Abstract
The rate of detection of chromosome abnormalities in T-cell proliferations is lower than that observed in B-cell malignancies. The former frequently involve the TCRα/δ locus at chromosome band 14q11. We have identified a YAC encompassing 70% of the TCRα/δ locus, which has been used as a fluorescence in situ hybridization probe to detect chromosome rearrangements involving 14q11, both at metaphase and within interphase nuclei, in patients with a variety of T-lymphoproliferative disorders. Its use allowed detection of previously unsuspected TCRα/δ rearrangements in 4/13 (30%) immature T-lineage acute leukemias, including two t(10; 14) and 2 minor inversion 14s. It also clarified interpretation of complex chromosome 14 abnormalities in mature T-cell proliferations (T-prolymphocytic leukemia and ataxia telangiectasia). Use of this probe will aid the detection and characterization of abnormalities involving the TCRα/δ locus, particularly in cases with normal or complex karyotypes and in those proliferations for which mitoses are difficult to obtain.
THE PROCESS OF somatic DNA recombination, which is necessary for the production of a functional antibody receptor gene also predisposes lymphoid cells to aberrant DNA rearrangements, which can result in chromosome translocation or inversion. As a consequence, chromosome translocations in T-lymphoid malignancies frequently involve the T-cell receptor (TCR) genes, with the TCRα/δ locus on chromosome 14q11 being most commonly affected. The incidence of rearrangements involving this locus depends on clinical subtype, occuring at a frequency of approximately 20% in T-acute lymphoblastic leukemia (T-ALL), 15% in T-non-Hodgkin's lymphoma (NHL),1,2 and 80% in T-prolymphocytic leukemia (PLL).3 Several nonrandom abnormalities involving 14q11 have been described, including the t(10; 14)(q24; q11), t(8; 14)(q24; q11), t(11; 14)(p13; q11), t(11; 14)(p15; q11), t(11; 14)(q23; q11), inv(14)(q11q32), t(14; 14) (q11; q32), and t(X; 14)(q28; q11).4
In addition to its role in T-cell malignancies, rearrangements involving the TCRα/δ locus are frequently found in ataxia telangiectasia (AT). AT is a rare autosomal recessive disorder, which until identification of the genetic basis,5 was defined by spontaneous chromosome breakage. The localization of the breaks are nonrandom and the rearrangements observed are principally reciprocal translocations and inversions occuring between the Ig and TCR genes. These patients are at an increased risk of developing malignancies, which are primarily of T-cell origin and frequently show a t(14; 14), an inv(14) or, more rarely, a t(X; 14).6
The TCRα/δ locus is unusual insofar as the TCRδ variable (V), diversity (D), joining (J), and constant region genes are situated within the TCRα locus, between the TCRαV and TCRαJ segments. TCRα V-J rearrangement therefore leads to deletion of the TCRδ locus on this allele. The TCRδ locus contains 3D segments and 4J segments7,8 whereas the TCRαJ region spans approximately 80 Kb and contains at least 61 J segments.9 The TCRα/δ locus (≈1 Mb) is transcribed in a centromere to telomere direction (Fig 1). Molecular analysis has shown that the majority of breakpoints are localized within the TCRδ gene, although they may also involve TCRαJ and rarely TCRαV.7 The limited complexity of the TCRδ locus renders detection of physiological V-D-J or pathological rearrangement by Southern blotting relatively easy. Detection of TCRα rearrangement is, however, hampered by the large number of J segments.
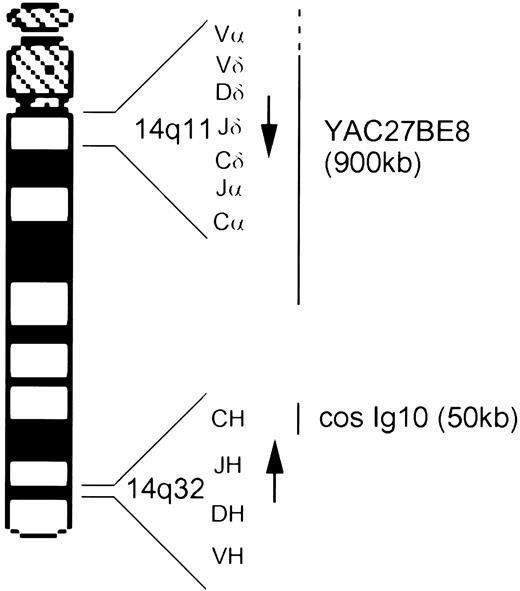
Ideogram of chromosome 14 showing the location and transcriptional orientation (arrow) of the TCRα/δ and IgH loci and the approximate localization of the two probes, cosmid Ig10 and YAC 27BE8.
Ideogram of chromosome 14 showing the location and transcriptional orientation (arrow) of the TCRα/δ and IgH loci and the approximate localization of the two probes, cosmid Ig10 and YAC 27BE8.
Fluorescence in situ hybridization (FISH) with probes for specific chromosomal regions is used increasingly in hematological cytogenetics. This technique allows the identification and verification of chromosome abnormalities, the analysis of poor quality metaphases, the detection of minor clones and of chromosome abnormalities within interphase nuclei. We have identified a 900-kb YAC, which encompasses at least 70% of the TCRα/δ locus, including the entire TCRδ locus. Use of this YAC allowed FISH detection of rearrangements into the TCRα/δ locus in both interphase nuclei and metaphase in a number of cell lines and in patients with T-cell malignancies, including in patients considered to have a normal karyotype by standard banding analysis.
MATERIALS AND METHODS
Patients and cell lines.Pathological samples were taken from blood, bone marrow (BM), and/or lymph nodes with informed consent. Diagnosis was based on morphological and immunophenotypic analysis by flow cytometric and immunocytochemical analysis, using standard techniques. Cases analyzed included: 10 T-ALL, 2 T-ALL/acute myeloid leukemia (AML), 1 large granular lymphocytic acute leukemia (LGL-AL), 2 T-PLL, 2 AT, and 6 cell lines. All cell lines have been previously described. The T-ALL 103.3 cell line expresses a myeloid (CD33+, CD15+, CD11b+) phenotype but on culture in the presence of interleukin-2 (IL-2) transforms to a surface CD3+, CD2+, CD7+, CD8+, TCRγδ+ phenotype. Both strains show a t(8; 14)(q24; q11).10 This line was generously provided by Daniella Santoli (Wistar Institute, Philadelphia, PA).
Isolation and characterization of YAC 27BE8.The TCRα/δ YAC 27BE8 was isolated from the ICRF human YAC library (ICRF Institute, London, UK).11 It is 900 kb in length with Cα located about 250 Kb from the telomeric end. Therefore, the YAC contains about 650 Kb centromeric to TCRαC, including the entire TCRδ locus. Part, but not all, of TCRαV is contained. The TCRαV families with at least one member present are: 1, 3, 6, 10, 12, 13, 14, 17, 18, 19, 21, 22, 23, 25, 26, 28, 30, 31, and 32 (F. Cornelis, unpublished data, June 1992).
Cytogenetic studies.Cytogenetic analysis was performed on BM, blood, or lymph node material from patients and on logarithmically growing T-cell lines, according to standard procedures. Cells were cultured for 24 and 48 hours without mitogenic stimulation and for 72 hours with phytohemaglutinin (PHA) and IL-2 (5 ng/mL, Valbiotech, Paris, France). PHA stimulated peripheral blood lymphocytes from 6 healthy individuals were used as control samples. Metaphase spreads were R and/or G banded and karyotypes designated according to the International System for Human Cytogenetic Nomenclature (ISCN) 1991.12
In situ hybridization.For each hybridization onto chromosome preparations 400 ng of biotinylated YAC DNA was combined with 10 μg Cot-l1 DNA in 50% formamide, 10% dextran sulphate, 2× SSC (0.3 mol/L NaCl, 0.03 mol/L trisodium citrate) pH 7. The IgH cosmid (Ig10,13 ) was kindly provided by T. Rabbitts (MRC Centre, Cambridge, UK). For each hybridization 50 ng of biotinylated cosmid DNA was combined with 2.5 μg of Cot-1 DNA in 10 μL of hybridization mix. For selected patients the YAC hybridization was repeated to include either 10 ng of chromosome 10 α-satellite centromere probe (Oncor, Gaithersburg, MD) or 50 ng of cosmid Ig10. The chromosome 14 paint (Cambio, Cambridge, UK) was used according to manufacturers instructions. Where chromosome painting was performed on previously hybridized slides the slides were treated as described by Heslop-Harrison14 with the exception that the coverslips were first removed in ethanol and that PN (0.1 mol/L Na2HPO4 , 0.1 mol/L NaH2PO4 ) buffer was used in place of 4× SSC. Hybridization was performed at 37°C overnight for cosmids and chromosome paints and for 2 nights for YACs. Immunological detection of biotin labeled probes was performed by incubating the slides in goat antibiotin antibody (1/50, Sigma, St Quentin-Fallavier, France), followed by rabbit antigoat conjugated to fluorescein isothiocyanate (FITC; 1/80, Sigma). For dual hybridization the slides were detected by Avidin-Texas Red (0.8/1000, Vector, Burlingame, CA) and mouse antidigoxigenin (1/250, Boehringer Mannheim, Meylan, France) followed by incubation in biotinylated goat antiavidin (1/100, Vector) and rabbitt antimouse FITC (1/500, Dako, Trappes, France) followed by Avidin Texas Red and swine antirabbitt-FITC (1/500, Dako). The slides were mounted in Vectorshield (Vector) containing either propidium iodide or 4′-6-diamidino-2-phenylindole (DAPI) and analyzed with a Leitz axiophot microscope attatched to an image analysis system (Applied Imaging; Perceptive Scientific Instruments, League City, TX). Some images were photographed directly onto Kodak Ektachrome Elite ASA400 film (Eastman Kodak, Rochester, NY). The band localization of the probe was determined by the examination of chromosome bands from DAPI stained chromosomes or conversion of the image to black and white G bands.
RESULTS
Analysis of normal T-cell mitoses.The 27BE8 YAC is known to be nonchimeric and hybridizes specifically to 14q11.11 Since the TCRα/δ locus undergoes somatic DNA recombination, with loop excision of intervening sequences, it was possible that this could result in the reduction or loss of the observed hybridization signal on at least one of the chromosome 14s in T lymphocytes. PHA stimulated circulating T lymphocytes from 6 healthy donors, however, showed 2 chromosome 14 signals of equal intensity in all mitoses examined (20 to 100 per sample). Because more than 90% of circulating T cells express the TCRαβ receptor, a large proportion of the peripheral blood lymphocyte (PBL) cells must have undergone TCRα rearrangement and TCRδ deletion on at least one allele. The majority of TCRα V-J rearrangements do not therefore lead to loss of the FISH hybridization signal.
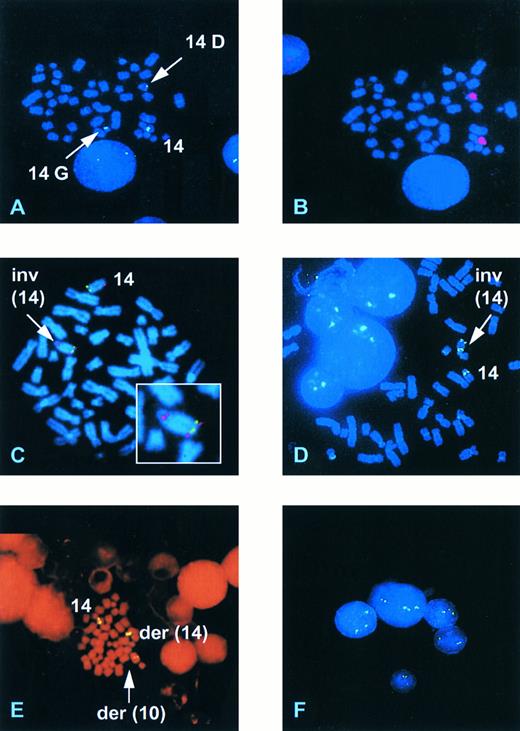
Detection of known 14q11 rearrangements.The ability of YAC 27BE8 to detect translocations into the TCRα/δ locus was then determined on four patients and two cell lines with known 14q11 rearrangements. The precise TCR breakpoints are not known for the individual patients, whereas those in T-ALL103.3 and RPMI.8402 involve TCRδ D-J.10 15 The results of FISH analyses are presented in Table 1. In all but one case a signal was observed on the normal chromosome 14 and on both derivative chromosomes involved in the rearrangement, showing that the YAC spans the breakpoint region. In one case (Bru) of AT only 2 signals were observed both on the derivative chromosome der(14)t(14; 14)(q11; q32) (14q+), one near the centromere and the other in the middle of the chromosome. However, this was consistent with the karyotype, since the other derivative chromosome resulting from the t(14; 14)(q11; q32), a 14q−, had been lost. In case (Deb) with T-PLL and a complex karyotype including an inv(14)(q11q32), FISH analysis revealed a more complex chromosome 14 rearrangement. Three signals were observed: one at 14q11 on the normal chromosome 14, one in a centromeric position on a G group sized chromosome (21-22, Y), consistent with a 14q−, and one in a telomeric position on a D group chromosome (13-15) (14G and 14D, Fig 2A). Chromosome 14 painting of the same mitoses previously hybridized with the YAC showed that the D group chromosome was indeed a chromosome 14 (Fig 2B), whereas the absence of a centromeric TCR α/δ signal confirmed that it was not a classical inv(14) nor a straight forward translocation. The small G group chromosome was not painted with the chromosome 14 paint but a similarly sized marker chromosome was identified when chromosome 14 painting was performed on fresh slides, presumably due to a reduced efficiency of painting on the rehybridized slides. These results are compatible with an inversion of chromosome 14 associated either with a concurrent or subsequent translocation to a G group size chromosome or with a deletion and insertion of part of 14q11, including the TCRα/δ locus, into a G group size chromosome.
Clinical Details of Cases With Karyotypic and FISH Results
| Case/UPN . | Diagnosis . | sCD3 . | Karyotype . | Metaphase YAC . | No. of Patients Interphase Signals . | No. Total of . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | Location . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | Interphase . |
| . | . | . | . | . | . | . | . | . | . | . | Cells . |
| . | . | . | . | . | . | . | . | . | . | . | Examined . |
| T-ALL 103.3 | Myeloid/pre T-cell line | — | 46,XY,t(8; 14)(q24; q11),−9,der(12)t(12; 13)(p11; q11), −13,+2mar [10] | 14, (der14), der(8) | 4 | 11 | 79 | 4 | 2 | — | 109 |
| RPMI 8402 | T-cell line | αβ | 82∼89,XXX,add(4)(q35)×2,del(6)(q?)×2,−8, t(11; 14)(p15; q11)×2,−13×2,−15×2,−18,−22,+2-4mar [cp8] | 14, der(14), der(11) | — | — | — | — | — | — | — |
| Pre | T-ALL | +* | 46,XY,t(11; 14)(p13; q11)[14] | 14, der(14), der(11) | 5 | 63 | 32 | — | — | — | 201 |
| 258 | T-PLL | αβ | 46,XX[1]/44-45,XX,del(1)(q?),−2,add(5)(p?),−6, −7,−8,add(9)(p?),del(11)(q21),add(12)(p?),t(14; 14) (q11; q32),add(16)(q?),+mar1,+mar2×2 [cp8] | der(14+)¶, der(14q−)# | 9.5 | 54 | 30 | 5 | 1 | 0.5 | 190 |
| Bru | AT | — | 46,XY[3]/45,XY −14,der(14)t(14; 14)(q11;q32), inc[22]ρ | der(14q+)¶ | — | — | — | — | — | — | — |
| Deb | T-PLL | αβ | 46,XY[1]/42-45,XY,−2,−3,i(8)(q10),del(11)(q21),−12, inv(14)(q11q32),+3mar[cp18] | 14, inv(14)**, mar# | 4 | 77.5 | 18 | 0.5 | — | — | 249 |
| 231 | T-ALL | — | 46,XX [1]/46,XX,add(10)(q?),−14,+mar[3]/46,id,i(14)(q10)[5] | 14, add(10), mar# | 0.5 | 17 | 71 | 11 | 0.5 | — | 205 |
| Gam | AT | — | 46,XX [26]/46,XX,der(14) [6] | 14, der(14)¶ | — | — | — | — | — | — | — |
| HP-BALL | T-cell line | αβ | 89∼91,XX,t(1; 5)(q31; q34)×2,−3,−7,−8,−13, add(14)(q32)×2,−16×2,+mar1×2,+mar2×2 [cp6] | 4 × 14 | — | — | — | — | — | — | — |
| 572 | T-ALL | γδ | 46, XX,−5,add(14)(q32),+mar [20] | 2 × 14 | 7.5 | 89 | 2.5 | 1 | — | — | 200 |
| Peer | T-cell line | γδ | 86∼90,XXX,−4×2,−5,+6,del(6)(q?)×2,+7×2,−8×2, del(9)(p?)×2,+i(9)(p10)×2,−12,−13,−15,−18,+3-5 mar [cp9] | 4 × 14 | — | — | — | — | — | — | — |
| Molt13 | T-cell line | γδ | 46,XX,i(17)(q10)[10] | 2 × 14 | — | — | — | — | — | — | — |
| Molt4 | T-cell line | — | 96∼100,XXYY,+4,del(6)(q?)×2,+7,i(7)(q10)×2, +8,+9,−13,+3mar [cp6] | 4 × 14 | — | — | — | — | — | — | — |
| 639 | T-ALL/AML† | — | 84∼86,−11,del(11)(q23),−14×1∼3,+10mar,inc [20] | 3 × 14 | 3 | 29 | 61.5 | 6 | 0.5 | — | 201 |
| 957 | T-ALL | γδ | 46,XX[40]/47,XX,+8[4]∥ | 14, inv(14) | 4 | 88.5 | 7.5 | — | — | — | 200 |
| Sh | T-ALL | — | 46,XY[20] | 2 × 14 | 6.5 | 92 | 1.5 | — | — | — | 203 |
| 226 | T-ALL/AML* | — | 46,XY[18] | 2 × 14 | 9 | 89 | 1 | 1 | — | — | 173 |
| Lin | T-ALL | — | 46,XY[20] | 2 × 14 | 6.5 | 92.5 | 1 | — | — | — | 200 |
| 951 | T-ALL | αβ | 46,XX[65] | 2 × 14 | 6 | 86.5 | 5.5 | 2 | — | — | 207 |
| 872 | T-ALL | αβ | 46,XY[16] | 2 × 14 | — | — | — | — | — | — | — |
| 634 | T-ALL | — | 46,XY[21] | 14, inv(14) | — | — | — | — | — | — | — |
| 586 | T-ALL* | — ‡ | 46,XY[20] | der(14), der(10) | 6 | 93 | 1 | — | — | — | 204 |
| 282 | LGL-AL* | — | 46,XX[23] | 14, der(14), der(10) | 3 | 46.5 | 36.5 | 8 | 5 | 1 | 202 |
| Control means (n = 6) | 5.7 | 91.8 | 2.3 | 0.2 | — | — | 198 | ||||
| Control SD | 2 | 3.25 | 1 | — | — | — | — | ||||
| Abbreviation: UPN, unique patient number. | |||||||||||
| * See text for full immunophenotypic details. | |||||||||||
| † TCR αβ and γδ not assessed. | |||||||||||
| ‡ CD3 weak. | |||||||||||
| ρ Includes the analysis of mitoses containing other structural abnormalities. | ∥ Total number of metaphases examined in blood and bone marrow cultures. | ||||||||||
| ¶ Two signals observed at positions corresponding to bands 14q11 and 14q32. | |||||||||||
| # One signal observed at a position corresponding to band 14q11. | |||||||||||
| ** One signal observed at a position corresponding to band 14q32. | |||||||||||
| Case/UPN . | Diagnosis . | sCD3 . | Karyotype . | Metaphase YAC . | No. of Patients Interphase Signals . | No. Total of . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | Location . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | Interphase . |
| . | . | . | . | . | . | . | . | . | . | . | Cells . |
| . | . | . | . | . | . | . | . | . | . | . | Examined . |
| T-ALL 103.3 | Myeloid/pre T-cell line | — | 46,XY,t(8; 14)(q24; q11),−9,der(12)t(12; 13)(p11; q11), −13,+2mar [10] | 14, (der14), der(8) | 4 | 11 | 79 | 4 | 2 | — | 109 |
| RPMI 8402 | T-cell line | αβ | 82∼89,XXX,add(4)(q35)×2,del(6)(q?)×2,−8, t(11; 14)(p15; q11)×2,−13×2,−15×2,−18,−22,+2-4mar [cp8] | 14, der(14), der(11) | — | — | — | — | — | — | — |
| Pre | T-ALL | +* | 46,XY,t(11; 14)(p13; q11)[14] | 14, der(14), der(11) | 5 | 63 | 32 | — | — | — | 201 |
| 258 | T-PLL | αβ | 46,XX[1]/44-45,XX,del(1)(q?),−2,add(5)(p?),−6, −7,−8,add(9)(p?),del(11)(q21),add(12)(p?),t(14; 14) (q11; q32),add(16)(q?),+mar1,+mar2×2 [cp8] | der(14+)¶, der(14q−)# | 9.5 | 54 | 30 | 5 | 1 | 0.5 | 190 |
| Bru | AT | — | 46,XY[3]/45,XY −14,der(14)t(14; 14)(q11;q32), inc[22]ρ | der(14q+)¶ | — | — | — | — | — | — | — |
| Deb | T-PLL | αβ | 46,XY[1]/42-45,XY,−2,−3,i(8)(q10),del(11)(q21),−12, inv(14)(q11q32),+3mar[cp18] | 14, inv(14)**, mar# | 4 | 77.5 | 18 | 0.5 | — | — | 249 |
| 231 | T-ALL | — | 46,XX [1]/46,XX,add(10)(q?),−14,+mar[3]/46,id,i(14)(q10)[5] | 14, add(10), mar# | 0.5 | 17 | 71 | 11 | 0.5 | — | 205 |
| Gam | AT | — | 46,XX [26]/46,XX,der(14) [6] | 14, der(14)¶ | — | — | — | — | — | — | — |
| HP-BALL | T-cell line | αβ | 89∼91,XX,t(1; 5)(q31; q34)×2,−3,−7,−8,−13, add(14)(q32)×2,−16×2,+mar1×2,+mar2×2 [cp6] | 4 × 14 | — | — | — | — | — | — | — |
| 572 | T-ALL | γδ | 46, XX,−5,add(14)(q32),+mar [20] | 2 × 14 | 7.5 | 89 | 2.5 | 1 | — | — | 200 |
| Peer | T-cell line | γδ | 86∼90,XXX,−4×2,−5,+6,del(6)(q?)×2,+7×2,−8×2, del(9)(p?)×2,+i(9)(p10)×2,−12,−13,−15,−18,+3-5 mar [cp9] | 4 × 14 | — | — | — | — | — | — | — |
| Molt13 | T-cell line | γδ | 46,XX,i(17)(q10)[10] | 2 × 14 | — | — | — | — | — | — | — |
| Molt4 | T-cell line | — | 96∼100,XXYY,+4,del(6)(q?)×2,+7,i(7)(q10)×2, +8,+9,−13,+3mar [cp6] | 4 × 14 | — | — | — | — | — | — | — |
| 639 | T-ALL/AML† | — | 84∼86,−11,del(11)(q23),−14×1∼3,+10mar,inc [20] | 3 × 14 | 3 | 29 | 61.5 | 6 | 0.5 | — | 201 |
| 957 | T-ALL | γδ | 46,XX[40]/47,XX,+8[4]∥ | 14, inv(14) | 4 | 88.5 | 7.5 | — | — | — | 200 |
| Sh | T-ALL | — | 46,XY[20] | 2 × 14 | 6.5 | 92 | 1.5 | — | — | — | 203 |
| 226 | T-ALL/AML* | — | 46,XY[18] | 2 × 14 | 9 | 89 | 1 | 1 | — | — | 173 |
| Lin | T-ALL | — | 46,XY[20] | 2 × 14 | 6.5 | 92.5 | 1 | — | — | — | 200 |
| 951 | T-ALL | αβ | 46,XX[65] | 2 × 14 | 6 | 86.5 | 5.5 | 2 | — | — | 207 |
| 872 | T-ALL | αβ | 46,XY[16] | 2 × 14 | — | — | — | — | — | — | — |
| 634 | T-ALL | — | 46,XY[21] | 14, inv(14) | — | — | — | — | — | — | — |
| 586 | T-ALL* | — ‡ | 46,XY[20] | der(14), der(10) | 6 | 93 | 1 | — | — | — | 204 |
| 282 | LGL-AL* | — | 46,XX[23] | 14, der(14), der(10) | 3 | 46.5 | 36.5 | 8 | 5 | 1 | 202 |
| Control means (n = 6) | 5.7 | 91.8 | 2.3 | 0.2 | — | — | 198 | ||||
| Control SD | 2 | 3.25 | 1 | — | — | — | — | ||||
| Abbreviation: UPN, unique patient number. | |||||||||||
| * See text for full immunophenotypic details. | |||||||||||
| † TCR αβ and γδ not assessed. | |||||||||||
| ‡ CD3 weak. | |||||||||||
| ρ Includes the analysis of mitoses containing other structural abnormalities. | ∥ Total number of metaphases examined in blood and bone marrow cultures. | ||||||||||
| ¶ Two signals observed at positions corresponding to bands 14q11 and 14q32. | |||||||||||
| # One signal observed at a position corresponding to band 14q11. | |||||||||||
| ** One signal observed at a position corresponding to band 14q32. | |||||||||||
Representative FISH analyses with the 14q11/TCRα/δ YAC and the 14q32/IgH cosmid. (A) Case Deb showing hybridization of a TCRα/δ YAC to the normal chromosome 14, a G group size chromosome (14G), and a D group size chromosome (14D). (B) The same mitosis as (A) rehybridized with a chromosome 14 paint. (C) Case Gam hybridized with the YAC (red) and the IgH Ig10 cosmid (green) showing a normal chromosome 14 and the presence of an inversion (arrow) with a breakpoint within 14q32 proximal to the IgH locus. (D) Case UPN 957 showing the presence of an inv(14). (E) Case UPN 282 showing hybridization of the TCRα/δ YAC to the normal chromosome 14, a der(14) and a der(10). (F ) Interphase nuclei from UPN 282 showing the presence of three hybridization signals in three of the five nuclei.
Representative FISH analyses with the 14q11/TCRα/δ YAC and the 14q32/IgH cosmid. (A) Case Deb showing hybridization of a TCRα/δ YAC to the normal chromosome 14, a G group size chromosome (14G), and a D group size chromosome (14D). (B) The same mitosis as (A) rehybridized with a chromosome 14 paint. (C) Case Gam hybridized with the YAC (red) and the IgH Ig10 cosmid (green) showing a normal chromosome 14 and the presence of an inversion (arrow) with a breakpoint within 14q32 proximal to the IgH locus. (D) Case UPN 957 showing the presence of an inv(14). (E) Case UPN 282 showing hybridization of the TCRα/δ YAC to the normal chromosome 14, a der(14) and a der(10). (F ) Interphase nuclei from UPN 282 showing the presence of three hybridization signals in three of the five nuclei.
Analysis of cases with ill-defined chromosome 14 rearrangements.Four additional cases with chromosome 14 structural abnormalities were analyzed. Case Gam, with AT, was classified karyotypically as an isolated der(14). FISH analysis showed a split signal on one of the chromosome 14s, with hybridization to bands 14q11 and 14q32, consistent with an inversion of chromosome 14 (Fig 2C). Dual hybridization with cos Ig10, from the IgH locus at 14q32 (Fig 1), indicated a breakpoint proximal to IgH (Fig 2C). The der(14) banding pattern was not, however, typical of a classical inv(14) and it is probable that further rearrangement had occurred in this case. In UPN231 FISH analysis clarified the karyotypic interpretation, initially classified as 46,XX, add(10)(q?),−14,+mar, insofar as it demonstrated that the marker chromosome was a 14q− and showed translocation of the TCRα/δ locus to a chromosome corresponding to the the add(10). It also confirmed the presence of an additional subclone bearing an isochromosome 14q, with hybridization signals observed close to the centromere on both long arms.
For both samples (HPB-ALL and UPN572) demonstrating an add(14)(q32) the TCRα/δ YAC remained intact. Further studies with the 14q32 IgH cos Ig10 (Fig 1) showed that in UPN572 the IgH cosmid remained in a telomeric position, consistent with an insertion or duplication of material proximal to 14q32. Karyotypically this case also showed loss of a chromosome 5 plus a marker chromosome, suggesting that some of the material inserted into this chromosome may have come from chromosome 5. The HPB-ALL cell line had a complex tetraploid karyotype that included a duplicated translocation t(1; 5)(q31; q34), a duplicated add(14)(q32), and two normal chromosome 14s. FISH studies with the IgH cosmid revealed that this locus had been translocated onto the der(1) chromosome. This was an unexpected finding since the t(1; 5) appeared balanced but evidently a complex translocation involving chromosomes 1, 5, and 14 had occured.
Karyotypically abnormal cases with no evident chromosome 14 rearrangement.The 27BE8 YAC was also used to screen for 14q11 abnormalities in 5 samples demonstrating clonal abnormalities not including any apparent chromosome 14 rearrangement. Diagnostic categories are shown in Table 1. The biphenotypic T-ALL/AML (UPN639) expressed CD2, CD7, CD5, cyt.CD3, CD34, CD33, and cyt.CD13 and had undergone clonal TCRγ rearrangement (data not shown). No structual abnormalities involving TCRα/δ were observed in the 3 cell lines and in 1 of the 2 patients. A 14q11 rearrangement was, however, detected by FISH in one patient (UPN957). No mitoses were obtained in the unstimulated cultures of either blood or BM and karyotypic and FISH analyses were performed on stimulated blood and BM cultures. A 47,XY,+8 clone was detected by karyotype analysis in 1/24 mitoses in the peripheral blood and 3/20 in the BM (3/44; 7% overall) while FISH identified a minor clone containing an inv(14)(q11q32) in 2/27 mitoses in the blood and in 1/93 mitoses in the BM culture (3/120; 2.5% overall) (Fig 2D). Repeat hybridization of the same FISH slides with a centromere 8 probe was uninterpretable. One of the mitoses containing the inv(14) contained less than 43 chromosomes, one 50 chromosomes, and one had 47 chromosomes with an extra C group chromosome (6-12, X). Although the inv(14) clone was minor, the finding of 3 mitoses with the same chromosomal abnormality would constitute the presence of a clone by standard cytogenetic nomenclature.
Analysis of patients with a normal karyotype.Eight patients with an apparently normal karyotype were studied. A rearrangement at 14q11 was detected in 3 cases (38%), including 2/6 T-ALL and in an unusual case of large granular lymphocytic acute leukemia (LGL-AL).The latter (UPN282) presented with acute clinical symptoms due to hepatosplenomegaly, lymphadenopathy, and leucoctosis (106 × 109/L), consisting of morphologically mature CD8+, CD2+, CD7+, CD5+, CD16+, surface CD3−, surface TCR−, cyt.CD3+, cyt.TCRβ+ circulating cells, 70% of which resembled LGL. Full details of this case have been reported elsewhere.16 Initial karyotypic analysis failed to detect any clonal abnormality in 23 mitoses from the BM sample. On FISH analysis 3 signals on a chromosome 14, a der(14) and a chromosome resembling a chromosome 10 were observed in 7% of mitoses (Fig 2E), along with occasional polyploid mitoses. FISH hybridization of the TCRα/δ YAC plus a chromosome 10 α-satellite probe showed that this abnormality was indeed a t(10; 14)(q24; q11). At relapse a karyotypic t(10; 14)(q24; q11) was observed, consistent with the initial FISH analysis
Karyotype analysis for UPN586 (T-ALL) was not performed at diagnosis but at first relapse, when the blast cells expressed CD2, CD5, CD7, CD4, and CD8 but were surface CD3 weak and TCRαβ and TCRγδ negative. On FISH analysis 20% of mitoses had two hybridization signals with an abnormal pattern of hybridization. Signals were observed on a G group size chromosome, consistent with a 14q−, and a second chromosome, suggesting the presence of a 14q11 rearrangement associated with loss of the normal chromosome 14. Chromosome 14 painting confirmed this interpretation. Archival slides from the cytogenetic preparation were re-examined and an abnormal clone containing a t(10; 14) (q24; q11) with loss of the normal chromosome 14 plus a marker chromosome was observed. Dual hybridization of the TCRα/δ YAC and a chromosome 10 α-satellite probe again confirmed that the abnormality was a t(10; 14)(q24; q11).
In patient UPN634 a minor clone with an inversion 14 was detected in 2 out of 54 metaphases analyzed. Both abnormal metaphases analyzed were aneuploid.
Detection of TCRα/δ translocation in interphase nuclei.The effectiveness of this YAC to detect abnormalities in interphase nuclei was assessed in a controlled study. Table 1 shows the results of the assessment of 109-200 nuclei in 6 control samples, 13 patients, and 1 cell line. In the normal controls 86% to 94% of interphase nuclei examined showed 2 hybridization signals with a mean value of 91.8% ± 3.2 (SD) and 1% to 4% of nuclei had 3 hybridization signals with a mean of 2.3 ± 1 (SD). The cut-off limit was set at 3 SDs for the detection of 3 signals (5.3%).
The presence of 3 hybridization signals could clearly be seen in the interphase nuclei of cells from patients carrying a 14q11 rearrangement (Fig 2F ). Eight patients with a 14q11 rearrangement were studied, these included 5 determined by karyotype and an additional 3 detected by metaphase FISH. In all cases except one a population of cells with 3 hybridization signals was present at a frequency higher than that observed for the normal controls (7.5 to 79%). It is noteworthy that the lowest incidence of abnormality observed in interphase nuclei was seen in UPN 937, in whom an inv(14) was only seen in 2.5% (3/120) of metaphases analyzed. In UPN 586 the interphase counts had a normal profile due to loss of the normal chromosome 14 from the clone containing the t(10; 14), and as such would not have been detected as a potential translocation if only interphase cells had been analyzed.
Six leukemic patients determined to have no 14q11 rearrangement by karyotype and metaphase FISH were also studied (Table 1). Four had a normal karyotype and two had clonal abnormalities, which included a trisomy 14 in one case. In 4 of these cases 1% to 2.5% of nuclei had 3 hybridization signals, within the normal range. In UPN 951 5.5% of interphase nuclei had 3 signals. This value is at the cut-off limit and may indicate the presence of a small clone not detected by metaphase FISH. UPN 639 with a trisomy 14 had 2 hybridization signals in 29% of nuclei and 3 in 61.5%, which would have suggested an underlying translocation had only interphase nuclei been examined.
DISCUSSION
In this study we have shown the effectiveness of a YAC which encompasses at least 70% of the TCRα/δ locus to detect chromosome rearrangements. Physiological TCRα rearrangement, as tested on αβ expressing peripheral T-lymphocytes and malignancies, did not result in loss or reduction of signal intensity. The YAC allowed detection of previously unsuspected TCRα/δ rearrangements in 4/13 (30%) immature T-lineage ALs, including in 3/10 T-ALLs, 1/1 LGL-AL, and 0/2 T-ALL/AML. It also clarified interpretation of all 4 mature T-cell proliferations analyzed.
YAC27BE8 can detect the presence of unsuspected abnormal clones.The YAC allowed detection of a 14q11 rearrangement in 3 of 8 (approximately 40%) patients with a normal karyotype; due to a t(10; 14)(q24; q11) in two and to an inv(14)(q11q32) in one. The relative minority of these clones and some bias in the selection of coexistant good quality normal metaphases for karyotypic analysis resulted in a failure to detect the abnormal clones in these cases. One advantage of using FISH to screen for the presence of specific abnormalities, as illustrated here, is that metaphases of poor quality can be easily examined.
The t(10; 14) has been reported to occur in approximately 5% in T-ALL but has also been described in T-NHL.17 The final incidence, following FISH analysis, in the present series was 3/12 AL (25%). The t(10; 14) involves the HOX-11 gene at 10q24.18 Molecular studies suggest that the frequency of aberrations involving HOX-11 in T-ALL is much higher than that detected by cytogenetic studies.19 In one series, 33% of pediatric T-ALLs showed HOX-11 mRNA expression, whereas only 8% demonstrated a t(10; 14).19 Our data would suggest that an abnormal clone may have been overlooked in some of these cases.
Hybridization of 5 cases (3 cell lines, 1 T-ALL, 1 T-ALL/AML) with an abnormal clone but no structural chromosomal 14 abnormalities only led to detection of a minor subclone containing an inv(14) in 3% of mitoses and 7.5% of interphase nuclei from the T-ALL. These results are not surprising since any abnormality involving this locus leads to rearrangement of virtually the whole of chromosome 14q, which is unlikely to be overlooked or misinterpreted on karyotypic analysis.
The significance of the minor inv(14) clones identified by FISH in 2/10 T-ALL is not clear, particularly since it was detected after PHA stimulation in one of the cases. Inversion of chromosome 14 is characteristically associated with AT and with T-PLL but it does occur rarely in apparent de novo T-ALL, with a frequency of less than 1%.2,20 It is the most frequently acquired sporadic rearrangement in human lymphocytes, occuring with a frequency between 1/1,000 to 1/10,000 and is also observed in the benign clones found in AT patients, with a frequency approximately 50 times higher than in the normal population.21 In the former situation the inv(14)(q11q32) is due to “illegitimate” rearrangement between the TCR α/δ and IgH loci whereas in the latter and in T-PLL the rearrangement involves TCR α/δ and the TCL1 locus, situated approximately 10 to 20 Mb centromeric to IgH at 14q32.6,22,23 Given the cytogenetic difficulty in distinguishing these two karyotypically similar inv(14)s with very different oncogenic potential, the use of the cos Ig10 probe described here may be useful for rapid partial characterization, since it will be moved to 14q11 by illegitemate rearrangement but not by TCRα/δ-TCL1 translocation. Unfortunately, insufficient material was available to determine which of these 2 possibilities occurred in the minor inv(14) subclones identified here. Their 2% to 3% incidence is much higher than for sporadic inversions but a diagnosis of AT is extremely unlikely, based on clinical criteria. The possible coexistence of a trisomy 8 and an inv(14) in one of the 2 T-ALLs is interesting in view of the known coexistence of these abnormalities in T-PLL.3 The clinical, morphological and immunological features of this case were, however, very different from T-PLL, insofar as the inv(14) and trisomy 8 occured in a 21-month-old child with TCRγδ+, morphologically immature blasts, whereas T-PLL classically presents as an accumulation of TCRαβ+ mature T cells in older individuals.
The 27BE8 YAC can be used to help characterization of complex chromosome 14 rearrangements.Hybridization with the 27BE8 YAC, often in conjuction with a centromeric IgH probe, clarified the karyotype in 5 cases. In one case an initial der(14) karyotypically distinct from an inv(14) (Gam) was found to be an inv(14), with a 14q32 breakpoint centromeric to IgH, thus probably implicating TCL1. A karyotypic inv(14) was shown to have undergone a more complex rearrangement. Analysis of 2 cases with karyotypic breakpoints at 14q32 showed that both involved insertion of unidentified material between TCR α/δ and IgH. Interestingly, the HPB-ALL line had undergone a probable complex translocation involving chromosomes 1, 5, and 14 and the T-ALL had lost one chromosome 5, suggesting that there may be similarities in these 2 abnormalities.
The 27BE8 YAC can be used for interphase analysis. Analysis of normal lymphocytes demonstrated a 5.3% normality (3 S.D.) cut-off for interphase analysis, in keeping with values determined for several FISH probes. In view of the aforementioned propensity for “illegitimate” TCR rearrangement it is unlikely that a lower cut-off could be achieved. The YAC confirmed the presence of 14q11 rearrangement in 7 of the 8 (87%) patients with 14q11 abnormalities detected by karyotype and/or metaphase FISH, with a good correlation observed between the incidence of abnormality in metaphase and interphase in all but two T-PLL cases. Both presented with characteristic extreme leucocytosis and demonstrated only one normal mitosis. In view of this, a lower than anticipated population of nuclei exhibiting 3 signals were observed. This may suggest the presence of nonproliferating subclones associated with loss of chromosome 14 material. Conversely, of the six cases without 14q11 rearrangement, four were normal by interphase analysis, one was borderline (5.5% of interphase nuclei), and one demonstrated 3 signals in more than 60% of nuclei. The false negative and the false positive cases were both due to chromosome 14 numerical abnormalities (monosomy 14 associated with a t(10; 14) in the false negative, and trisomy 14 in the false positive). These results show the need for caution in the interpretation of results of interphase analysis in the absence of karyotypic infomation. In these situations, interphase analysis should be confirmed by the use of probes from other regions of chromosome 14 to distinguish between aneuploidy and translocation. This would also potentially clarify borderline cases. If these limitations are taken into account, the use of this YAC for interphase analysis will allow assessment of other T-cell malignancies, particularly the mature malignancies, where routine karyotypic analysis is difficult due to poor growth, minor clonal populations, and consequent difficulty in distinguishing clonal from reactive mitoses. When sucessful, abnormalities of 14q11 are frequently observed in these disorders.24
In conclusion, use of the 27BE8 YAC doubled the detection rate and/or improved characterization of TCRα/δ rearrangements in the 23 cases analyzed. The seven additional TCRα/δ rearrangements identified were found in three cases with a normal karyotype, one with an isolated trisomy 8, and three with an ill-defined chromosome 14 abnormality. They included three t(10; 14) and four inv(14), which were minor in the 2 T-ALLs (see above) and complex in the AT and T-PLL cases. It is perhaps not surprising that inv(14) were commonly underestimated, given their relatively subtle nature, their occurrence in minor subclones and/or in cases with complex abnormalities. Only larger scale screening for these abnormalites will determine the molecular type and the significance of inv(14) in de novo T-ALL. The highest increment in detection of chromosome 14q11 abnormalities (approximately 40%) was observed in the 8 T-lineage AL with an apparently normal karyotype. Overall, 25% of T-ALL are reported to have a normal karyotype. Our results suggest that it is in these cases that FISH analysis with the 27BE8 YAC will have the greatest impact. The relatively high frequency of TCRα/δ rearrangements in T-cell malignancies overall indicates that this YAC will also be useful as a first line approach in the analysis of all T-lymphproliferative disorders. These approaches may well lead to the detection of previously unsuspected translocations and thus, in concertation with the rapid expansion of human genome data availability, to the identification of novel genes involved in T-lymphoid oncogenesis.
ACKNOWLEDGMENT
We thank Sylvie Nusbaum, Gaelle LeGuyader, and Corrine Millien for their technical assistance and Martine Netter for photographic assistance.
Supported by the Direction de la Recherche Clinique de L'Assistance Publique-Hôpitaux de Paris (Contract A0A94061); the Fondation de France/Fondation Contre la Leucémie, Paris, France; the Fondation pour la Recherche Médicale, Paris, France; the Ligue Nationale contre le Cancer (Comité de Paris); and the Leukemia Research Fund of Great Britain, London.
Address reprint requests to Elizabeth A. Macintyre, MD, PhD, Laboratoire d'Hématologie, Tour Pasteur, Hôpital Necker-Enfants Malades, 149, rue de Sèvres, 75743 Paris, Cédex 15, France.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal