Abstract
The aims of this study were twofold: (1) to assess the marrow of patients with T-lineage acute lymphoblastic leukemia (T-ALL) for the presence of molecular residual disease (MRD) at different times after diagnosis and determine its value as a prognostic indicator; and (2) to compare the sensitivity, rapidity, and reliability of two methods for routine clinical detection of rearranged T-cell receptor (TCR). Marrow aspirates from 23 patients with T-ALL diagnosed consecutively from 1982 to 1994 at the Division of Pediatric Hematology and Oncology, University of Catania, Italy, were obtained at diagnosis, at the end of induction therapy (6 to 7 weeks after diagnosis), at consolidation and/or reinforced reinduction (12 to 15 weeks after diagnosis), at the beginning of maintenance therapy (34 to 40 weeks after diagnosis), and at the end of therapy (96 to 104 weeks after diagnosis). DNA from the patients' marrow was screened using the polymerase chain reaction (PCR) for the four most common TCR δ rearrangements in T-ALL (Vδ1Jδ1, Vδ2Jδ1, Vδ3Jδ1, and Dδ2Jδ1) and, when negative, further tested for the presence of other possible TCR δ and TCR γ rearrangements. After identification of junctional rearrangements involving V, D, and J segments by DNA sequencing, clone-specific oligonucleotide probes 5′ end-labeled either with fluorescein or with [γ-32P]ATP were used for heminested PCR or dot hybridization of PCR products of marrows from patients in clinical remission. For 17 patients with samples that were informative at the molecular level, the estimated relapse-free survival (RFS) at 5 years was 48.6% (±12%). The sensitivity and specificity for detection of MRD relating to the outcome were 100% and 88.9% for the heminested fluorescence PCR and 71.4% and 88.9% for Southern/dot blot hybridization, respectively. Predictive negative and positive values were 100% and 90.7% for heminested fluorescence PCR, respectively. The probability of RFS based on evidence of MRD as detected by heminested fluorescence PCR at the time of initiation of maintenance therapy was 100% and 0% for MRD-negative and MRD-positive patients, respectively. Thus, the presence of MRD at the beginning of maintenance therapy is a strong predictor of poor outcome, and the molecular detection of MRD at that time might represent the basis for a therapeutic decision about such patients. By contrast, the absence of MRD at any time after initiation of treatment strongly correlates with a favorable outcome. The heminested fluorescence PCR appears to be more accurate and more rapid than other previously used methods for the detection of residual leukemia.
POLYMERASE CHAIN REACTION (PCR) analysis of the clone-specific junctional regions of rearranged genes for both IgH and the T-cell receptor (TCR) has been widely exploited as a sensitive test for detection of minimal residual disease (MRD) in acute lymphoblastic leukemia (ALL).1-5
These techniques have proven valuable because they allow the monitoring of almost all cases of ALL, despite several shortcomings such as cumbersome methodology and instability of the rearrangements. Several small clinical studies have indicated that detection of residual leukemia in ALL correlates with impending relapse.6-13 This finding contrasts with observations in chronic myelogenous leukemia, where such correlation is poor.14 The most promising results so far with respect to identifying early prognostic indicators of disease outcome have been obtained by monitoring cytoreduction during therapy. Indeed, studies in B-lineage ALL indicate that rapid cytoreduction below the threshold of molecular detection during the induction phase of treatment is correlated with long-term survival.12,13 However, neither study enabled prediction of exactly which patients would relapse after the end of maintenance therapy, most likely reflecting the heterogeneity of the ALL population.15 In a fraction of patients in those studies, the positivity or negativity of the results does not correlate with disease outcome. That observation, together with the complexity of the methodology used, does not justify the use of this approach for routine patient monitoring.
T-cell ALL (T-ALL) is a clinically homogeneous disease with a high frequency of treatment failure, and the T-cell immunophenotype is associated with high-risk features in about 75% of cases and has been considered an adverse prognostic factor.16 However, present protocols for intensive treatment yield results similar to those in patients with non–T-ALL.17-19 When patients relapse, chemotherapy is ineffective, and allogeneic bone marrow transplantation probably represents the only chance of cure. A more timely alternative treatment might be initiated if it were possible to identify the population at risk of relapse. In this context, TCR δ and TCR γ rearrangements represent an optimal target for monitoring MRD by PCR in patients with T-ALL, since at least one allele is rearranged in 95% of the cases and remains stable in more than 90% of the cases.20 Moreover, TCR δ rearrangement, which occurs in 70% of T-ALL patients, exhibits limited potential combinatorial diversity, allowing the use of simple procedures to screen positive patients, and the extensive junctional diversity permits preparation of patient-specific probes.20 21
We monitored MRD status in children with T-ALL throughout the course of treatment to the end of therapy in an effort to identify a time point after diagnosis at which MRD represents a more accurate prognostic indicator of disease outcome. We compared the sensitivity and applicability of two different PCR-based methods for the routine molecular detection of MRD.
SUBJECTS AND METHODS
A total of 23 patients with T-ALL diagnosed consecutively from 1982 to 1994 at the Division of Pediatric Hematology and Oncology of the University of Catania (Italy) were studied.
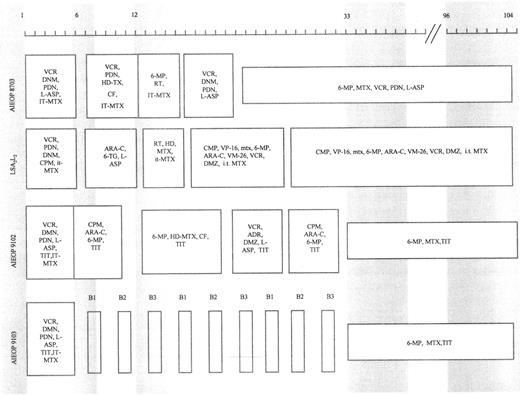
Patients were treated according to different protocols being tested at the time of their diagnosis. Figure 1 shows a schematic representation of the treatment protocols together with the time of marrow sampling. Marrow aspirates were obtained at diagnosis, at the end of induction therapy (6 to 7 weeks after diagnosis), at consolidation and/or reinforced reinduction (12 to 15 weeks after diagnosis), at the beginning of maintenance therapy (33 to 40 weeks after diagnosis), and at cessation of therapy (96 to 104 weeks after diagnosis) from 16 patients who were studied retrospectively and from four patients diagnosed from January 1994 who participated in a prospective study. The remaining three patients did not have adequate stored samples suitable for study.
Schematic representation of protocols LSA2L2 , AIEOP 8703, 9102, and 9103. Shaded areas indicate the time (in weeks) of marrow sampling. ADR, adriamycin; ARA-C, cytarabine; PDN, prednisone; CF, citrovorum factor; 6-MP, 6-mercaptopurine; CPM, cyclophosphamide; MTX, methotrexate; DNM, daunorubicin; RT, radiotherapy; DMZ, dexamethasone; 6-TG, 6-thioguanine; HD, high-dose; IT, intrathecal; TIT, triple IT therapy (PDN, ARA-C, MTX); HD-L-ASP, high-dose L-asparaginase; VCR, vincristine; VM-26, teniposide; VP-16, etoposide; L-ASP, L-asparaginase. B1, block 1 (6-MP, DMZ, VCR, HD-MTX, HD-ARA-C, HD-L-ASP, and TIT); B2, block 2 (6-TG, DMZ, VDS, HD-MTX, DNM, CPM, HD-L-ASP, and TIT); B3, block 3 (DMZ, VP-16, HD-ARA-C, HD-L-ASP, and TIT).
Schematic representation of protocols LSA2L2 , AIEOP 8703, 9102, and 9103. Shaded areas indicate the time (in weeks) of marrow sampling. ADR, adriamycin; ARA-C, cytarabine; PDN, prednisone; CF, citrovorum factor; 6-MP, 6-mercaptopurine; CPM, cyclophosphamide; MTX, methotrexate; DNM, daunorubicin; RT, radiotherapy; DMZ, dexamethasone; 6-TG, 6-thioguanine; HD, high-dose; IT, intrathecal; TIT, triple IT therapy (PDN, ARA-C, MTX); HD-L-ASP, high-dose L-asparaginase; VCR, vincristine; VM-26, teniposide; VP-16, etoposide; L-ASP, L-asparaginase. B1, block 1 (6-MP, DMZ, VCR, HD-MTX, HD-ARA-C, HD-L-ASP, and TIT); B2, block 2 (6-TG, DMZ, VDS, HD-MTX, DNM, CPM, HD-L-ASP, and TIT); B3, block 3 (DMZ, VP-16, HD-ARA-C, HD-L-ASP, and TIT).
Because frozen cells were not available, in most cases DNA was extracted from bone marrow smears of stained or unstained slides.22
Continued complete remission (CCR) was defined as less than 5% of lymphoblasts detectable by morphologic observation of bone marrow aspirates.
PCR amplification and sequencing of rearranged TCR δ and γ chains in diagnostic marrow.Diagnostic marrow DNA was screened by PCR for the presence of the four most common combinations of TCR δ rearrangements in T-ALL (Vδ1Jδ1, Vδ2Jδ1, Vδ3Jδ1, and Dδ2Jδ1).7,21 When these rearrangements were undetectable, marrow DNA was tested for the presence of other possible TCR δ rearrangements and subsequently for TCR γ rearrangements.21-23 PCR was performed essentially by the method of Saiki et al.24 Each reaction mixture (100 μL) contained 1 μg genomic DNA, 6 pmol of each 5′ and 3′ oligonucleotide primer, 0.2 mmol/L each dNTP, 10 μL 10× reaction buffer (Perkin Elmer-Cetus, Norwalk, CT), and 2.5 U Taq DNA polymerase (Perkin Elmer-Cetus). Synthetic oligonucleotide primers for PCR were produced by Integrated DNA Technologies (Coraville, IA), based on published sequences.7,21 2536 Reaction mixtures containing TCR δ primers were denatured at 94°C, 55°C, and 72°C for 3 minutes each and amplified for a total of 35 cycles at 94°C, 55°C, and 72°C for 1, 2, and 3 minutes, respectively, in a thermocycler (model 480; Perkin Elmer-Cetus). Reaction mixtures containing TCR γ primers were heated at 94°C for 2 minutes and subjected to 35 cycles at 94°C, 60°C, and 72°C for 1, 2, and 3 minutes each, respectively. Clonally amplified bands were excised with a sterile scalpel, and the DNA was eluted with a Microcon 30 filter. Two-microliter samples were analyzed on a 4% Nusieve gel (FMC BioProducts, Rockland, ME) to quantify DNA in the bands, and sequence analysis was performed using the dye-terminator ready-reaction cycle sequencing kit (no. 401384; ABI, Foster City, CA). For PCR-amplified fragments of 150 to 250 bp, 10 ng amplified DNA, and 1 to 2 pmol primer in a 20-mL reaction volume were used. Two sequencing reactions, one for the forward and one for the reverse primer, were performed for each sample. The thermocycling profile for DNA sequencing was 95°C for 30 seconds, 50°C for 15 seconds, and 60°C for 4 minutes for a total of 25 cycles. Products were assayed on a 6% acrylamide gel in an ABI sequencer model no. 373 and analyzed using Navigator software from ABI.
DNA sequencing data were automatically analyzed for rearranged V, D, and J segments using software provided by The Wistar Institute Cancer Center Bioinformatics Facility (Philadelphia, PA). Probes specific for each rearranged TCR, spanning the joined segments, were designed and checked for self-annealing, primer dimer formation, and melting point.
Detection of MRD.Clone-specific oligonucleotide probes were synthesized and 5′ end-labeled with fluorescein for heminested fluorescence PCR. The same probes were also 5′ end-labeled with [γ-32P]ATP for standard hybridization of PCR products. The sensitivity of heminested fluorescence PCR was examined using the ALL2 leukemia cell line,26 which has two alleles with a rearranged IgH locus and one allele with a rearranged δ locus (Vδ2Dδ3). Briefly, a two-step PCR amplification, in which the first amplification was made using standard primers and the second amplification using a heminested PCR with one fluorescence primer spanning the junctional segments and a cold primer in the D segment, was able to detect a single ALL2 leukemic cell among 100,000 normal lymphocytes.
For MRD screening of clinical remission samples, 1 μg marrow DNA was amplified using a cold primer/probe at the junction of VD or DJ segments and a fluorescent primer in the J segment. When results were negative, amplification was repeated in two steps, using cold V and J primers first for PCR followed by reamplification of 1 μL of the first PCR product using a cold primer/probe and a fluorescent J primer. PCR products (10 μL) were assayed on a 10% polyacrylamide gel on a vertical Mighty Small II electrophoresis system (Hoefer, San Francisco, CA) at 200 V for 80 minutes. The gel was scanned at 700 V in a Fluorimager SI and analyzed using ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Dot blot hybridization with 32P-labeled clone-specific probes.Amplified product (10 μL) was spotted on a nylon membrane (Zetabind; Cuno Laboratories Products, Brorad, UK), UV cross-linked, and prehybridized for 3 hours at 42°C in a stock solution of 6× SSC, 1× Denhardt buffer, 0.2% sodium dodecyl sulfate (SDS), and 0.05% sodium pyrophosphate. Hybridization with clone-specific oligonucleotides 5′ end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (GIBCO-BRL, Paisley, UK) and separated from unincorporated radioisotope on Push Columns (Stratagene, La Jolla, CA) was conducted overnight at 42°C in a solution containing 6× SSC, 1× Denhardt buffer, 0.05% sodium pyrophosphate, 2 mg tRNA, and 400 ng radiolabeled probe.26 Filters were washed twice in 2× SSC plus 1% SDS at room temperature for 20 minutes. A final high-stringency wash was performed in 6× SSC plus 1% SDS at an optimal temperature (3°C below the calculated melting point) predetermined for each individual probe. Autoradiography was performed on XAR-5 film (Kodak, Rochester, NY) with intensifying screens for 12 hours to 5 days at −70°C.
Statistical analysis.Fisher's exact test and the Cox proportional hazards modeling technique27 were used to explore prognostic factors associated with relapse-free survival (RFS). The following variables were included in the analysis: white blood cell (WBC) count (<50,000 v ≥ 50,000/μL), age (>1 and ≤ 10 years v <1 and ≥ 10 years), gender, and MRD status at 13 to 15 weeks and versus 35 to 40 weeks as determined by heminested fluorescence PCR. Five-year estimated survival rates were calculated using the method of Kaplan and Meier.28
RESULTS
Table 1 lists the sex, age, peripheral blood cell count, immunophenotype, and protocol treatment of 23 patients included in the study. The estimated RFS at 5 years was 48.6% (±12%) for 17 patients with samples that were informative at the molecular level. The follow-up study was updated in June 1996. The median follow-up period for patients in first CCR was 33 months (range, 18 to 125). The relative risk of relapse was not significantly associated with any variable considered at the time of diagnosis. These findings may be due to the fact that, based on uniform age/WBC count criteria, 13 of 17 (76%) children had high-risk features at diagnosis. Furthermore, the intensive treatment protocols that patients received may have nullified the prognostic value of factors at diagnosis.
Clinical Characteristics of 23 Patients With T-Cell ALL at Diagnosis
| Patient No. . | Date of Diagnosis . | Gender . | Age . | WBC Count (per μL) . | Immunophenotype . | Treatment Protocol* . |
|---|---|---|---|---|---|---|
| . | . | . | (yr) . | . | . | . |
| CT1 | 20/9/91 | F | 7 | 36,000 | CD7+ TdT+ CD2− CD5+ CD3− | 9103 |
| CT2 | 18/7/91 | F | 14 | 17,200 | CD7+ TdT− CD2+ CD5+ CD3+ | 9103 |
| CT3 | 13/1/86 | F | 8 | 33,000 | T-rosette+ | LSA2L2 |
| CT4 | 26/1/94 | F | 14 | 1,720 | CD7+ TdT+ CD2− CD5− CD3+ CD4+ CD8+ | 9103 |
| CT5 | 15/9/93 | M | 9 | 34,800 | CD7+ TdT+ CD2+ CD5+ CD3− | 9102 |
| CT6 | 17/2/87 | M | 10 | 1,000 | CD7+ TdT− CD2− CD5+ CD3+ CD4+ CD8+ | 8703 |
| CT7 | 11/11/93 | M | 10 | 3,000 | CD7+ TdT− CD2+ CD5+ CD3+ CD4+ CD8+ | 9102 |
| CT8 | 23/7/87 | M | 5 | 175,200 | CD7+ TdT+ CD2+ CD5+ CD3− | 8703 |
| CT9 | 12/9/94 | M | 12 | 81,200 | CD7+ TdT+ CD2+ CD5+ CD3+ | 9102 |
| CT10 | 18/10/84 | M | 4 | 40,000 | T-rosette+ | LSA2L2 |
| CT11 | 29/4/89 | F | 3 | 158,000 | CD7+ TdT+ CD2+ CD5+ CD3− CD4+ | 8703 |
| CT12 | 8/5/94 | M | 15 | 45,700 | CD7+ TdT+ CD2− CD5+ CD3+ | 9103 |
| CT13 | 21/8/85 | M | 8 | 78,500 | T-rosette+ (CD7+ TdT+ CD2+ CD5+ CD3− at relapse) | LSA2L2 |
| CT14 | 21/5/93 | F | 7 | 209,000 | CD7+ TdT− CD2+ CD5+ CD3+ | 9103 |
| CT15 | 17/10/90 | F | 2 | 90,000 | CD7+ TdT+ CD5+ CD3− | 9103 |
| CT16 | 26/4/93 | M | 9 | 57,600 | CD7+ TdT+ CD2+ CD5+ CD3− | 9102 |
| CT17 | 1/5/89 | M | 10 | 52,800 | CD7+ TdT+ CD2+ CD5+ CD4+ CD8− | 8703 |
| CT18 | 31/10/91 | F | 4 | 173,600 | CD7+ TdT+ CD2+ CD5+ CD3− | 9103 |
| CT19 | 15/9/93 | F | 1 | 137,000 | CD7+ TdT+ CD2+ CD5+ CD3− | 9102 |
| CT20 | 19/3/91 | M | 8 | 178,000 | CD7+ TdT+ CD2+ CD5+ CD3+ | 9103 |
| CT21 | 2/9/93 | M | 5 | 4,500 | CD7+ TdT+ CD2− CD5+ CD3+ CD4+ CD8+ | 9102 |
| CT22 | 18/7/94 | M | 2 | 150,000 | CD7+ TdT+ CD2+ CD5+ CD3+ | 9102 |
| CT23 | 17/9/90 | M | 4 | 34,200 | CD7+ TdT+ CD2+ CD5+ CD3− | 8703 |
| Patient No. . | Date of Diagnosis . | Gender . | Age . | WBC Count (per μL) . | Immunophenotype . | Treatment Protocol* . |
|---|---|---|---|---|---|---|
| . | . | . | (yr) . | . | . | . |
| CT1 | 20/9/91 | F | 7 | 36,000 | CD7+ TdT+ CD2− CD5+ CD3− | 9103 |
| CT2 | 18/7/91 | F | 14 | 17,200 | CD7+ TdT− CD2+ CD5+ CD3+ | 9103 |
| CT3 | 13/1/86 | F | 8 | 33,000 | T-rosette+ | LSA2L2 |
| CT4 | 26/1/94 | F | 14 | 1,720 | CD7+ TdT+ CD2− CD5− CD3+ CD4+ CD8+ | 9103 |
| CT5 | 15/9/93 | M | 9 | 34,800 | CD7+ TdT+ CD2+ CD5+ CD3− | 9102 |
| CT6 | 17/2/87 | M | 10 | 1,000 | CD7+ TdT− CD2− CD5+ CD3+ CD4+ CD8+ | 8703 |
| CT7 | 11/11/93 | M | 10 | 3,000 | CD7+ TdT− CD2+ CD5+ CD3+ CD4+ CD8+ | 9102 |
| CT8 | 23/7/87 | M | 5 | 175,200 | CD7+ TdT+ CD2+ CD5+ CD3− | 8703 |
| CT9 | 12/9/94 | M | 12 | 81,200 | CD7+ TdT+ CD2+ CD5+ CD3+ | 9102 |
| CT10 | 18/10/84 | M | 4 | 40,000 | T-rosette+ | LSA2L2 |
| CT11 | 29/4/89 | F | 3 | 158,000 | CD7+ TdT+ CD2+ CD5+ CD3− CD4+ | 8703 |
| CT12 | 8/5/94 | M | 15 | 45,700 | CD7+ TdT+ CD2− CD5+ CD3+ | 9103 |
| CT13 | 21/8/85 | M | 8 | 78,500 | T-rosette+ (CD7+ TdT+ CD2+ CD5+ CD3− at relapse) | LSA2L2 |
| CT14 | 21/5/93 | F | 7 | 209,000 | CD7+ TdT− CD2+ CD5+ CD3+ | 9103 |
| CT15 | 17/10/90 | F | 2 | 90,000 | CD7+ TdT+ CD5+ CD3− | 9103 |
| CT16 | 26/4/93 | M | 9 | 57,600 | CD7+ TdT+ CD2+ CD5+ CD3− | 9102 |
| CT17 | 1/5/89 | M | 10 | 52,800 | CD7+ TdT+ CD2+ CD5+ CD4+ CD8− | 8703 |
| CT18 | 31/10/91 | F | 4 | 173,600 | CD7+ TdT+ CD2+ CD5+ CD3− | 9103 |
| CT19 | 15/9/93 | F | 1 | 137,000 | CD7+ TdT+ CD2+ CD5+ CD3− | 9102 |
| CT20 | 19/3/91 | M | 8 | 178,000 | CD7+ TdT+ CD2+ CD5+ CD3+ | 9103 |
| CT21 | 2/9/93 | M | 5 | 4,500 | CD7+ TdT+ CD2− CD5+ CD3+ CD4+ CD8+ | 9102 |
| CT22 | 18/7/94 | M | 2 | 150,000 | CD7+ TdT+ CD2+ CD5+ CD3+ | 9102 |
| CT23 | 17/9/90 | M | 4 | 34,200 | CD7+ TdT+ CD2+ CD5+ CD3− | 8703 |
See Fig 1.
Analysis of TCR gene rearrangements.Table 2 summarizes rearrangements of the TCR gene detected in the patient population studied. Fifty-eight samples of bone marrow from 20 patients were analyzed. In all but seven samples (12%), less than 5% of lymphoblasts were detectable by morphologic observation. Fifteen of 20 patients (75%) had at least one allele with a nondeletional TCR δ rearrangement [Vδ1Jδ1 in five cases (33%), Vδ2Jδ1 in two cases (13%), Vδ3Jδ1 in two cases (13%), Dδ2Jδ1 in two cases (13%), Vδ2Jδ3 in two cases (13%), Vδ1Jδ2 in one case (6.6%), and Vδ2Dδ3 in one case (6.6%)]. Thus, initial screening showed that 11 of 20 patients (55%) have one of the four most common TCR δ rearrangements. Among five patients who did not show a TCR δ rearrangement, samples from two cases did contain TCR γ rearrangements (Vγ6-Jγ1.3 and Vγ2-Jγ1.3) and samples from three patients were not informative at the molecular level.
Detection of MRD Using Heminested Fluorescent PCR or Dot Blot Assay
| Patient No. . | TCR Rearrangement . | Patient's Leukemia-Specific Probe† . | End of Induction Therapy, 6-7 Weeks . | 12-15 Weeks . | 35-40 Weeks . | MRD at End of Therapy . | Length of First CCR (mo) . | Status of Disease . |
|---|---|---|---|---|---|---|---|---|
| CT1 | Vδ1Jδ1 | ctATTGCGCTTAGTGg | +/+ | +/− | −/− | −/− | +57 | First CCR |
| CT2 | Vδ1Jδ1 | ggatacgCTCTACTTcaccg | +/+ | +/− | +/− | NP | 12 | Relapse BM |
| CT3 | Vδ3Jδ1 | tagGGCCTGGAGCTAcc | +/+ | +/− | −/− | −/− | +125 | First CCR |
| CT4 | Vδ1Jδ1 | gggCCCCATCGGTacac | +/+* | +/+ | +/+ | NP | 11 | Relapse BM |
| CT5 | Vδ3Jδ1 | gcctCTACCGTAGGtag | −/− | −/− | −/− | −/− | +33 | First CCR |
| CT6 | Vγ2-Jγ1.3 | gggCTAGATCCCTAgat | +/+ | +/+ | +/+ | NP | 20 | Relapse BM |
| CT7 | Vδ3Jδ3 | GGTTATGTTGTTGAAGTTCGT | +/+ | +/+ | +/+ | NP | 20 | Relapse BM |
| CT8 | Vγ6-Jγ1.3 | gataggTCCGTTACGTAg | +/+* | +/+* | +/+ | NP | 10 | Relapse BM |
| CT9 | Vδ1Jδ2 | gaactGCATTGGCCTCTAc | +/+ | +/+ | −/− | TE | +21 | First CCR |
| CT11 | Vδ3Jδ1 | AGCAGATCCCGACGGGTA | +/+ | +/− | +/− | NP | 18 | Relapse BM-CNS |
| CT12 | Vδ2Dδ3 | gacACCGGGAGAAATGact | +/+* | +/+ | +/+ | TE | +18 | First CCR |
| CT14 | Vδ1Jδ1 | ggAATTGccactggg | +/+* | +/+ | +/+ | NP | 12 | Relapse BM |
| CT15 | Dδ2Jδ1 | cttcttacTAGCCCAGCG | +/+* | +/+* | NP | NP | 8 | Relapse BM |
| CT16 | Dδ2Jδ1 | cttacGTACCGGATaca | +/+ | +/− | −/− | −/− | +38 | First CCR |
| CT18 | Vδ2Jδ1 | GGGGACTAACTATGCCCC | +/+ | +/− | −/− | −/− | +55 | First CCR |
| CT21 | Vδ2Jδ1 | ctacCCCGAAGCGCCGAG | +/+ | +/− | −/− | −/− | +33 | First CCR |
| CT22 | Vδ1Jδ1 | gggTTAGGTATTTATacac | +/+ | +/+ | −/− | −/− | +20 | First CCR |
| Patient No. . | TCR Rearrangement . | Patient's Leukemia-Specific Probe† . | End of Induction Therapy, 6-7 Weeks . | 12-15 Weeks . | 35-40 Weeks . | MRD at End of Therapy . | Length of First CCR (mo) . | Status of Disease . |
|---|---|---|---|---|---|---|---|---|
| CT1 | Vδ1Jδ1 | ctATTGCGCTTAGTGg | +/+ | +/− | −/− | −/− | +57 | First CCR |
| CT2 | Vδ1Jδ1 | ggatacgCTCTACTTcaccg | +/+ | +/− | +/− | NP | 12 | Relapse BM |
| CT3 | Vδ3Jδ1 | tagGGCCTGGAGCTAcc | +/+ | +/− | −/− | −/− | +125 | First CCR |
| CT4 | Vδ1Jδ1 | gggCCCCATCGGTacac | +/+* | +/+ | +/+ | NP | 11 | Relapse BM |
| CT5 | Vδ3Jδ1 | gcctCTACCGTAGGtag | −/− | −/− | −/− | −/− | +33 | First CCR |
| CT6 | Vγ2-Jγ1.3 | gggCTAGATCCCTAgat | +/+ | +/+ | +/+ | NP | 20 | Relapse BM |
| CT7 | Vδ3Jδ3 | GGTTATGTTGTTGAAGTTCGT | +/+ | +/+ | +/+ | NP | 20 | Relapse BM |
| CT8 | Vγ6-Jγ1.3 | gataggTCCGTTACGTAg | +/+* | +/+* | +/+ | NP | 10 | Relapse BM |
| CT9 | Vδ1Jδ2 | gaactGCATTGGCCTCTAc | +/+ | +/+ | −/− | TE | +21 | First CCR |
| CT11 | Vδ3Jδ1 | AGCAGATCCCGACGGGTA | +/+ | +/− | +/− | NP | 18 | Relapse BM-CNS |
| CT12 | Vδ2Dδ3 | gacACCGGGAGAAATGact | +/+* | +/+ | +/+ | TE | +18 | First CCR |
| CT14 | Vδ1Jδ1 | ggAATTGccactggg | +/+* | +/+ | +/+ | NP | 12 | Relapse BM |
| CT15 | Dδ2Jδ1 | cttcttacTAGCCCAGCG | +/+* | +/+* | NP | NP | 8 | Relapse BM |
| CT16 | Dδ2Jδ1 | cttacGTACCGGATaca | +/+ | +/− | −/− | −/− | +38 | First CCR |
| CT18 | Vδ2Jδ1 | GGGGACTAACTATGCCCC | +/+ | +/− | −/− | −/− | +55 | First CCR |
| CT21 | Vδ2Jδ1 | ctacCCCGAAGCGCCGAG | +/+ | +/− | −/− | −/− | +33 | First CCR |
| CT22 | Vδ1Jδ1 | gggTTAGGTATTTATacac | +/+ | +/+ | −/− | −/− | +20 | First CCR |
Results of heminested fluorescent PCR and dot blot assay are reported before and after the slash, respectively.
Abbreviations: NP, not performed (patients relapsed before this time point); BM, bone marrow; TE, too early.
Greater than 5% of lymphoblasts detectable by morphologic observation of BM aspirates.
Capital letter indicates the junction sequence of the rearranged segments.
Detection of MRD.MRD was detected in 16 of 17 patients (94%) after 6 to 7 weeks of chemotherapy by either dot blot or heminested fluorescence PCR (Table 2). Differences in the detection sensitivity of the two assays were observed after 12 to 15 weeks of chemotherapy and thereafter. Dot blot assay performed on samples obtained after 12 to 15 weeks of chemotherapy indicated persistence of leukemic cells in nine of 17 patients (64%), and six of the nine positive patients subsequently relapsed; three are in first CCR after 18, 21, and 20 months, respectively. Among eight patients with negative results at 12 to 15 weeks, two relapsed and six are in first CCR after 33, 33, 38, 55, 57, and 125 months, respectively.
By contrast, heminested fluorescence PCR of the same sample obtained after 12 to 15 weeks of chemotherapy showed persistence of leukemic cells in 16 of 17 patients (94%) regardless of treatment protocol. Eight of the positive patients subsequently relapsed, and eight are in first CCR after 18, 20, 21, 33, 38, 55, 57, and 125 months, respectively. The negative patient is in first CCR. Thus, analysis of MRD at the time of consolidation or reinforced induction did not enable discrimination between patients likely to respond to therapy and patients likely to relapse, with the exception of a single patient with negative results by the heminested fluorescence PCR.
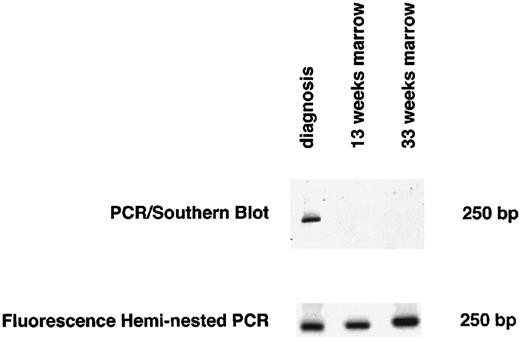
Marrow samples obtained at the beginning of maintenance therapy showed no evidence of MRD by heminested fluorescence PCR in eight of 15 patients who could be monitored (one patient, no. CT15, relapsed before this time). All eight patients are currently in first CCR after 20, 21, 33, 33, 38, 55, 57, and 125 months, respectively. Of eight patients who were positive, seven relapsed. The dot blot assay indicated the absence of MRD in 10 patients, yet two of these patients relapsed after 12 and 18 months, respectively. Figure 2 shows an example of screening for MRD using both heminested fluorescence PCR and Southern blot detection assay for one of these two patients (no. CT11 in Tables 1 and 2).
Detection of MRD by both the heminested fluorescence PCR and the PCR/Southern blot assay 13 and 33 weeks after diagnosis for 1 representative patient with discordant results for MRD (no. CT11 in Tables 1 and 2).
Detection of MRD by both the heminested fluorescence PCR and the PCR/Southern blot assay 13 and 33 weeks after diagnosis for 1 representative patient with discordant results for MRD (no. CT11 in Tables 1 and 2).
Sensitivity and specificity for detection of MRD were 100% and 88.9% for the heminested fluorescent PCR and 71.4% and 88.9% for dot blot detection, respectively. Predictive negative and positive values were 100% and 90.7% for heminested fluorescence PCR, respectively.
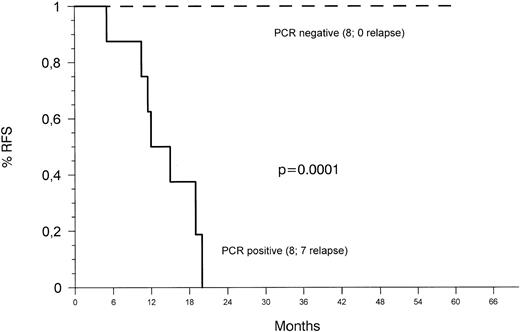
The probability of RFS based on evidence of MRD (Fig 3) as detected by heminested fluorescence PCR 34 to 40 weeks after diagnosis was 100% and 0% for MRD-negative and MRD-positive patients, respectively. Differences between the two groups were statistically significant (P < .0001). Analysis of samples at the termination of therapy revealed no evidence of leukemic clones in any of six patients tested.
Kaplan-Meier plots of RFS according to the presence or absence of MRD detected with the heminested fluorescence PCR assay 34 to 40 weeks after diagnosis. Patients who relapsed before this time point were censored.
Kaplan-Meier plots of RFS according to the presence or absence of MRD detected with the heminested fluorescence PCR assay 34 to 40 weeks after diagnosis. Patients who relapsed before this time point were censored.
DISCUSSION
PCR-based techniques that target rearranged antigen receptors for detection of MRD in children with ALL appear to be extraordinarily sensitive and specific. However, these assays are based on genetic rearrangements that are clone-specific rather than leukemia-specific, so that clonal evolution, clonal selection, emergence of an independent new clone, and secondary leukemias can all hamper reliability of the detection of MRD. In B-lineage ALL, IgH and cross-lineage TCR δ and TCR γ rearrangements can evolve during overgrowth of the leukemic clone, leading to biclonality or oligoclonality at diagnosis and to clonal evolution.23 29-31 It has been suggested that studies of MRD should involve monitoring gene rearrangements at multiple loci to minimize false-negative results, but such precautions might not be necessary if the detection assays are performed relatively early during chemotherapy, before any mutated clones reexpand.
MRD studies, mainly based on B-lineage ALL, have indicated considerable variation in the persistence of ALL during remission. In fact, MRD has been detected 1 to 3 months and up to 24 months6,7,10 after diagnosis in patients who have remained in long-term remission. By contrast, patients with negative results for detection of MRD at different intervals after diagnosis can relapse.11 Therefore, in B-lineage ALL, the absence of detectable MRD is not a sufficient criterion for prediction of cure. It is possible that the kinetics of the decrease in MRD differ among different cytogenetic or immunophenotypic subsets of childhood ALL. A study32 reported that five of six patients with t(1; 19)-positive ALL in first CCR achieved molecular remission within 3 months, and one patient displayed MRD after 84 months of CCR. Another study33 reports that all of five long-term survivors with t(1; 14)-ALL were consistently negative for MRD after completion of induction therapy, whereas all of seven patients with consistently positive MRD had resistant disease or had relapsed.
In the present study of a subgroup of children with T-ALL, we find that the persistence or absence of MRD at a specific time during treatment, ie, 34 to 40 weeks after diagnosis when the patient enters maintenance therapy, best correlates with subsequent bone marrow relapse or absence of relapse, respectively. This result does not change if one, two, or three of eight MRD-negative patients with the shortest follow-up times eventually relapse. Results for sensitivity of the heminested fluorescence PCR should be interpreted more cautiously, since one, two, or three relapses in this study would significantly affect its predictive negative and positive values.
Our results are consistent with those of several recent studies. Neale et al,34 who studied MRD retrospectively in five T-ALL patients (adults and children) in morphologic remission by examining TCR δ rearrangements, found no evidence of MRD in any bone marrow samples from four patients who were in long-term remission, whereas they found MRD in every sample from one patient who relapsed 2 years after diagnosis. Similarly, Hansen-Hagge et al,3 who studied MRD in seven children with T-ALL, detected the persistence of a leukemic clone in all patients who subsequently relapsed, but did not detect MRD in samples obtained after 6 to 10 months of therapy from patients in long-term remission. Cavè et al35 reported three children with T-ALL in whom MRD was undetectable 6 to 8 months after diagnosis: two were in CCR 12 and 18 months after diagnosis, respectively, and one had a relapse that involved the central nervous system 10 months after diagnosis.
Our study, the only one presently available that systematically examines the extent of MRD at different milestone events during therapy, also showed that almost all patients (94%) have PCR-based evidence of MRD up to 12 to 15 weeks after completion of induction therapy, regardless of treatment protocol. Thus, it therefore seems likely that prolonged induction therapy and early intensification of treatment play a critical role in the treatment of these patients. The role of maintenance therapy is less clear, since it was ineffective, at least in the group with persistence of leukemic cells after 34 to 40 weeks of treatment.
A potential limitation in the routine of monitoring MRD rests in the labor-intensive and time-consuming nature of current detection procedures. We find that the rapid and sensitive heminested fluorescence PCR, once probes specific for the most common TCR γ rearrangements were available, yields informative results in 8 hours as compared with the usual 5 days and simplifies detection of MRD in children with T-ALL. Standardization of techniques would greatly facilitate comparison of results among different studies, as well as assessment of MRD in prospective clinical trials.
In conclusion, our analysis of MRD in children with T-ALL using a rapid and reliable PCR-based method shows that the presence of MRD 34 to 40 weeks after diagnosis is predictive of a poor outcome and might represent the basis for a therapeutic decision aimed at hematopoietic rescue in such patients. By contrast, the absence of MRD any time after diagnosis is correlated with a favorable outcome.
Supported in part by grants from the National Cancer Institute (CA10815), the American Cancer Society (DHP-94), the Parker Hughes Foundation (G.R.), and the Associazione Italiana Ricerca sul Cancro.
Address reprint requests to Salvatore P. Dibenedetto, MD, Divisione di Ematologia ed Oncologia Pediatrica Dell'Università di Catania, Viale Andrea Doria 6, 95100 Catania, Italy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal