Abstract
The onset of IgH transcription and rearrangement is a defining characteristic of the progenitor population in which B-lineage commitment occurs. These features were used to better define the earliest stage of B-cell commitment in humans and to determine if these stages differ as a function of human ontogeny. Fetal and adult bone marrow mononuclear cells were sorted into B-lineage subpopulations on the basis of surface expression of the stem cell marker CD34, the pan-B–cell marker CD19, and IgM and analyzed for transcription and rearrangement of the IgH locus. The locus was found to be transcriptionally active before surface expression of CD19, as indicated by the presence of germline Iμ, Cμ, and DHQ52 transcripts in the CD34+ CD19− subpopulation. Transcripts from IgH alleles that had undergone DJCμ rearrangements were also detected in the CD34+ CD19− subpopulation. Within this subpopulation, low levels of DXP-containing DJCμ transcripts were detected in both fetal and adult cells. Although DHQ52 DJCμ transcripts were abundant in fetal CD34+ CD19− cells, they were not detected in cells of the same phenotype derived from adult bone marrow. In both fetus and adult, VH3-and VH6-containing VDJCμ transcripts were detected only in the CD19+ subpopulations. These data indicate that transcription of DHQ52-JH and DXP-JH rearrangements differs during fetal and adult B lymphopoiesis. Moreover, in both fetus and adult, transcription of unrearranged components of the IgH locus and DJ rearrangements can proceed before the surface expression of CD19.
THE HALLMARK OF B-lineage development is the ordered rearrangement of the V, D, and J gene segments that encode the variable domains of Ig. This B-lineage–restricted rearrangement process typically initiates in the heavy chain locus (IgH), beginning in the two DH loci, which together include approximately 30 DH gene segments. All but 1 of the functional DH gene segments have been mapped to a locus 20 to 60 kb upstream of the JH gene segments; the exception is the DHQ52 gene segment that is located less than 100 bp 5′ of JH 1. The DH locus can be subdivided into four 9-kb subregions, each of which contains at least one member of the seven remaining DH families (DXP, DLR, DM, DN, DK, DA, and DIR).1 Rearrangement of one of these 30 DH gene segments to one of the six JH elements creates a DJ join, which is capable of being transcribed. Next, 1 of approximately 50 functional VH gene segments,2 located 5′ of the DH locus, rearranges to the newly formed DJ join. In-frame rearrangement of a V to the DJ join creates a complete Ig heavy chain variable domain. After in-frame rearrangement of one of the two light(L) chain loci (κ or λ), translation of the heavy and light chain mRNA allows production of IgM molecules that, when expressed on the surface of the developing lymphocyte, define the B cell.
Use of DH gene segments is developmentally regulated during human ontogeny. In early fetal tissues, DHQ52 is found in nearly 40% of IgH transcripts, whereas DHQ52 is found in less than 1% of adult IgH transcripts.3-6 In contrast, members of the DXP and DLR families are found in less than 10% of fetal IgH transcripts, but contribute to more than 40% of IgH transcripts from adult B cells. It is unclear if these differences in DH use are the product of differential activation of the JH proximal DHQ52 locus, the result of developmentally regulated differences in DH rearrangement frequencies, or the product of antigen receptor influenced selection of the expressed repertoire.6
Although early B-lineage progenitor cells are most easily distinguished by the rearrangement status of their Ig loci, it is difficult to isolate normal subpopulations of B-lineage progenitors solely on this basis, because few rearrangement intermediates are expressed as protein products on the cell surface. However, a number of other surface molecules that are expressed at discrete stages of B-cell development can be used to separate B-cell precursors into a series of subpopulations via flow cytometry.7
The surface coexpression of the pan-B–cell marker CD19 and IgM defines B lymphocytes. Cells expressing CD19 and very low levels of surface IgM comprise a population of late pre-B cells, defined by lack of terminal deoxynucleotidyl transferase (TdT) expression in the nucleus and the presence of the surrogate light chain complex on the cell surface.8 Early pre-B cells, which express cytoplasmic μ-chain and nuclear TdT, can be identified by surface expression of CD19 in the absence of surface IgM. Coexpression of surface CD19 and the stem cell marker CD34 characterizes a population of bone marrow pro-B cells that are interleukin-7 responsive9,10 and are the earliest known committed B-lineage cells. Finally, cells with forward and side light scatter characteristics of lymphocytes that express CD34 but lack any lineage-specific markers such as CD19 comprise a heterogeneous population of lymphocyte precursors of multiple lineages. The level of lineage commitment in these cells is not well defined.11
We have analyzed IgH transcription and rearrangement in sorted fetal and adult bone marrow B-lineage cells to define the commitment status of normal human bone marrow B-lineage subpopulations. Each subpopulation was tested by reverse transcriptase-polymerase chain reaction (RT-PCR) for the presence of a set of germline and rearranged IgH transcripts representative of different stages in IgH rearrangement, and a subset of DJ rearrangements was examined at the DNA level. Our studies indicate that germline IgH transcription and DJCμ rearrangements occur in CD34+ cells that do not express surface CD19, whereas VDJCμ transcription correlates with CD19 surface expression. In addition, our studies indicate that fetal and adult CD34+ B-lineage progenitors differ in terms of the pattern of transcription of the IgH locus.
MATERIALS AND METHODS
Fluorescence-activated cell sorting (FACS) and separation of B-lymphocyte progenitor subpopulations.Fetal bone marrow (FBM) cells were flushed from the long bone specimens of four fetuses of 18 to 22 weeks of gestation (determined by fetal foot length), and adult bone marrow (ABM) was derived from the resected ribs of three kidney donors (30 to 48 years old) in accordance with the University of Alabama at Birmingham Human Use Institutional Review Board. Each sample was independently sorted and analyzed. The mononuclear cell population was first purified by Ficoll-Hypaque centrifugation and then divided into two aliquots of equal numbers of cells. One aliquot was stained with 4G7 anti-CD19 (γ1κ) (Becton Dickinson, Mountain View, CA) and 8G12anti-CD34 (γ1κ) (Becton Dickinson). The other aliquot was stained with the anti-CD19 and anti-sIgM (Southern Biotechnology Associates, Birmingham AL). The following subpopulations were collected using a FACS Star Plus instrument (Becton Dickinson): CD34+CD19−, CD34+ CD19+, CD19hi IgM−, CD19+ IgMlo, and CD19+ IgM+. Postsort analysis was performed using a FACS Scan instrument (Becton Dickinson).
Isolation of total RNA and cDNA synthesis.Total RNA was isolated from each bone marrow subpopulation using Tri-Reagent (Molecular Research Center, Inc, Cincinnati, OH) according to the manufacture's protocol.12 Using standard protocols,13 one third of each RNA sample was used to synthesize first-strand cDNA with AMV reverse transcriptase (Boehringer Mannheim, Indianapolis, IN). For each cDNA preparation, a control synthesis reaction was performed without reverse transcriptase to rule out contamination of the RNA or cDNA reagents. Two separate cDNA preparations were made with the RNA from each sorted sample. Each cDNA preparation was analyzed by PCR as described below.
PCR amplification for IgH transcripts.Iμ,14 germline Cμ,3 germline DHQ52,15 rearranged DHQ52-JH-Cμ, rearranged DXP-JH-Cμ, VH6-DJCμ, and VH3-DJCμ transcripts were amplified from 1 μL of the cDNA reaction product from each lymphocyte progenitor subpopulation. PCR was performed using a Perkin-Elmer Cetus model 480 thermal cycler (Perkin-Elmer, Norwalk, CT). Each PCR amplification was performed under the following reaction conditions: 38.5 μL dH2O, 5 μL of 10× PCR buffer (GIBCO/BRL, Gaithersburg, MD), 1.5 μL of 50 mmol/L MgCl2, 1 μL of 10 mmol/L dNTPs, 1 μL of 20 μmol/L each primer, and 2.5 U Taq DNA polymerase (GIBCO/BRL). Each reaction underwent 30 cycles of amplification composed of denaturation at 95°C for 1 minute, primer annealing for 1 minute, and extension for 1 minute at 72°C. A final extension was performed at 72°C for 7 minutes. The primer combinations used to amplify cDNA, in separate reactions, were as follows: Iμ (5′-TCT GAT AGA GTG GCC TTC ATT-3′ and 5′-AAT TCT CAC AGG AGACGC GAG-3′ ),14 germline Cμ (5′-GGC TTC ACA TTC AGG TAT GCA A-3′ and 5′-GCCCAG ACT GTC ATG GCT ATC A-3′ ),3 DHQ52 (5′-GAG CTG AGAACC ACT GTG-3′ and 5′-GGA GAA AGT GAT GGA GTC GGG-3′ ),15,16 DXP 1′ (5′-AATGAA TTC TGT GTC ACT GTG GTA TTA-3′ and 5′-GGA GAA AGT GAT GGA GTC GGG-3′ ),17 VH6 family (5′-AGA ATT CTC GTC TCC TTCCTC ATC-3′ and 5′-GGA GAA AGT GAT GGA GTC GGG-3′ ),15 and VH3 family (5′-AGA ATT CGT TTG GGC TGA GCT GGC T-3′ and 5′-GGA GAA AGT GAT GGA GTC GGG-3′ ).3 Actin mRNA was amplified as a control for the reliability of the cDNA synthesis, using the primers (5′-GTG GAC TTG GGA GGA GGA CTC TGG G-3′ and 5′-GCGGGA AAT CGT GCG TGA CAT T-3′ ).18 PCR conditions were such that the amplifications were performed in the linear range, and each primer pair, in control experiments, exhibited similar limits of detection (6 pg of cDNA). For each cDNA preparation, at least two PCR reactions for each primer pair were performed. The relative amplification efficiency of each transcript was similar in each experiment.
PCR amplification of genomic rearrangements.Cells from sorted subpopulations of fetal bone marrow mononuclear cells were lysed directly in PCR buffer containing 10 mmol/L Tris ⋅ HCl, pH 8.0, 1.5 mmol/L MgCl2 , 50 mmol/L KCl, 0.45% NP-40, and 0.45% Tween-20 at a concentration of 5,000 cells per 50 μL of buffer. This preparation was treated with 10 mg/mL of proteinase K (Boehringer Mannheim) at 56°C for 1 hour, followed by heat-inactivation at 90°C for 15 minutes.19 Fifty microliters of this preparation was used for PCR amplification (Perkin-Elmer model 480). PCR reaction conditions were as described above. The primers used for amplification of genomic DNA were DHQ52 (5′-CCC CCTACC AGC CGC AGG GTT T-3′ and 5′-GCA GAA AAC AAA GGC CCT TAGA-3′ )16 and DXP 1′ (5′-AAT GAA TTC TGT GTC ACTGTG GTA TTA-3′ and 5′-GCA GAA AAC AAA GGC CCT TAG A-3′ ).17
Southern analysis.Ten microliters of each PCR reaction product was separated on a 2% agarose gel, followed by transfer to a nylon membrane (Nytran; Schleicher and Schule, Keene, NH) using standard protocols.13 The membranes were probed with 32P-γ-ATP end-labeled oligonucleotides complementary to sequences internal to the PCR primer set used. These probes were as follows: for DHQ52, DXP1′ VH6 and VH3 cDNA amplifications (5′-AAT TCT CAC AGG AGA CGA G-3′ ),15 Iμ (5′-GGT AGT CCG GGA GAC CCAAA-3′ ),14 and germline Cμ (5′-GTGCAG CAG GGC AGC CAG CTG AAT-3′ )3; for DHQ52 and DXP genomic DNA amplifications (5′-CCC TTG GCC CCA GAC GTC C-3′ )16; and for actin (5′-CCG GCC CCT CCA TCG TCC AC-3′ ).18 The membranes were washed in 1× SSC and 0.1% sodium dodecyl sulfate, followed by exposure to x-ray film (Eastman Kodak, Rochester, NY) for 24 hours, unless otherwise indicated.
Quantitation of PCR products.The density of each amplification detected by autoradiography was determined by a Pharmacia LKB Ultrascan XL densitometer (Pharmacia LKB Biotechnology, Uppsala, Sweden) using the Gelscan XL software, version 2.1. To correct for differences in mRNA concentrations, the density was normalized by dividing the density of the actin band for each sample. To correct for differences in exposure, the relative expression of each message was determined by dividing the density of band corresponding to each individual developmental stage (ie, CD34+ CD19−; CD34+ CD19+, etc) by the density of the band with the highest density, leaving that band with an arbitrary relative expression of 1 and and each band of lesser density with a fractional value.
RESULTS
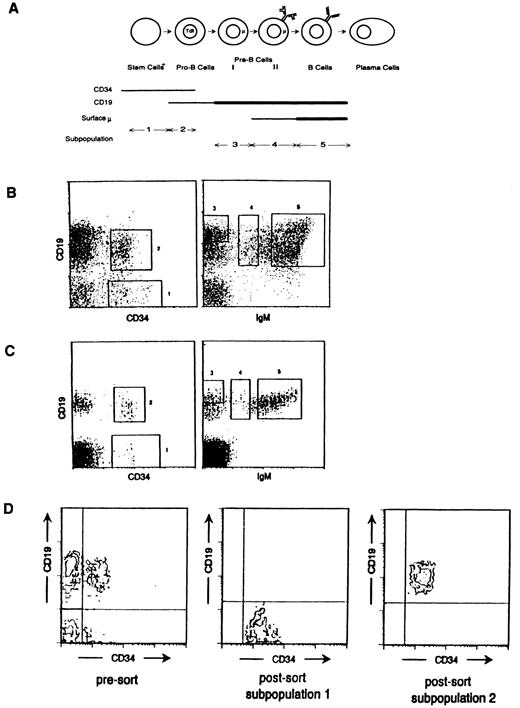
Germline transcription of the IgH locus occurs before CD19 expression.FBM and ABM was sorted into five B-lineage subpopulations on the basis of CD34, CD19, and IgM surface expression (Fig 1A). Cells with light scatter characteristics of lymphocytes expressing CD34, but not CD19, comprised subpopulation 1. This subpopulation contains a heterogeneous mixture of lymphoid progenitor cells, some of which may be committed to the B lineage.11 Subpopulation 2, expressing both CD19 and CD34, is comprised primarily of pro-B cells. The first pre-B–cell compartment, subpopulation 3, was defined as CD19hi, IgM−, and CD34−, although only two-color sorts were performed. The second pre-B–cell compartment, subpopulation 4, was defined as CD34−, CD19+, and IgMlo and contains cells that express the surrogate light chain receptor complex.8 Finally, mature B cells, subpopulation 5, are comprised of cells that coexpress CD19 and high levels of surface IgM. Typical collection gates for the sorted bone marrow subpopulations are shown in Fig 1B and C. Postsort FACS analysis of each of the collected populations consistently showed 99% purity as illustrated for FBM subpopulations 1 and 2 in Fig 1D.
Bone marrow B-lineage subpopulations collected on the basis of CD34, CD19, and IgM surface expression. (A) Cartoon illustrating each of the five subpopulations collected and the B-lineage developmental stage that each represents. Thick lines indicate higher levels of surface expression of the respective B-cell marker. (B) Typical collection gates used to separate the FBM subpopulations are shown in these FACS profiles of 22-week-old FBM stained with anti-CD34, anti-CD19, and anti-IgM as described in the Materials and Methods. (C) Typical collection gates used to separate the ABM subpopulations are shown in these FACS profiles of 30-year-old ABM stained with anti-CD34, anti-CD19, and anti-IgM as described in the Materials and Methods. (D) Typical postsort FACS analysis of the fetal CD34+ CD19− subpopulation illustrating 99% purity in each subpopulation. Postsort analysis of the other fetal and adult subpopulations collected also indicated 99% purity (data not shown).
Bone marrow B-lineage subpopulations collected on the basis of CD34, CD19, and IgM surface expression. (A) Cartoon illustrating each of the five subpopulations collected and the B-lineage developmental stage that each represents. Thick lines indicate higher levels of surface expression of the respective B-cell marker. (B) Typical collection gates used to separate the FBM subpopulations are shown in these FACS profiles of 22-week-old FBM stained with anti-CD34, anti-CD19, and anti-IgM as described in the Materials and Methods. (C) Typical collection gates used to separate the ABM subpopulations are shown in these FACS profiles of 30-year-old ABM stained with anti-CD34, anti-CD19, and anti-IgM as described in the Materials and Methods. (D) Typical postsort FACS analysis of the fetal CD34+ CD19− subpopulation illustrating 99% purity in each subpopulation. Postsort analysis of the other fetal and adult subpopulations collected also indicated 99% purity (data not shown).
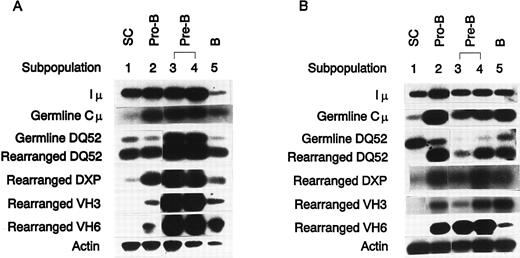
Each of the five B-lineage subpopulations collected was analyzed for Iμ, Cμ, and DHQ52 germline transcripts (Fig 2) using an RT-PCR assay as described in the Materials and Methods. All three transcripts were detected in every B-lineage subpopulation tested, including the CD34+ CD19− subpopulation 1 (Fig 3), from both FBM and ABM.
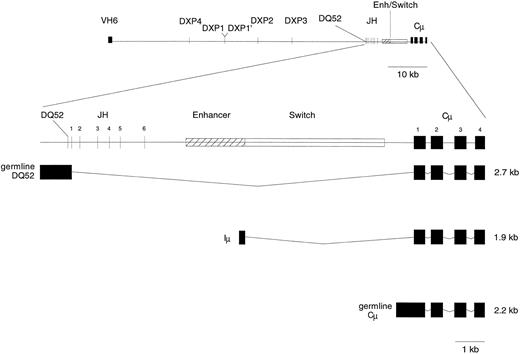
Map of the human VH6-DJ-Cμ locus. Shown is a 400-kb region extending from VH6 through Cμ. The approximate positions of the DXP gene segments are denoted within each of the four DH tandem repeats in the distal DH locus, labeled as DXP1 (D21/7) and DXP1′ (D21/0.5), DXP2, DXP3, and DXP4. The expanded portion indicates the location of DHQ52 within the proximal DH locus, the six JH gene segments, and the intronic sequence between JH and Cμ that includes the proximal IgH enhancer. The composition of germline transcripts from DHQ52, Iμ, and germline Cμ is illustrated. For the transcripts shown, boxes indicate transcribed regions and lines indicated areas that are removed due to RNA processing.
Map of the human VH6-DJ-Cμ locus. Shown is a 400-kb region extending from VH6 through Cμ. The approximate positions of the DXP gene segments are denoted within each of the four DH tandem repeats in the distal DH locus, labeled as DXP1 (D21/7) and DXP1′ (D21/0.5), DXP2, DXP3, and DXP4. The expanded portion indicates the location of DHQ52 within the proximal DH locus, the six JH gene segments, and the intronic sequence between JH and Cμ that includes the proximal IgH enhancer. The composition of germline transcripts from DHQ52, Iμ, and germline Cμ is illustrated. For the transcripts shown, boxes indicate transcribed regions and lines indicated areas that are removed due to RNA processing.
IgH transcripts in human bone marrow B-lineage subpopulations. Iμ, germline Cμ, germline DHQ52, and rearranged DHQ52 DJ, DXP DJ, VH3, and VH6 transcripts were detected as described in the Materials and Methods and the Results. (A) FBM IgH transcripts. (B) ABM IgH transcripts. Shown are results representative of repeated PCR analysis of four independently sorted FBM samples and three independently sorted ABM samples.
IgH transcripts in human bone marrow B-lineage subpopulations. Iμ, germline Cμ, germline DHQ52, and rearranged DHQ52 DJ, DXP DJ, VH3, and VH6 transcripts were detected as described in the Materials and Methods and the Results. (A) FBM IgH transcripts. (B) ABM IgH transcripts. Shown are results representative of repeated PCR analysis of four independently sorted FBM samples and three independently sorted ABM samples.
DJH rearrangement occurs before CD19 surface expression.Rearranged DHQ52 transcripts were detected in all of the sorted FBM B-lineage subpopulations, including the CD34+ CD19− subpopulation 1 (Fig 3A). In contrast, DHQ52 DJ-rearranged transcripts were not detected in ABM-derived subpopulation 1 cells. Instead, the onset of these transcripts coincided with surface expression of CD19 (Fig 3B). Using a similar PCR-based assay, we were able to detect abundant DXP DJ transcripts in subpopulations 2 through 5, and a very low level of these transcripts persisted in the CD34+ CD19− subpopulation 1 cells regardless of gestational age.
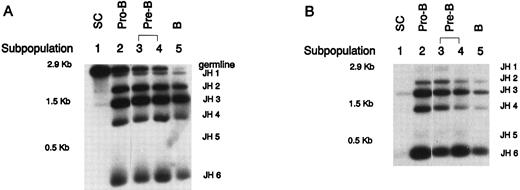
DHQ52 and DXP gene segments rearrange to different JH segments in fetal CD34+ CD19− cells.Genomic DNA isolated from each FBM subpopulation was subjected to a PCR assay designed to detect DHQ52 or DXP to JH rearrangements. Such an assay yields a ladder of different-sized PCR products depending on the specific JH element involved in the rearrangement. As can be seen in Fig 4, both DXP and DHQ52 rearrangements were detected in genomic DNA from fetal subpopulation 1 cells. Interestingly, within this CD34+ CD19− population, DHQ52 rearranged with JH1, JH2, and JH3, whereas DXP appears to be restricted to JH3 and JH6 rearrangements. These two DH segments rearrange with the JH segment in a stochastic manner in more mature B-lineage subpopulations; except for DHQ52, which appears to rearrange to JH5 infrequently throughout fetal B-lineage development.
PCR assay to detect IgH DJ rearrangements in sorted FBM B-lineage subpopulations. (A)FBM DHQ52 DJ rearrangements. (B) FBM DXP DJ rearrangements. No germline band is observed in the DXP rearrangements lane because the primers anneal to sequences that are widely separated (20 kb) in unrearranged DNA. Shown are results representative of repeated PCR analysis of three independently sorted FBM samples and two independently sorted ABM samples.
PCR assay to detect IgH DJ rearrangements in sorted FBM B-lineage subpopulations. (A)FBM DHQ52 DJ rearrangements. (B) FBM DXP DJ rearrangements. No germline band is observed in the DXP rearrangements lane because the primers anneal to sequences that are widely separated (20 kb) in unrearranged DNA. Shown are results representative of repeated PCR analysis of three independently sorted FBM samples and two independently sorted ABM samples.
Because the PCR for DHQ52-JH DNA rearrangement will produce an approximately 3-kb germline product in the absence of rearrangement, it is possible to use this as a measure of the extent of D to JH rearrangement within cells. As indicated by the decreasing intensity of the germline band with B-lineage maturation, the relative number of cells within a given subpopulation bearing DHQ52 to JH rearrangements on at least one allele increases with maturation of the B-lineage.
Transcription of VDJCμ rearrangements was detected only in cells expressing surface CD19.The VH6 gene segment, V6-1, is the most JH proximal gene segment in the human genome and is used more frequently in the fetus than in the adult.15 The VH3 family is the largest family in humans, representing approximately 40% to 45% of the functional repertoire. Individual VH3 gene segments, such as V3-23 and V3-30.3, are also overrepresented in fetal tissues.6 Each bone marrow B-lineage subpopulation was tested for the presence of VH3 and VH6 family VDJCμ transcripts (Fig 3). VH3 and VH6 VDJCμ transcripts were only detected in subpopulations 2 through 5, all of which are characterized by the surface expression of CD19.
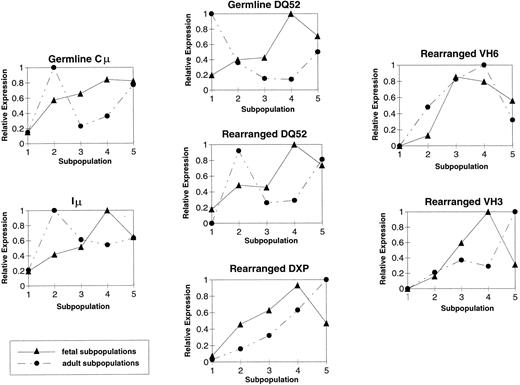
IgH transcript levels differ between fetal- and adult-derived B-lineage precursors.Although the PCR conditions were within the linear range of amplification, the limited number of cells available precluded the repetitive amplifications that would have been necessary to perform a rigorous quantitative analysis of mRNA expression. However, it is possible to derive a semiquantitative overview of the relative changes in mRNA abundance by first measuring the density of each band, adjusting the density for total mRNA abundance by taking into account the density of the actin band, and then comparing the density of each band in each series of amplifications (ie, fractions 1 through 5 for each mRNA type) relative to the most intense band of that series. In the case of the amplification of germline DHQ52 transcripts from ABM, for example, the relative intensity for the amplified product from fraction 1 would be 1.0, and each band thereafter would represent a fraction of that value.
By this measure, in addition to the qualitative differences in DHQ52-containing transcripts, the pattern of relative abundance of the other IgH transcripts was also observed to differ between the fetal and adult B-lineage subpopulations (Fig 5). In the fetus, the abundance of the germline Cμ transcripts gradually increased as the B-lineage cells matured. Fetal Iμ RNA abundance steadily increased between the stem cell stage (fraction 1) and the late pre-B stage (fraction 4), only to decline at the B-cell stage (fraction 5). This pattern of steady increase with a decline in fraction 5 also characterized fetal germline DHQ52, rearranged DHQ52, and DXP-containing Cμ transcripts. In contrast, in the adult, the highest level of sterile DHQ52 RNA abundance was in the sample enriched for stem cells (fraction 1), followed by a burst of germline Cμ, Iμ, and rearranged DHQ52 RNA abundance in fraction 2, the pro-B–cell stage (Fig 5). These differing patterns may reflect alterations in IgH locus accessibility between fetal- and adult-derived cells.
Quantitation of RT-PCR IgH products in FBM and ABM. Shown are graphs depicting the average relative abundance of individual IgH transcripts detected in the fetal (N = 3) and adult (N = 2) subpopulations. (▴) Fetal samples. (•) Adult samples.
Quantitation of RT-PCR IgH products in FBM and ABM. Shown are graphs depicting the average relative abundance of individual IgH transcripts detected in the fetal (N = 3) and adult (N = 2) subpopulations. (▴) Fetal samples. (•) Adult samples.
The pattern of VH3 abundance also differed between fetal and adult B-lineage development. Fetal VH3 transcripts appeared to increase sharply and to peak at the late pre-B–cell stage (fraction 4), following the same pattern seen for the IgH transcripts discussed above. However, in the adult, VH3 transcripts were of low abundance in the early stages when compared with the B-cell stage (fraction 5). In contrast, the relative abundance of VH6 mRNA steadily increased through the pre-B–cell stage only to decrease at the B-cell stage (fraction 5) in both the fetus and adult, suggesting either a reduction in the expression of VH6 mRNA per cell or a loss of cells that express VH6 mRNA.
DISCUSSION
The progenitor subpopulation in which B-lineage commitment first occurs has yet to be identified, but it is presumed that a defining characteristic of this subpopulation will be the onset of IgH rearrangement. It has been postulated that activation of the IgH locus for rearrangement may begin with activation of the IgH enhancer.20 Under this hypothesis, germline transcription of the IgH locus could target recombinase proteins to gene segments accessible for rearrangement.1
One indication of the transcriptional activation of the IgH locus is the appearance of unrearranged μ-containing transcripts, one of which initiates within the IgH enhancer itself (Iμ)14 and the second that initiates 5′ of the first Cμ exon (germline Cμ).3 A third germline transcript has been described that initiates upstream of the DHQ52 gene segment, reads through the intron between the DH and JH1, and then is spliced to the CH1 exon of Cμ.15 We were able to detect all three unrearranged transcripts in all of the subpopulations of cells that expressed surface CD19, including cells that also expressed surface IgM. Thus, germline transcription of Iμ and Cμ appears to be an active process throughout normal human B-cell development provided that the necessary promoter elements have not been deleted by rearrangement of the locus. In contrast to the mouse, some B cells in human retain one allele in the germline configuration.21-23 Germline transcription of the third sterile product, germline DHQ52, also appears to persist in B-lineage cells that have maintained one heavy chain allele in an unrearranged state.
Detection of Iμ, germline Cμ, and germline DHQ52 transcripts in the CD34+ CD19− subpopulation raised the possibility that at least a subset of the CD19− progenitor cells in FBM and ABM may be capable of initiating IgH gene segment rearrangement. The presence of DJCμ rearrangements and detection of both DHQ52 and DXP rearranged DJCμ transcripts in this CD34+ CD19− compartment confirm that this is the case. In related studies, Rag1, Rag2, and TdT transcripts have been detected in this early compartment, indicating that genes necessary for recombination are transcribed during the CD34+ CD19− stage.24 The finding of DJCμ rearrangements and IgH transcripts suggests that a percentage of CD34+CD19− cells may be committed to the lymphoid lineage. However, the precise level of commitment is difficult to determine because DJCμ rearrangements could either reflect one of the earliest stages of B-lineage commitment or DJCμ rearrangements could occur in non–T-lineage or non–B-lineage restricted lymphoid progenitors.25 In companion studies, IgH germline and DJCμ transcripts were not detected in populations of fetal human thymocytes depleted of CD34+- and/or CD19+-bearing cells and sorted on the basis of CD4 and CD8 expression. However, the DNA in each thymocyte subpopulation (CD4−CD8−, CD4+CD8+, CD4+CD8−, and CD4−CD8+) contained both DHQ52 and DXP DJCμ rearrangements.26 27 These findings suggest that CD34+ CD19− cells actively transcribing DJH joins may be restricted to the B-lineage pathway.
PCR analysis of the genomic DNA from fetal CD34+CD19− subpopulations showed that the pattern of JH usage was restricted within this subpopulation. Rearrangements involving DHQ52 predominantly involved JH1, JH2, or JH3, whereas most, if not all, DXP family gene segment rearrangements involved JH3 or JH6. This bias was not seen in cells representing later stages of B-lineage maturation. The observed pattern of JH usage may reflect preferential association between the recombination signal sequences of the respective DH families, as has been postulated in the mouse.28 29 Alternatively, either the proximity of the DHQ52 gene segment to the 5′ most JH gene segments or the state of accessibility of the DJ locus may influence the likelihood of gene segment rearrangement in this early subpopulation of B-lineage cells.
CD19 is a B-lineage associated surface molecule that has been shown to play a role in modulating signal transduction through the B-cell receptor complex. Its role in early B lymphopoiesis before the expression of surface IgM is not well defined.9,30-33 The surface expression of CD19 in transformed cell lines that have not undergone IgH rearrangement and the ability of CD19+ progenitor cells to undergo differentiation into B cells in vitro has led to the hypothesis that the appearance of CD19 on the surface membrane is a major marker for B-lineage commitment.7,34 The presence of DJ rearrangements in bone marrow progenitors that lack CD19 suggests that surface CD19 expression is not essential for the initial activation of the human IgH locus. This finding is in agreement with studies in mice in which the CD19 gene has been inactivated by gene targeting. In these mice, there is no apparent impairment of early B-lineage differentiation and gene segment rearrangement.35,36 It has also been shown that CD19 acts in conjunction with interleukin-7 to modulate the expression of recombinase associated genes in human CD34+ CD19+ B-progenitor cells.10 In this present study, VDJ transcripts were detected only in cells that expressed surface CD19. Thus, it is possible that CD19 may have a role in modulating the completion of IgH rearrangement.
Our studies were designed to measure major differences in mRNA abundance; thus, the quantitation of IgH transcripts by densitometry represents only a rough estimate of the changes in RNA abundance during B-lineage development. With this caveat in mind, quantitative analysis of changes in transcript abundance indicates additional differences in the activity of the IgH locus between fetal and adult B-lineage cells. First, the striking difference in rearranged DHQ52 expression between FBM and ABM was complemented by differences in the relative expression of germline DHQ52 transcripts. In the adult, germline DHQ52 transcription is most intense in the subpopulation enriched for stem cells (fraction 1). In these cells, rearranged DHQ52 transcripts could not be detected. Although transcription of germline DHQ52 appears to be an early step in B-lineage differentiation, the absence of rearrangements indicates that accessibility of the locus to the transcription machinery is not sufficient to induce recombination.
In the fetus, the relative abundance of the sterile IgH constant region transcripts (germline Cμ and Iμ) increased gradually with lineage maturation, peaking at the late pre-B cell stage (fraction 4). In the adult, there was a striking increase in constant region transcript abundance between the stem cell (fraction 1) and the pro-B–cell stage (fraction 2). This could reflect a more general difference between fetal and adult B-lineage cells in the accessibility of the region surrounding the enhancer elements between JH and Cμ.37 38 Fetal cells may gradually activate the IgH enhancer, opening the locus slowly during B-lineage development; whereas, in the adult, the locus might open more abruptly at the pro-B–cell stage (fraction 2), responding more rapidly to full activation of the enhancer at an earlier stage of development. Differential activation of the JH-Cμ locus versus the major DH locus could explain why the burst of Iμ and germline Cμ transcripts in the adult was accompanied by a similar increase in abundance of JH-proximal rearranged DHQ52-containing transcripts, whereas rearranged DXP-containing transcripts did not follow this pattern.
In both fetal and adult cells, transcripts containing VH6 Cμ rearrangements increase in abundance through the pre-B–cell stage and then decline in cells that express surface Ig. This loss of mRNA abundance of the developmentally regulated VH6 gene segment6 is reminiscent of the pattern defined by the loss of another developmentally regulated VH gene segment, murine VH81X, which tends to encode self-reactive antibodies.6,39 To date, many VH6-containing antibodies have also been found to be self reactive40 and the loss of VH6 expression may reflect the deletion of self-reactive B cells.
Rearranged VH3 transcripts increase in both fetal and adult B-lineage cell populations. However, in the fetus, VH3 RNA abundance decreases in fraction 5, the sIgM+ stage, following the same pattern seen in the other IgH transcripts. In the adult, VH3 abundance is highest in the mature B-cell stage. Recent studies have documented an accumulation of a population of sIgM+ B cells in the adult that has the characteristics of a memory B-cell population, including evidence of somatic mutation in the Ig variable domains.41 It is possible that the increase in VH3 abundance seen in fraction 5 reflects the contribution of memory cells that have been selected by antigen.
Previous studies in a number of species have shown that the composition of the heavy chain CDR3 (formed by VDJ joining and N region addition) is regulated in such a way as to limit the diversity of the heavy chain CDR3 interval during fetal life.6 In humans, the repertoire of fetal heavy chain CDR3 intervals is enriched for the use of a unique DH gene segment, DHQ52, whereas this gene segment is rarely used in adults. The mechanism(s) that regulates the use of specific DH gene segments is unknown. One hypothesis holds that the differences in DH use are the product of antigen-receptor–based selection. Alternatively, the differences in DH use may be operating at the level of rearrangement. In this study, we compared IgH gene segment transcription and rearrangement as a function of developmental age within the same primary lymphoid organ, the bone marrow. The most striking differences in IgH transcription were detected within the CD34+ CD19− bone marrow subpopulation, in which IgH rearrangement intermediates are not thought to be expressed on the cell surface. We detected high levels of rearranged DHQ52 DJ transcripts in mid-second trimester fetal CD34+ CD19− cells, whereas these transcripts were undetectable in the equivalent subpopulation in ABM. In contrast, rearranged DXP DJ transcripts were present at a low level in this subpopulation in both fetal and adult samples. These data indicate that the developmental differences in the use of the DHQ52 gene segment are already apparent in the earliest stages of B-cell development before surface expression of the antigen receptor. Early rearrangement of the DHQ52 gene segment may serve as a marker for the first wave of fetal B-lineage progenitors in the human.
ACKNOWLEDGMENT
We thank Dr M.D. Cooper for helpful discussions and Dr G.E. Wu for critical reading of the manuscript. F.E.B. thanks The Wellesley Hospital Research Institute for support during the final preparation of this manuscript.
Supported in part by National Institutes of Health Grants No. AI34568, AI07051, AI23694, AI39816, and AI34568.
Address reprint requests to F.E. Bertrand III, The Wellesley Hospital Research Institute, 160 Wellesley St E, Toronto, Ontario M4Y 1J3, Canada.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal