Abstract
Glanzmann's thrombasthenia (GT) is a recessive autosomal bleeding disorder characterized by abnormal platelet aggregation due to a qualitative or quantitative defect of the glycoprotein (GP) IIb-IIIa complex (integrin αIIbβ3). We describe a new mutation in the GPIIIa gene responsible for type I GT in a consanguineous Algerian family. A discordance between phenotyping and genotyping of the GPIIIa-related HPA-1 platelet alloantigen system in three family members heterozygous for the disease suggested a genetic defect in the GPIIIa gene and a normal GPIIb gene. Sequence analysis of amplified genomic DNA fragments showed a 6-bp deletion in exon 7 of the GPIIIa gene resulting in the amino acid deletion/substitution (Ile325Pro326Gly327 → Met) and creating a new BspHI restriction site. Expression of the mutated integrin β3 subunit cDNA in Chinese hamster ovary cells showed that the cDNA gene was transcribed into a full-length β3 protein with an apparent molecular weight identical to wild-type β3 and accumulated as a single-chain molecule in the cell cytoplasm. The absence of heterodimeric complex formation of the mutant β3 protein with endogeneous αv was shown by immunoprecipitation experiments, intracellular immunofluorescent labeling, and a semiquantitative enzyme-linked immunosorbent assay using the αvβ3 complex-specific monoclonal antibodies LM609 and 23C6. Substitution of the methionine residue by a proline, present at position 326 of wild-type β3, did not restore the ability of the recombinant mutant β3 protein to associate with αv, suggesting that the Ile-Pro-Gly motif is located in a β3 domain important for integrin subunit interaction. The association of a BspHI restriction site with this newly identified mutation has allowed allele-specific restriction analysis of Algerian GT individuals and the identification of two new unrelated type I patients exhibiting the same mutation, suggesting that the described mutation might be significant in this population and that BspHI restriction analysis will provide a useful screening assay for antenatal diagnosis and genetic counselling.
PLATELET GLYCOPROTEINS (GP) IIb and IIIa form a calcium-dependent heterodimeric complex and function as the major fibrinogen receptor of the platelet membrane.1 Upon platelet activation, the GPIIb-IIIa complex becomes competent to bind several RGD-containing plasma proteins such as fibrinogen, fibronectin, von Willebrand factor, and vitronectin and thus plays a critical role in platelet aggregation and adhesion.2-4 Hereditary defects of the GPIIb-IIIa receptor cause Glanzmann's thrombasthenia (GT), an autosomal recessive bleeding disorder characterized by life-long cutaneous bleeding, due to the failure of GT platelets to aggregate in response to physiologic agonists such as adenosine diphosphate, thrombin, or collagen.5 GT patients are grouped into three types according to the amount of GPIIb-IIIa complexes expressed on their platelet surface, with their platelets being functionally undistinguishable: less than 5% of GPIIb-IIIa for type I patients, 10% to 20% for type II patients, and 50% to 100% for type III (variant) patients.6
Platelet GPIIb-IIIa belongs to the gene family of integrins that are noncovalent αβ heterodimeric adhesion receptors that mediate many of the cell-cell and cell-matrix interactions essential for cell adhesion, aggregation, migration, growth, and differentiation.7,8 The extracellular domains of the integrin α and β subunits interact with each other to form a ligand binding site,9 whereas the cytoplasmic tails interact with components of the cytoskeleton, allowing integrin clustering in focal adhesion plaques.10 GPIIb-IIIa, also known as integrin αIIbβ3, shares a common β3 subunit with the vitronectin receptor αvβ3, present in a large variety of cells and also expressed as a minor constituent in the platelet membrane.11 In GT, the level of platelet αvβ3 receptor expression has been used as a marker to differentiate patients with a genetic defect located either on αIIb or β3, as normal to increased amounts of αvβ3 are found in patients with a GPIIb gene defect, whereas both GPIIb-IIIa and αvβ3 are undetectable in patients with a genetic defect of GPIIIa.12 With the isolation of cDNA and genomic clones for GPIIb and GPIIIa, identification of the genetic defects responsible for GT have become possible and, over the last few years, more than 30 different mutations have been described. Analysis of these molecular defects is of considerable importance, because they contribute to a better knowledge of the structural requirements necessary for αIIbβ3 and αvβ3 integrin subunit biogenesis and dimerization, as well as receptor maturation, cell surface expression, and ligand binding function.
We have previously reported the preliminary biochemical characterization of a patient with type I GT.13 We now report the molecular analysis of the genetic defect and provide evidence that the newly identified small deletion/substitution (Ile325Pro326Gly327 → Met) in the GPIIIa gene of patient HS is responsible for type I GT, because this mutation prevents αβ heterodimerization and surface expression of the β3 receptor complex without affecting the biogenesis of the mutant β3 subunit.
MATERIALS AND METHODS
Case report.Family H was referred to our laboratory as the youngest 1-year-old daughter (HS) presented defective platelet aggregation to adenosine diphosphate and collagen in a context of epistaxis and bleeding tendency. In this consanguineous Algerian family, with the parents being first cousins, the propositus HS was diagnosed as type I GT because her platelets were deficient in GPIIb and GPIIIa both on the platelet surface and in α-granules and had undetectable levels of intragranular fibrinogen as shown by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and periodic acid schiff (PAS) staining, Western blot, crossed immuno-electrophoresis, and Scatchard analysis.13 Moreover, the patient developed an iso-immunization directed against GPIIb-IIIa after the first platelet transfusion. Preliminary identification of the defective gene was based on phenotyping and genotyping of the human platelet alloantigen systems HPA-1 (GPIIIa) and HPA-3 (GPIIb) in the available family members heterozygous for GT.14 The patient's mother was found heterozygous for the HPA-3 alloantigenic system, suggesting normal expression of the two GPIIb genes and possibly a defective GPIIIa gene. These data were further confirmed by the discordance observed between the heterozygous HPA-1a/1b genotype and the homozygous HPA-1a phenotype in three family members heterozygous for GT. These results suggested defective surface expression of the HPA-1b allele, due to a GPIIIa gene alteration.
Monoclonal antibodies (MoAbs).The MoAb P37 (anti-β3, CD61) was kindly provided by Dr J. Gonzalez-Rodriguez (Instituto de Quimica Fisica, Madrid, Spain). The MoAb 4D10G3 (anti-β3) was a generous gift of Dr D.R. Phillips (COR Therapeutics, South San Francisco, CA). The MoAb VNR139 (antihuman αv, CD51), cross-reacting with human and hamster αv, was from GIBCO-BRL (Merelbeke, Belgium). The MoAb LM609, reacting with β3 when complexed to αv, was from Chemicon International (Temecula, CA) and the MoAb 23C6 (anti-αvβ3) was a generous gift of Dr M. Horton (Imperial Cancer Research Fund, London, UK).
Single-strand conformation polymorphism analysis (SSCP). Genomic DNA was extracted from peripheral blood leukocytes according to the standard salting out procedure described by Miller et al.15 DNA from healthy volunteer blood donors was used as a control. Oligonucleotides used for SSCP analysis of each β3 exon were those reported by Jin et al.16 The specific primers used for exon 7 and 9 amplification were the following: primer 7a (sense), 5′-TTTGGTAAGCTCTGGACATCT-3′; primer 7b (antisense), 5′-GACTCTCCGCGGGACTATT-3′; primer 9a (sense), 5′-GGGCCCAACTGTGTCTAAAT-3′; and primer 9b (antisense), 5′-AAGGGCGATAGTCCTCCTC-3′. Initial denaturation at 95°C for 5 minutes was followed by 40 cycles of 15 seconds at 98°C, 30 seconds at 56°C, and 1 minute at 72°C; a final elongation step was performed at 72°C for 7 minutes. The length of each amplified fragment was suitable for screening of point mutations using the SSCP method. The polymerase chain reaction (PCR) products were subjected to electrophoresis on a 12.5% polyacrylamide gel (exon 7, 4°C and 100 Vh; exon 9, 15°C and 250 Vh). After migration, the gels were stained with silver nitrate to visualize the separated DNA fragments according to the PhastSystem development procedure (Pharmacia, Guyancourt, France).
Nucleotide sequence analysis of genomic DNA.Amplified DNA fragments were purified by preparative agarose gel electrophoresis using the Magic PCR Preps DNA purification system (Promega, Lyon, France). Direct sequencing was performed with the fmol DNA sequencing System (Promega) using a Taq DNA polymerase (sequencing grade) according to the instructions of the manufacturer.
Site-directed mutagenesis of β3 cDNA.The Altered Sites in vitro mutagenesis kit (Promega) was used to generate two different mutations in full-length β3 cDNA constructs: the deletion/insertion mutation identified in patient HS (β3-met) and a mutation introducing a proline instead of a methionine at position 326 in the patient's β3 cDNA (β3-pro). Briefly, the full-length cDNA-encoding wild-type β3 (generous gift of Dr D. Phillips) was cloned into the phagemid p-ALTER-1 (Promega). For the β3-met construct, a mismatched primer corresponding to nucleotides 1054-1074/1079-1098 of the β3 cDNA allowed the replacement of 9 nucleotides encoding Ile325Pro326Gly327 by 3 nucleotides encoding Met and generating a new BspHI restriction site T ↓ CATGA (primer, 5′-CAGAACTATAGTGAGCTCATGACCACAGTTGGGGTTCTG-3′). For the β3-pro construct, the mismatched primer 5′-CTATAGTGAGCTCCCGACCACAG-3′ from bases 1059 to 1087 was used, allowing a replacement of Met by Pro in the mutant β3-met cDNA. Mutagenesis was performed according to the manufacturer's instructions. The full-length mutated β3 cDNAs were excised from the p-ALTER phagemid with 5′-Xba I/HindIII-3′ and inserted into the Xba I/HindIII site of the pBJ1 mammalian cell expression vector as previously described,17 and the full-length β3 constructs were completely sequenced after subcloning into the pBJ1 vector.
Cell culture.The Chinese hamster ovary (CHO) cell line CRL 9096, defective in the dehydrofolate reductase gene (CHO dhfr-) was purchased from the American Type Culture Collection (Rockville, MD). The cells were grown in Iscove's medium, supplemented with glutamine, penicilline, and streptomycine, 10% heat-inactivated fetal bovine serum, hypoxanthine (10−4 mol/L), and thymidine (10−5 mol/L; GIBCO-BRL), and were routinely passaged with EDTA buffer, pH 7.4 (1 mmol/L EDTA, 126 mmol/L NaCl, 5 mmol/L KCl, and 50 mmol/L HEPES).
Transfection of CHO cells with human β3 integrin and selection of stable cell clones.Wild-type or mutant β3 full-length cDNA in pBJ1 vector (20 μg) and 1 μg of dhfr plasmid (pMDR901) were cotransfected into CHO cells using lipofectamine (GIBCO-BRL) as previously described.17 Fourty-eight hours after transfection, the cells were grown in nucleoside-free α-minimal essential medium (GIBCO-BRL) used as selective medium. Dhfr-positive cell clones were selected and expression of the recombinant human β3 integrin was determined by reverse transcription-PCR (RT-PCR) of β3 mRNA and by Western blot analysis of the recombinant protein, using the anti-β3 MoAb 4D10G3.
RT-PCR of β3 mRNA isolated from transfected CHO cells.Total RNA was isolated from 5 × 106 transfected cells according to the method of Chomczynski and Sacchi.18 First-strand cDNA synthesis from 2 μg of total RNA was performed with the Perkin-Elmer RNA-PCR kit (Perkin Elmer-Cetus, Norwalk, CT) using the β3 antisense 17mer 5′-CAGGTGGCATTGAAGGA-3′ as a primer. Exon 7 of the β3 subunit was amplified using the β3 sense primer 5′-GATGCATCCCACTTGCTG-3′ and the previously used antisense primer. Amplified products were submitted to BspHI digestion and analyzed by agarose gel electrophoresis.
Intracellular detection of recombinant β3 integrin.Immunofluorescence staining of the selected transfectants for intracellular expression of human β3 was performed on microscope glass coverslips. Briefly, cultured cells were harvested with EDTA buffer and washed with serum-free Iscove's medium. The cells were then seeded onto microscope glass coverslips precoated with fetal calf serum and allowed to attach and spread overnight. The cells were fixed for 15 minutes at 4°C with 3% paraformaldehyde, 60 mmol/L sucrose in phosphate-buffered saline (PBS), pH 7.4, rinsed twice with PBS, and permeabilized with labeling buffer (PBS, pH 7.4, 0.5% Triton X-100, 0.5% bovine serum albumin) for 15 minutes at room temperature. For immunofluorescence staining of β3 integrin, the coverslips were incubated 30 minutes with the primary mouse MoAb P37 (anti-β3) or LM609 (anti-αvβ3) diluted in labeling buffer, washed twice, and incubated for another 30 minutes with fluorescein isothiocyanate (FITC)-conjugated goat antimouse IgG (Tebu, Le Perray en Yvelines, France). The coverslips were finally washed three times in labeling buffer and mounted in Mowiol 40-88 (Aldrich, Steinheim, Germany). The specimens were examined with a Leica-DMRB fluorescence microscope (Leica, Wetzlar, Germany) using a 63× oil immersion objective. Microphotographs were taken using Kodak Tmax 400 films (Eastman Kodak Co, Rochester, NY).
Analysis of surface expression of transfected β3 integrin in CHO cells.Surface expression of the transfected human β3 integrin was analyzed by flow cytometry using the MoAb P37 (anti-β3). Positive transfectants were detached from culture plates with EDTA buffer, pH 7.4, and washed twice in PBS. The cells (5 × 105) were then incubated for 30 minutes on ice with the primary antibody, washed with PBS, and further incubated for 30 minutes on ice with an FITC-conjugated goat antimouse secondary antibody (Tebu). Cells were washed and resuspended in PBS and then analyzed on an Epics flow cytometer (Coultronics, Miami, FL).
Western blot analysis.CHO cell extracts, undigested or α-chymotrypsin–digested platelet samples, and immuno-precipitates were electrophoresed under nonreducing or reducing conditions on a 7% SDS-polyacrylamide gel using a Mini-PROTEAN II electrophoresis system (Bio-Rad, Nazareth, Belgium) and transferred onto nitrocellulose using a semidry transblot apparatus (LKB-Pharmacia, Roosendaal, The Netherlands). The membranes were blocked for 1 hour in blotting buffer (20 mmol/L Tris, pH 7.4, 150 mmol/L NaCl, 5% nonfat dry milk, 0.1% Tween-20) and incubated overnight with the anti-β3 MoAb 4D10G3 and the anti-αv MoAb VNR139 diluted in blotting buffer. After several 5- to 10-minute washes in blotting buffer, membranes were incubated for 1 hour with goat antimouse IgG conjugated to horseradish peroxydase (Amersham, Gent, Belgium). Membranes were then washed in TBS, pH 7.4 (20 mmol/L Tris,137 mmol/L NaCl) and developed using the chemiluminescence ECL kit (Amersham) according to the manufacturer's instructions.
Immunoprecipitation analysis of recombinant human β3 integrin expressed in CHO cells.Transfected cells were harvested with EDTA buffer, washed in cold PBS buffer, pH 7.4, and lysed for 30 minutes in 500 μL of ice-cold lysis buffer pH 7.5 (10 mmol/L Tris, 150 mmol/L NaCl, 2.5 mmol/L phenylmethylsulfonyl fluoride, and 1% Triton X-100). Lysates were precleared by centrifugation at 10,000 rpm for 10 minutes at 4°C. Transfected cell lysates were incubated with the anti-β3 MoAb P37 for 3 hours at 4°C. Protein-A Sepharose beads (50 μL of a 50% suspension in lysis buffer) were added and incubated for 2 hours at 4°C. The protein-A Sepharose beads were washed 6 times with lysis buffer and boiled in 30 μL of SDS sample buffer (2% SDS, 10% glycerol, 25 μg/mL bromophenol blue in 15,625 mmol/L Tris-HCl, pH 6.8). Immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting as described below.
Quantitation of intracellular β3 integrin by enzyme-linked immunosorbent assay (ELISA).The wells of a microtiter plate were coated overnight at 4°C with 0.6 μg/100 μL of goat antimouse IgG in 64 mmol/L sodium carbonate buffer, pH 9.6, washed three times with TBS-ELISA buffer, pH 7.4 (10 mmol/L Tris, 150 mmol/L NaCl, 0.5 mmol/L CaCl2 , containing 0.5% Nonidet P40 and 0.05% Tween-20) and saturated 30 minutes at 4°C with the same buffer. Murine MoAbs directed to αvβ3 (23C6) or β3 (P37) were added at a concentration of 0.6 μg/100 μL to each well. After 90 minutes at 4°C, the plate was washed three times and CHO cell lysates diluted in TBS-ELISA buffer (50 μg protein/100 μL) were added to each well. After 90 minutes of incubation at 4°C, the wells were washed three times and 100 μL of anti-HPA-1a (PlA1) alloantiserum diluted 1:40 in TBS-ELISA buffer was added to each well. The plate was incubated for 90 minutes at room temperature, washed 3 times, and further incubated for 90 minutes at 4°C with 100 μL/well of goat antihuman IgG conjugated to horseradish peroxidase. After 6 washes, 100 μL of H2O2 + ortho-phenylene diamine in citrate/phosphate buffer, pH 5, were added as a substrate to measure the relative amount of conjugated antibody bound to each well. After 45 minutes of incubation in the dark, the enzymatic reaction was stopped by adding 50 μL of 1 mol/L H2SO4 and the optical densities were measured at 490 nm.
RESULTS
GPIIIa DNA analysis of patient HS.Because of the very young age of infant HS and the small amount of leukocyte DNA available, initial SSCP analysis was performed with DNA of the patient's father as well as control DNA. The 14 exons encoding GPIIIa were amplified by PCR, and the fragments were analyzed by SSCP. The length and migration of all amplified genomic DNA fragments appeared identical to control samples, except for the DNA fragments corresponding to exons 7 and 9 (Fig 1). This result was confirmed with the patient's genomic DNA for exons 7 and 9, and sequence analysis of these exons showed known polymorphisms in exon 9 described by Zimrin et al19 and Wang et al.20 In contrast, sequencing of exon 7 showed a 6-nucleotide deletion resulting in the substitution of Ile325Pro326Gly327 by a Met and giving rise to a new BspHI restriction site (Fig 2).
PCR-SSCP analysis of exon 7 of the GPIIIa gene. A 165-bp DNA fragment corresponding to exon 7 of the GPIIIa gene was amplified by PCR using genomic DNA as a template and subjected to SSCP. Lane 1, control DNA; lane 2, father of the patient HS heterozygous for GT. The positions of the shifted bands observed with DNA of the patient's father are indicated.
PCR-SSCP analysis of exon 7 of the GPIIIa gene. A 165-bp DNA fragment corresponding to exon 7 of the GPIIIa gene was amplified by PCR using genomic DNA as a template and subjected to SSCP. Lane 1, control DNA; lane 2, father of the patient HS heterozygous for GT. The positions of the shifted bands observed with DNA of the patient's father are indicated.
Nucleotide sequence analysis of the genomic DNA fragment encompassing exon 7 of the GPIIIa gene. Genomic DNA from patient HS or control DNA was amplified using primers complementary to intronic sequences flanking exon 7 of the GPIIIa gene. The amplified 165-bp cDNA fragment was directly submitted to DNA sequence analysis. Nucleotides 1318 to 1323 encoding amino acids 325 to 327 of the normal GPIIIa gene are displayed on the left and the substitution of Ile325Pro326Gly327 by Met in the patient gene is shown on the right.
Nucleotide sequence analysis of the genomic DNA fragment encompassing exon 7 of the GPIIIa gene. Genomic DNA from patient HS or control DNA was amplified using primers complementary to intronic sequences flanking exon 7 of the GPIIIa gene. The amplified 165-bp cDNA fragment was directly submitted to DNA sequence analysis. Nucleotides 1318 to 1323 encoding amino acids 325 to 327 of the normal GPIIIa gene are displayed on the left and the substitution of Ile325Pro326Gly327 by Met in the patient gene is shown on the right.
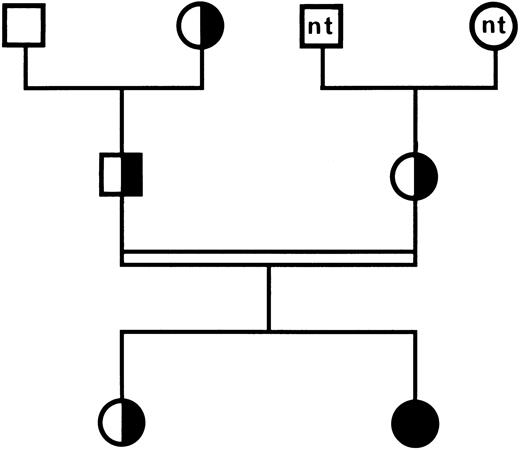
Genotype analysis in family members.These data were confirmed by analyzing the inheritance of the mutation in family members by amplification on genomic DNA encoding exon 7 of the GPIIIa gene, followed by BspHI restriction analysis. The normal allele was characterized by a 165-bp fragment and the mutated allele by two fragments of 108 and 51 bp, respectively (data not shown). As displayed in the family tree in Fig 3, the patient was homozygous for the mutant allele and her parents and her sister were heterozygous, whereas the paternal grandfather was homozygous for the normal allele.
Family tree of patient HS. (□) Normal individual; (╞, ◑) heterozygous for GT; (•) homozygous for GT; nt, not tested. The parents of the propositus are first cousins. Consanguinity is indicated by a double line.
Family tree of patient HS. (□) Normal individual; (╞, ◑) heterozygous for GT; (•) homozygous for GT; nt, not tested. The parents of the propositus are first cousins. Consanguinity is indicated by a double line.
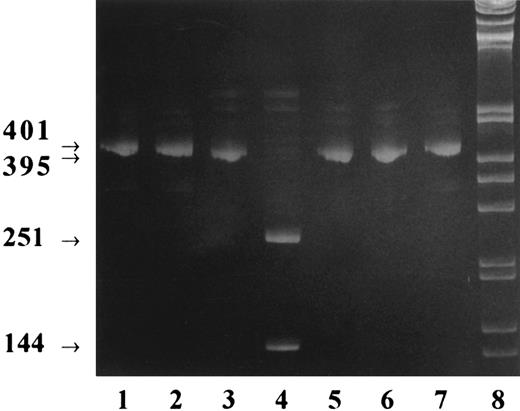
Recombinant expression of the mutant β3 integrin subunit in CHO cells.To determine how the small 3 amino acid deletion in GPIIIa identified in patient HS affected the biogenesis of the β3 integrin complex generating the type I GT phenotype, we investigated heterologous expression of the recombinant mutant β3 subunit in CHO cells. Previous data have provided evidence that, in CHO cells, the recombinant human β3 integrin subunit associates with an endogenous αv hamster subunit and is exposed on the cell surface as a chimeric αv (hamster )-β3 (human ) complex.21 Similar xenoassociations of human β3 with the integrin αv subunit have also been demonstrated in transfected Cos cells.22 Expression vectors containing the full-length cDNA encoding either mutant β3 (β3-met) or wild-type β3 (β3-wt) were constructed and cotransfected with the selectable marker gene dhfr into dhfr-negative CHO cells. Also, to ascertain whether the absence of the proline residue at position 326 of wild-type β3 could be responsible for incorrect folding of the mutant β3 protein and subsequent retention and degradation in the endoplasmic reticulum, a third construct was generated in which the methionine in the mutant β3 cDNA was replaced by a proline (β3-pro). Resistant cell clones were selected and tested for recombinant β3 integrin mRNA expression by RT-PCR using β3-specific primers. Amplified fragments were further submitted to BspHI restriction analysis. As shown in Fig 4, the smaller size (395 bp) of the cDNA fragments β3-met and β3-pro, visualized by their slightly increased electrophoretic mobility, was clearly apparent when compared with the 401-bp fragment obtained from cells expressing wild-type β3. Furthermore, as expected, BspHI restriction analysis of the amplified fragments showed two fragments of 251 and 144 bp exclusively for the cell clone expressing the β3-met mutation. Finally, direct sequence analysis of the fragments β3-met and β3-pro confirmed the expected mutation (data not shown).
BspHI restriction analysis of cDNA after RT-PCR of transfected CHO cell mRNA. Total RNA was purified from the transfected cell clones CHO β3-wt, CHO β3-met, and CHO β3-pro; reverse transcribed; and amplified by RT-PCR using specific primers. The amplified cDNA was digested by BspHI, and for each cell clone, undigested and digested cDNA was submitted to acrylamide gel electrophoresis. Lanes 1 and 2, β3-wild-type cDNA fragment of 401 bp before (lane 1) and after (lane 2) BspHI digestion; lanes 3 and 4, β3-met 395-bp cDNA fragment before (lane 3) and after (lane 4) digestion; lanes 5 and 6, β3-pro 395-bp cDNA fragment before (lane 5) and after (lane 6) digestion; lane 7, undigested wild-type β3 cDNA fragment; lane 8, 1-kb DNA ladder (GIBCO).
BspHI restriction analysis of cDNA after RT-PCR of transfected CHO cell mRNA. Total RNA was purified from the transfected cell clones CHO β3-wt, CHO β3-met, and CHO β3-pro; reverse transcribed; and amplified by RT-PCR using specific primers. The amplified cDNA was digested by BspHI, and for each cell clone, undigested and digested cDNA was submitted to acrylamide gel electrophoresis. Lanes 1 and 2, β3-wild-type cDNA fragment of 401 bp before (lane 1) and after (lane 2) BspHI digestion; lanes 3 and 4, β3-met 395-bp cDNA fragment before (lane 3) and after (lane 4) digestion; lanes 5 and 6, β3-pro 395-bp cDNA fragment before (lane 5) and after (lane 6) digestion; lane 7, undigested wild-type β3 cDNA fragment; lane 8, 1-kb DNA ladder (GIBCO).
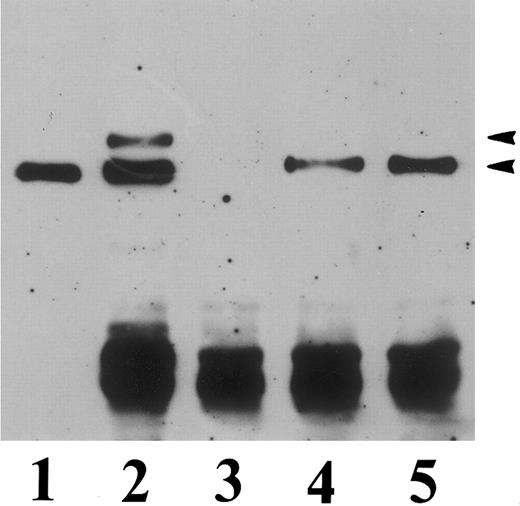
To determine whether the mutant β3 mRNA was transcribed into a detectable β3 protein, Western blot analysis was performed using the anti-β3 MoAb 4D10G3. As shown in Fig 5, a single band having an apparent molecular weight of 90 kD and comigrating with recombinant wild-type β3 as well as native platelet β3 could be visualized in the cell clones β3-met and β3-pro, thus providing evidence for normal transcription of the mutant mRNA into a full-length β3 protein. Furthermore, no evidence for rapid degradation of the mutant β3 gene product was apparent, because the anti-β3 MoAb 4D10G3, which reacts with the 110- and the 60-kD chymotryptic fragments of platelet GPIIIa, only identified a single band corresponding to the mature 90-kD form of the β3 subunit in the cell clones β3-met and β3-pro.
Immuno-blot analysis of CHO cells transfected with human β3 integrin. Triton X-100 lysates of transfected CHO cells (100 μg protein) or SDS-solubilized platelet lysates (5 μg protein) were run in a 7% polyacrylamide gel and electrophoresed under nonreducing conditions, transferred to nitrocellulose, and incubated with the antihuman β3 MoAb 4D10G3. Lane 1, CHO cells transfected with wild-type β3; lane 2, mock-transfected CHO cells; lane 3, CHO cells transfected with the mutant β3-met cDNA construct; lane 4, CHO cells transfected with the mutant β3-pro construct; lane 5, α-chymotrypsin–digested platelet lysate; lane 6, control undigested platelet lysate.
Immuno-blot analysis of CHO cells transfected with human β3 integrin. Triton X-100 lysates of transfected CHO cells (100 μg protein) or SDS-solubilized platelet lysates (5 μg protein) were run in a 7% polyacrylamide gel and electrophoresed under nonreducing conditions, transferred to nitrocellulose, and incubated with the antihuman β3 MoAb 4D10G3. Lane 1, CHO cells transfected with wild-type β3; lane 2, mock-transfected CHO cells; lane 3, CHO cells transfected with the mutant β3-met cDNA construct; lane 4, CHO cells transfected with the mutant β3-pro construct; lane 5, α-chymotrypsin–digested platelet lysate; lane 6, control undigested platelet lysate.
Effect of the mutation on surface exposure of the recombinant mutant β3 protein.To analyze intracellular localization or surface exposure of the transfected β3 protein, we performed indirect immunofluorescence labeling of the cells using the anti-β3 MoAb P37. As shown in Fig 6, fluorescence-activated cell sorting (FACS) analysis of the transfected cell clones showed surface labeling of the cells expressing wild-type β3, whereas no surface fluorescence could be detected for the cell clones expressing β3-met or β3-pro. Intracellular labeling of the transfected cells was then performed after adhesion of the cells to microtiter wells precoated with fetal calf serum. For all cell clones studied, intracellular staining of the β3 subunit could be detected. However, interestingly, whereas staining of the cells expressing wild-type β3 showed a punctuate fluorescence corresponding to focal adhesion localization of the β3 integrin subunit, a diffuse staining was observed for the cells expressing either β3-met or β3-pro. Furthermore, intracellular labeling of these cells with the complex specific MoAb LM609, reacting with human αvβ3 as well as the chimeric αv(hamster )- β3(human ) receptor, gave a negative result, demonstrating a defective αβ dimerization of the mutant β3 subunits. In contrast, this antibody labeled the wild-type αvβ3 complex in adhesion plaques as well as a mutant αvβ3 ▵ 744 complex,21 unable to localize in focal adhesion plaques and exhibiting a diffuse cytoplasmic staining. Identical results were also obtained with the complex-specific αvβ3 MoAb 23C6 (data not shown).
Immunofluorescence analysis of surface expression and intracellular localization of recombinant β3 integrin in transfected CHO cells. (A) FACS analysis of CHO cells in suspension after indirect immunofluorescence surface labeling with the anti-β3 MoAb P37. (B) Intracellular labeling of human β3 integrin in transfected CHO cells. The cells were grown on coverslips precoated with fetal calf serum, fixed, permeabilized, and processed for indirect immunofluorescence using the anti-β3 MoAb P37 and the complex-specific anti-αvβ3 LM609. (1) Mock-transfected CHO cells. (2) CHO cells transfected with wild-type β3 integrin. (3) CHO cells transfected with mutant β3-met. (4) CHO cells transfected with mutant β3-pro. (5) CHO cells transfected with mutant β3Δ744.
Immunofluorescence analysis of surface expression and intracellular localization of recombinant β3 integrin in transfected CHO cells. (A) FACS analysis of CHO cells in suspension after indirect immunofluorescence surface labeling with the anti-β3 MoAb P37. (B) Intracellular labeling of human β3 integrin in transfected CHO cells. The cells were grown on coverslips precoated with fetal calf serum, fixed, permeabilized, and processed for indirect immunofluorescence using the anti-β3 MoAb P37 and the complex-specific anti-αvβ3 LM609. (1) Mock-transfected CHO cells. (2) CHO cells transfected with wild-type β3 integrin. (3) CHO cells transfected with mutant β3-met. (4) CHO cells transfected with mutant β3-pro. (5) CHO cells transfected with mutant β3Δ744.
The absence of heterodimeric complex formation was further demonstrated by immunoprecipitation experiments using the anti-β3 MoAb P37. The immunoprecipitates were resolved by SDS-PAGE under reducing conditions and transferred to nitrocellulose, and the precipitated proteins were visualized with the anti-β3 MoAb 4D10G3, reacting exclusively with human β3, and the anti-αv MoAb VNR-139, reacting with hamster αv. As shown in Fig 7, the anti-β3 MoAb precipitated the chimeric αv(hamster )-β3(human ) complex from cells transfected with wild-type β3, whereas the same antibody precipitated exclusively the β3 subunit from the cells transfected with the mutant β3 subunits.
Immunoprecipitation of recombinant human β3 integrin expressed in transfected CHO cells. Detergent extracts of mock-transfected CHO cells and positive transfectants were incubated with anti-β3 MoAb P37. The immunoprecipitates were resolved by 7% SDS-PAGE under reducing conditions, transferred to nitrocellulose, and visualized with an MoAb to αv (VNR139), reacting with hamster αv, and MoAb 4D10G3, reacting exclusively with human β3. The strong band visualized with goat antimouse IgG conjugated to horseradish peroxydase corresponds to precipitated MoAb P37. Lane 1, total platelet lysate serving as a positive control; lane 2, CHO β3-wild-type cells; lane 3, mock-transfected CHO cells; lane 4, CHO β3-met cells; lane 5, CHO β3-pro cells. The position of αv and β3 is indicated.
Immunoprecipitation of recombinant human β3 integrin expressed in transfected CHO cells. Detergent extracts of mock-transfected CHO cells and positive transfectants were incubated with anti-β3 MoAb P37. The immunoprecipitates were resolved by 7% SDS-PAGE under reducing conditions, transferred to nitrocellulose, and visualized with an MoAb to αv (VNR139), reacting with hamster αv, and MoAb 4D10G3, reacting exclusively with human β3. The strong band visualized with goat antimouse IgG conjugated to horseradish peroxydase corresponds to precipitated MoAb P37. Lane 1, total platelet lysate serving as a positive control; lane 2, CHO β3-wild-type cells; lane 3, mock-transfected CHO cells; lane 4, CHO β3-met cells; lane 5, CHO β3-pro cells. The position of αv and β3 is indicated.
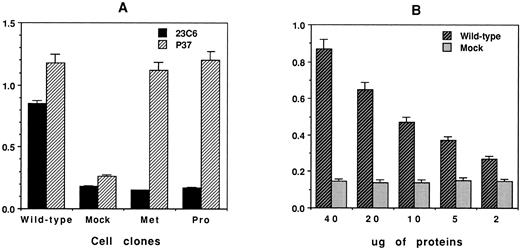
Finally, to ascertain that the absence of αβ coprecipitation observed for the cell clones β3-met and β3-pro was not due to an excess of recombinant monomeric free β3 competing with αvβ3 complexes during the immunoprecipitation experiment, a microtiter plate ELISA assay was developed to evaluate the amount of free versus complexed β3 subunits in each transfected cell clone. Free β3 or αβ heterodimers present in the cell lysate samples of identical protein concentration (50 μg) were captured with the MoAb P37 or 23C6 immobilized at the bottom of the microtiter well, and bound β3 antigen was detected with a polyclonal human anti–HPA-1a (PLA1) antibody. As shown in Fig 8A, similar amounts of the β3 antigen were detected with the MoAb P37 in each cell clone. In contrast, the MoAb 23C6 allowed detection of the complexed β3 antigen only in the cell clone expressing wild-type β3. Finally, to evaluate the sensitivity of the assay with respect to complexed β3 antigen, a serial dilution titration assay was performed with cell lysate containing wild-type β3. The data in Fig 8B provide evidence that the assay allowed the detection of complexed β3 antigen in CHO-β3wt cell samples containing as low as 2 μg of total cell extract.
Detection of free and complexed β3 antigen in CHO cells transfected with wild-type or mutant β3 cDNA. A microtiter plate was precoated with 0.6 μg/100 μL of goat antimouse IgG before the addition of either anti-β3 MoAb P37 or anti-αvβ3 MoAb 23C6. The plate was then incubated with transfected CHO cell lysate (50 μg protein). The captured β3 antigen in the wells was detected by the addition of human anti–HPA-1a IgG alloantibodies, and anti–HPA-1a binding was quantitated using peroxidase-labeled goat antihuman IgG according to established ELISA methodology. The optical density (OD) was measured at 490 nm. (A) Quantitation of free versus complexed β3 antigen in CHO cells transfected with β3-wt, β3-met, and β3-pro. (B) Titration curve demonstrating the sensitivity of the ELISA assay with MoAb 23C6 used for the detection of αvβ3 complexes in CHO β3-wt cell lysate.
Detection of free and complexed β3 antigen in CHO cells transfected with wild-type or mutant β3 cDNA. A microtiter plate was precoated with 0.6 μg/100 μL of goat antimouse IgG before the addition of either anti-β3 MoAb P37 or anti-αvβ3 MoAb 23C6. The plate was then incubated with transfected CHO cell lysate (50 μg protein). The captured β3 antigen in the wells was detected by the addition of human anti–HPA-1a IgG alloantibodies, and anti–HPA-1a binding was quantitated using peroxidase-labeled goat antihuman IgG according to established ELISA methodology. The optical density (OD) was measured at 490 nm. (A) Quantitation of free versus complexed β3 antigen in CHO cells transfected with β3-wt, β3-met, and β3-pro. (B) Titration curve demonstrating the sensitivity of the ELISA assay with MoAb 23C6 used for the detection of αvβ3 complexes in CHO β3-wt cell lysate.
DISCUSSION
GT is the most common inherited platelet disorder; over the last few years, more than 30 distinct genetic defects responsible for GT have been characterized at the molecular level.23 The majority of these defects cause type I GT, characterized by a virtual absence of the GPIIb-IIIa receptor, due to gene rearrangements,24-28 nonsense mutations,29-34 frameshift mutations, and mutations that interfere with pre-mRNA splicing, resulting in unstable mRNA transcripts or the production of unstable or truncated protein products.29,35-38 Small deletions or single amino acid substitutions that do not affect the open reading frame have been shown to produce essentially type II and type III (variant) thrombasthenia, in which low levels or normal numbers of nonfunctional GPIIb-IIIa heterodimers are expressed on the platelet surface. Interestingly, for all the mutations studied so far, it appears that point mutations in GPIIIa permit normal or nearly normal GPIIb-IIIa expression,39-44 whereas point mutations or small deletions in GPIIb prevent GPIIb-IIIa expression on the platelet surface, without affecting heterodimer formation.45-47 Mutations causing GT have therefore been classified into two categories based on the defect in the biosynthetic pathway of GPIIb-IIIa, the preassembly defects and the postassembly defects.48 However, surprisingly, no mutations inducing an absence of subunit assembly have been characterized so far.
In this study, we have characterized the molecular genetic defect causing type I GT in a 1-year-old patient of an Algerian consanguineous family. Discordance between HPA-1 genotyping and phenotyping for the three heterozygous family members suggested that the genetic defect was related to the GPIIIa gene.14 Based on these results, the 14 coding exons of GPIIIa were submitted to PCR-SSCP and a particular pattern of migration was found for exon 7 in the patient and obligate carriers but not in control DNA. Sequencing of exon 7 showed a 6-nucleotide deletion resulting in a Ile325Pro326Gly327 → Met substitution, suggesting that this deletion was the defect responsible for GT.
Because the mutation identified in the GPIIIa gene of patient HS conserved the entire open reading frame, we were eager to assess how this small deletion/substitution was able to interfer with the biosynthetic pathway and surface exposure of the GPIIb-IIIa complex. Introduction of this mutation into wild-type recombinant β3 integrin expressed in CHO cells resulted in the same defective surface expression as observed with the patient's platelets, thus confirming that the identified mutation in the GPIIIa gene was responsible for the type I GT phenotype. However, interestingly, in contrast to all previously described GPIIIa mutations causing type I GT, the mutant β3 cDNA was transcribed into stable mRNA allowing β3 protein synthesis. The mutant β3 protein was easily detectable by Western blot analysis and migrated in SDS-PAGE gels with an apparent molecular weight identical to wild-type β3. The mutation did not particularly affect the stability of the recombinant mutant protein, because similar amounts of mutant and wild-type β3 protein could be identified in the cell lysates either by Western blot analysis or by semiquantitative ELISA assay. And finally, proteolytic cleavage products were undetectable by Western blot analysis, as shown with the MoAb 4D10G3 reacting equally well with native and chymotrypsin-digested platelet GPIIIa. However, despite the intracellular pool of the mutant β3-met protein, no association with endogenous hamster αv integrin subunit could be demonstrated using intracellular immunofluorescence labeling, immunoprecipitation experiments, and a semiquantitative ELISA assay. In contrast, wild-type β3 was readily expressed on the cell surface as a heterodimer associated with hamster αv. The β3-met protein thus failed to associate with the integrin αv subunit in transfected CHO cells and proved to be a useful model to define precise structural domains of the β3 integrin subunit involved in αβ subunit association and αβ complex maturation.
Domains in GPIIb and GPIIIa involved in αβ integrin dimerization have been identified by Calvete et al49 by proteolytic dissection of isolated resting GPIIb-IIIa, showing that sequences within the GPIIIa loop region between the cysteine-rich domains (residues [217-235]S-S[262-298], 324-366, and 403-421) are involved in the GPIIb-IIIa heterodimer intersubunit surface. Interestingly, these domains correspond to exon boundaries within the gene structure of GPIIIa, the domain 324-366 being encoded by exons 7 and 8 (residues 320-396). The Ile325Pro326Gly327 → Met deletion/insertion identified in patient HS is located in the 324-366 domain, suggesting that an alteration within this contact area critically destabilizes the optimal conformation of the GPIIIa chain necessary for assembly of the α and β subunits into a stable αβ complex. Because proline residues are important for the proper folding of proteins and influence their secondary and tertiary structures, we wondered whether the deletion of Pro326 was essentially responsible for the defective heterodimerization of the β3 subunit in patient HS. Several proline residues are indeed highly conserved among all human integrin β subunits (with the exception of integrin β8) and searching of the GenBank showed that Pro326 of human GPIIIa, which is located 15 amino acids downstream of a highly conserved IFAVT motif, is conserved in all integrin β subunits cloned so far, including such highly divergent species as caenorhabditis elegans, oryctolagus cuniculus, xenopus laevis, mouse, chicken, or monkey. The only exception was found for drosophila integrin βnu, in which a methionine is present at position 15 downstream of the IFAVT motif. To determine the precise functional role of Pro326 in β3 integrin complex formation, we investigated whether replacing the patient's methionine with a proline would restore complex formation and surface exposure of the recombinant receptor. However, similar to the β3-met protein, the β3-pro protein, still defective of the adjacent Ile325 and Gly327 residues, was unable to form a complex with the endogenous αv subunit, suggesting that not proline by itself, but rather the three deleted amino acids Ile-Pro-Gly are located in a structurally important motif necessary for αβ dimerization. Recently, a Cys-Tyr mutation at position 374 of GPIIIa causing GT has been reported.43 This mutation did not prevent GPIIb-IIIa heterodimerization, despite the fact that the loss of the cystein residue was most likely responsible for abnormal intrachain cystein bonding and abnormal folding of the GPIIIa subunit. Similarly, major deletions in the extracellular domain of GPIIb affecting the Ca2+ binding domains have been reported.45-48 Although all of these deletions impaired intracellular processing and cell surface expression of GPIIb-IIIa, they did not prevent GPIIb-IIIa subunit association. To our knowledge, the mutation described here is quite exceptional as it is the first deletion/insertion in the GPIIIa gene described so far that causes type I GT by interfering with integrin αβ heterodimerization, despite regular transcription of the mutant GPIIIa gene into a full-length protein.
The 6-nucleotide deletion in exon 7 (CCCAGG) of the GPIIIa gene in patient HS gives rise to a new BspHI restriction site, allowing easy allele-specific restriction analysis of Algerian or North-African individuals. We have recently identified two new unrelated Algerian GT patients having the same mutation (C. Kaplan, unpublished data), suggesting that the described deletion is not only family specific, but might also be population specific, as previously shown for the Iraqi-Jewish and Arab populations in Israel.12 29 The association of a new BspHI restriction site with this mutation will allow screening of potential GT carriers in this population and will provide a useful screening assay for antenatal diagnosis and genetic counselling.
In conclusion, we have characterized the molecular genetic defect in patient HS and shown that the deletion/insertion Ile325Pro326Gly327 → Met in the GPIIIa gene is responsible for type I GT. Characterization of the biosynthetic pathway of the recombinant mutant β3 protein in heterologous cells has provided evidence that this defect prevents heterodimerization of the integrin β3 subunit. These data contribute to a better understanding of the structural requirements necessary for αβ integrin complex formation and highlight the importance of the Ile325Pro326Gly327 motif in the 324-366 GPIIIa domain involved in integrin subunit association.
ACKNOWLEDGMENT
The authors thank Drs J. Gonzalez-Rodriguez, M. Horton, and D.R. Phillips for their generous gift of MoAbs.
Supported by grants from Centre de Recherche Public-Santé, Luxembourg; Centre National de la Recherche Scientifique (CNRS), France; and EC Biomed Grant No. CT931685. M.-C.M.-K. is supported in part by an EC Marie Curie fellowship (ERBFMBICT 961531).
Address reprint requests to Nelly Kieffer, PhD, Laboratoire Franco-Luxembourgeois de Recherche Biomédicale, Centre Universitaire, 162 A, avenue de la Faı̈encerie, L-1511 Luxembourg (Grand Duchy of Luxembourg).





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal