer and A. D'Andrea, unpublished observation, January 1997). Interestingly, mutant FAC polypeptide derived from FA(C) patients fails to bind FAA.
CLONING OF FA GENES: IMPLICATIONS FOR DIAGNOSIS
The cloning of the FA genes has already had a considerable impact on the diagnosis and treatment of patients with FA. Although the DEB test is highly sensitive and specific for the diagnosis of FA,18 it fails to distinguish among FA patients in different complementation groups and fails to identify heterozygote carriers of mutant FA genes. The availability of the FAA and FAC genes will allow the rapid and accurate diagnosis of most FA patients and will form the basis of a prenatal diagnostic test for known families with FA. The identification of “common” mutant alleles of the FAA and FAC gene will also aid in rapid diagnosis. As discussed above, two specific mutant alleles of the FAC gene (IVS4+4 A > T and 322delG) account for 75% of all FAC mutations.80,82 115 Exon 14 mutations account for most of the remaining FA(C) patients. Specific FAA mutations may also be associated with specific ancestral backgrounds, although the relative prevalence of these FAA mutations among FA(A) patients remains to be tested.
The phenotypic variation of FA patients makes the diagnosis of specific FA genotypes critical. For instance, in one complementation group, FA(C), patients with the IVS4+4 A > T mutation have more severe disease, with severe congenital abnormalities and early onset of hematopoietic disease, while patients with the 322delG mutation have a relatively mild disease, with few congenital abnormalities and later onset of hematopoietic disease.82 Similar genotype/phenotype analysis is now possible for FAA patients. Such correlations are important because they guide FA patient management. For instance, if a more severe phenotype correlates with a particular FAA genotype, these FAA patients may be treated more aggressively, with the early use of BM transplantation or gene therapy protocols.
Molecular diagnosis is not entirely dependent on the knowledge of both mutant alleles. Once a family has been assigned to a complementation group by a functional assay or by the detection of a specific mutant allele, chromosome linkage analysis will be convenient and useful. Highly polymorphic microsatellite markers are located in close proximity to both FAA and FAC genes and can be used for the detection of carriers and for prenatal diagnosis.
CLONING OF FA GENES: IMPLICATIONS FOR THERAPY
The availability of the FA genes will profoundly influence the treatment of FA. Regardless of complementation group, most FA patients die from complications of their BM failure.1 Since most patients do not have a histocompatible donor, an alternative is retroviral transduction of an FA gene. For several reasons, retroviral transfer of an FA gene into the hematopoietic stem cells of an FA patient is a plausible option. For example, the FAA gene can effectively correct the cross-linking agent sensitivity and chromosomal instability of FA-A cells.109 Also, retroviral transduction of the FAC cDNA into FA(C) primary cells improves their survival and longevity, compared with uninfected cells.116 The survival advantage of corrected FA cells in vitro may translate into a survival advantage of the cells for in vivo studies. Gene transfer studies with the FA genes will be limited by the small number of hematopoietic stem cells that can be isolated from FA patients. For this reason, many centers have initiated programs to collect and cryospreserve hematopoietic stem cells from FA patients before the onset of anemia. Although useful, gene transfer studies, like BM transplant, will not improve the developmental abnormalities or cancer risk in nonhematopoietic tissues of FA patients.
Finally, the cloning of the FA genes will provide molecular insight into their role in other clinical features of FA, such as susceptibility to cancer. Some genotypic variants of FAA may correlate with a higher incidence of cancer. There exists no clear evidence for an increase in cancer incidence among heterozygote carriers of FA.117-120 Still, somatic mutation or loss of FA genes could theoretically contribute to oncogenesis in some heterozygote carriers or in some individuals who do not harbor germline mutations in FA genes. Accordingly, a systematic survey of the FA genes in primary tumors and tumor cell lines may reveal FA mutations.
The increased incidence of AML in FA suggests that FA genes may play a specific role in AML in the general (non-FA) patient population. Several reports of interstitial deletions of the long arm of chromosome 9 (9q-) associated with AML have been reported.121 122 In primary leukemic cells from some AML patients, these interstitial deletions overlap the FAC locus (G. Kupfer and A. D'Andrea, unpublished observation, December 1996). Taken together, these results suggest that the somatic loss of one or both FA genes may predispose to leukemic transformation in non-FA patients or heterozygous carriers of FA.
SOMATIC MOSAICISM AND IN VIVO SELECTION IN FANCONI ANEMIA
The phenomenon of somatic reversion of disease has recently been described in hereditary disorders of blood and liver.123 If a genetic disease causes cells to grow poorly, then spontaneously corrected cells, resulting from new mutations that revert mutant genes, may have a substantial growth advantage. Patients who improved clinically after somatic reversion and selection have previously been reported for both adenosine deaminase deficiency124 and Bloom's syndrome.45 46 Recently the same phenomenon has been reported in FA. Approximately 15% of FA patients have somatic mosaicism in their peripheral blood (Hans Joenje, personal communication, May 1997). In these patients, a mixture of MMC-sensitive and -resistant cells are found. In one patient, mitotic recombination was documented as the molecular mechanism of somatic reversion. Somatic mosaicism may make the diagnosis of FA difficult because of inconclusive chromosome breakage studies. If FA is strongly suspected in a patient despite normal breakage studies in peripheral blood, a definitive test can be performed on primary skin fibroblasts.
The high percentage of patients with somatic mosaicism indicates a strong selective advantage for cells that have lost the FA phenotype. This raises the possibility that gene therapy in FA may be aided by in vivo selection. At this time it is unclear whether any special physiologic circumstances are required for this selection to occur. Somatic reversion also suggests that some mutant FA genes have frame-shift mutations. These mutations may be “corrected” by new mutations that correct the open reading frame of the FA gene.
Somatic mosaicism may also cause some complications for FA patients. A high incidence of graft rejection has been noted in the BM transplantation of FA patients. Because of their increased sensitivity to bifunctional alkylating agents, FA patients typically receive a much less aggressive ablative treatment before BMT. If the recipient FA patient has somatic mosacism (particularly in T cells), some endogenous cells may be resistant to the ablative regimen and cause graft rejection. Further studies will be required to confirm this hypothesis.
FUTURE DIRECTIONS
The cloning of two FA genes (FAC and FAA) has provided an unprecedented opportunity to understand the molecular basis of FA. With these genes, new approaches can be taken to explore the cellular role of FAC and FAA. First, the mammalian genes may be used to identify FAC or FAA homologues in yeast or Drosophila. A genetic dissection of the FA gene pathway in one of these organisms may implicate the pathway in a basic cellular function such as cell growth, cell cycle, or genomic stabilization. Second, the FA genes may be used for the generation of additional mouse models of FA. As described above, cells derived from mice deficient in the FAC gene product exhibit genomic instability, similar to that observed in cell lines derived from FA patients. Similarly, knockouts of the FAA gene are now possible. Genetic interactions of FAC and FAA could be further analyzed by crossing different knockout strains. Third, antibodies may be generated against the FAC and FAA proteins. These antibodies can be used to identify a (possible) interaction between FAC and FAA and can be used to identify other binding partners of these proteins.
Also, as described, the availability of the FAA and FAC genes will directly impact the diagnosis and therapy of FA. Mutations in FA genes can be directly screened as an adjunct diagnostic procedure to the DEB test. Retroviruses that transduce the FA genes can be used for rapid diagnosis and complementation analysis of primary cells from suspected FA patients. Gene therapy studies with autologous peripheral blood CD34+ stem cells can be initiated for patients who do not have sibling-matched histocompatible donors.116 The observation of somatic reversion in FA provides some reason for optimism regarding the efficacy of gene therapy. Finally, once the molecular basis of the disease is established, more rational therapies or preventative measures may be devised for patients and families with FA.
ACKNOWLEDGMENT
The authors thank H. Joenje, A. Savoia, and M. Kelley for providing unpublished data.
Address reprint requests to Alan D. D'Andrea, MD, Dana-Farber Cancer Institute, Pediatric Oncology, 44 Binney St, Boston, MA 02115.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal