Abstract
Bone marrow (BM) CD34+ cells residing in the G0 phase of cell cycle may be the most suited candidates for the examination of cell cycle activation and proliferation of primitive hematopoietic progenitor cells (HPCs). We designed a double simultaneous labeling technique using both DNA and RNA staining with Hoechst 33342 and Pyronin Y, respectively, to isolate CD34+ cells residing in G0(G0CD34+ ). Using long-term BM cultures and limiting dilution analysis, G0CD34+ cells were found to be enriched for primitive HPCs. In vitro proliferation of G0CD34+ cells in response to sequential cytokine stimulation was examined in a two-step assay. In the first step, cells received a primary stimulation consisting of either stem cell factor (SCF), Flt3-ligand (FL), interleukin-3 (IL-3), or IL-6 for 7 days. In the second step, cells from each group were washed and split into four or more groups, each of which was cultured again for another week with one of the four primary cytokines individually, or in combination. Tracking of progeny cells was accomplished by staining cells with PKH2 on day 0 and with PKH26 on day 7. Overall examination of proliferation patterns over 2 weeks showed that cells could progress into four phases of proliferation. Phase I contained cytokine nonresponsive cells that failed to proliferate. Phase II contained cells dividing up to three times within the first 7 days. Phases III and IV consisted of cells dividing up to five divisions and greater than six divisions, respectively, by the end of the 14-day period. Regardless of the cytokine used for primary stimulation, G0CD34+ cells moved only to phase II by day 7, whereas a substantial percentage of cells incubated with SCF or FL remained in phase I. Cells cultured in SCF or FL for the entire 14-day period did not progress beyond phase III but proliferated into phase IV (with <20% of cells remaining in phases I and II) if IL-3, but not IL-6, was substituted for either cytokine on day 7. G0CD34+ cells incubated with IL-3 for 14 days proliferated the most and progressed into phase IV; however, when SCF was substituted on day 7, cells failed to proliferate into phase IV. Most intriguing was a group of cells, many of which were CD34+, detected in cultures initially stimulated with IL-3, which remained as a distinct population, mostly in G0 /G1 , unable to progress out of phase II regardless of the nature of the second stimulus received on day 7. A small percentage of these cells expressed cyclin E, suggesting that their proliferation arrest may have been mediated by a cyclin-related disruption in cell cycle. These results suggest that a programmed response to sequential cytokine stimulation may be part of a control mechanism required for maintenance of proliferation of primitive HPCs and that unscheduled stimulation of CD34+ cells residing in G0 may result in disruption of cell-cycle regulation.
HEMATOPOIETIC STEM cells, and most likely primitive hematopoietic progenitor cells (HPCs), are believed to reside in a metabolically and mitotically dormant state within the bone marrow (BM) microenvironment.1-3 In the context of the cell cycle, these cells, therefore, lie dormant within the specialized resting stage of G1 known as G0 . Cells residing in G0 are metabolically inactive almost to the extent of a complete shut-down of protein synthesis.4 Cellular activation induces exit of cells from G0 and entry into G1 , a phase of the cell cycle characterized by initiation of transcription and accumulation of RNA. Although progression into active phases of cell cycle and subsequent proliferation is a normal process for most active somatic cells, such a process may be detrimental for the hematopoietic potential of primitive HPCs because activation and proliferation of these cells may proceed along two different paths.5 The first is believed to result in the generation of progeny cells possessing identical functional properties to the original cells, a process known as self-renewal. Cells proceeding along the second path undergo proliferation-associated differentiation and lineage commitment, an irreversible process resulting in the loss of these cells from the pool of primitive HPCs. Mechanisms leading to activation of stem cells and their subsequent decision to commit to either self renewal or to differentiation remain unresolved. Paramount in the lack of our understanding of these mechanisms is the difficulty to obtain functionally and mitotically homogeneous populations of marrow cells highly enriched for primitive HPCs.
We and others have isolated groups of human and murine marrow cells identified with the DNA stain Hoechst 33342 (Hst) and the mitochondrial dye rhodamine 123 as metabolically inactive quiescent cells.6-8 Although such cells were proven to be enriched for primitive HPCs, at the time of isolation, these cells were mitotically heterogeneous. Mitotic heterogeneity stems from the fact that inspite of more than 90% of isolated cells being quiescent, these cells resided in two distinct phases of cell cycle, namely G0 and G1 . As such, studies aiming at examining cytokine-mediated activation and progression of dormant cells into active phases of cell cycle were not possible because the target population consisted of cells at different levels of activation and cytokine responsiveness. Still, examination of the effects of hematopoietic growth factors and cytokines on proliferation and differentiation of HPCs allowed for a general classification of these mediators as early- or late-acting cytokines.5 We speculated that to examine hematopoietic cell activation and the direct effects of individual cytokines on cell cycle progression of dormant HPCs, it was essential to obtain mitotically homogeneous populations of cells, preferably in G0 .
Discrimination between G0 and G1 is possible if DNA staining is coupled with RNA labeling.9 Such an approach allows for the identification of cells beginning to transcribe and accumulate RNA as they progress from G0 into G1 . Pyronin Y (PY) is an RNA-specific dye that has been successfully used for two parameter cell-cycle analyses.10 We combined Hst and PY staining to identify human marrow CD34+ cells residing in G0 at the time of isolation. In this report we show that these cells were highly enriched for progenitor cells displaying primitive hematopoietic properties. In addition, G0CD34+ cells were successfully used in activation and proliferation experiments in which it was possible to identify the need for an organized sequential delivery of activation signals to ensure maximum proliferation potential. Furthermore, unscheduled activation signals appeared to arrest the proliferation of these cells even after resumption of scheduled signals.
MATERIALS AND METHODS
Collection and fractionation of human bone marrow CD34+ cells.Human BM aspirates were collected from normal adult volunteers after obtaining informed consent according to guidelines established by the Human Investigation Committee of the Indiana University School of Medicine. Low density BM cells were separated over Ficoll/Hypaque (Pharmacia, Piscataway, NJ) and then enriched by immunomagnetic selection for CD34+ cells, as previously described.11 All reagents for the immunomagnetic separation procedure were kindly provided by Baxter Healthcare, Santa Ana, CA. Enriched CD34+ cells were stained on ice for 20 minutes with fluorescein isothiocyanate (FITC)-conjugated anti-CD34 (Becton Dickinson Immunocytometry Systems [BDIS], San Jose, CA), or isotype-matched nonspecific myeloma proteins for controls. Cells were washed and resuspended for flow cytometric cell sorting in phosphate-buffered saline (PBS) supplemented with 1% human serum albumin.
Hst and PY staining.CD34+ cells sorted to homogeneity as described below were stained with Hst (Molecular Probes, Eugene, OR) and PY (Sigma Chemical Company, St Louis, MO, catalog number 9172) according to the following procedure. Hst was prepared at 1.6 μmol/L in Hst buffer consisting of Hanks Balanced Salt Solution supplemented with 20 mmol/L HEPES, 1 g/L glucose, and 10% fetal bovine serum (FBS; Hyclone, Logan, Utah) with the pH adjusted to 7.2 while PY was prepared in the same buffer to deliver a final concentration of 1 μg/mL. Sorted CD34+ cells were washed twice in Hst buffer and resuspended in 1.5 mL of 1.6 μmol/L Hst solution (up to 5 × 106 cells). Cells were incubated in a 37°C water bath for 45 minutes, at which time PY was added to the cells without any prior washing and the incubation continued for another 45 minutes. Cells were washed once in excess Hst buffer before sorting. One sample stained with Hst only and another stained with PY only were prepared as control.
Flow cytometric analysis and cell sorting.Immunomagnetically selected CD34+ cells were stained with FITC-conjugated CD34 (BDIS). Cells were sorted as previously described6 on a FACStarplus flow cytometer (BDIS) to yield total CD34+ cells. Sorted CD34+ cells were immediately stained with Hst and PY, as described previously, and sorted on a FACStarplus. The front laser of the FACStarplus provided 40 mW of 488 nm light to excite PY while the back Krypton laser was tuned to deliver 50 mW of 350 nm ultraviolet (UV) light to excite Hst. Both signals were collected on a linear scale and were used to generate a dual parameter dot plot (Fig 1). With the Hst signal displayed on the X-axis, a typical DNA histogram could be generated with cells residing in G0 /G1 phases of the cell cycle appearing in a discrete peak. Within this peak, cells in deep G0 displayed little RNA staining and were, therefore, identified as those displaying dim PY staining along the Y axis. Cells in G1 , which had more RNA, stained brighter and were higher than G0 cells along the Y axis (Fig 1). A small sorting window, constructed to contain approximately 5% of the cells (Fig 1), was established for the collection of G0 cells taking care to exclude dead cells. Another window containing 5% to 10% of the cells was constructed at the top of the G0 /G1 peak to collect cells in the G1 phase of cell cycle. Viability and purity of sorted cells always exceeded 98% and 95%, respectively. During staining, sorting, plating, and during at least the next 2 days, cells stained with Hst and PY were kept away from direct light. Isolation of BM CD34+ HLA-DR− CD15− cells was accomplished by staining cells with allophycocyanin-conjugated CD34 (Caltag Laboratories, Burlingame, CA), FITC-conjugated HLA-DR, and phycoerythrin (PE)-conjugated CD15 (BDIS). A FACScan (BDIS) was used for flow cytometric analysis.
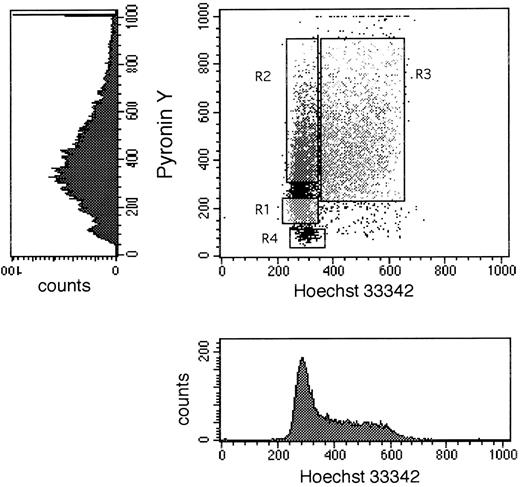
Dot plot and single parameter histograms of a typical Hoechst 33342 (X axis) and Pyronin Y (Y axis) staining of bone marrow CD34+ cells. CD34+ cells were first sorted to homogeneity, then stained with Hst and PY as described in Materials and Methods. The single parameter histograms are shown here to elucidate the contribution of each stain to the formation of the dot plot. The DNA/RNA staining shown in this figure was derived from a cultured (7 days with SCF, IL-3, and IL-6) BM sample of CD34+ cells to elaborate the S/G2 + M population for better definition of all three phases of cell cycle. Sort windows R1, R2, and R3 shown in the dot plot represent the windows used to collect G0 , G1 , and S/G2 + M cells, respectively. R4 designates the area in such a dot plot where dead cells are located.
Dot plot and single parameter histograms of a typical Hoechst 33342 (X axis) and Pyronin Y (Y axis) staining of bone marrow CD34+ cells. CD34+ cells were first sorted to homogeneity, then stained with Hst and PY as described in Materials and Methods. The single parameter histograms are shown here to elucidate the contribution of each stain to the formation of the dot plot. The DNA/RNA staining shown in this figure was derived from a cultured (7 days with SCF, IL-3, and IL-6) BM sample of CD34+ cells to elaborate the S/G2 + M population for better definition of all three phases of cell cycle. Sort windows R1, R2, and R3 shown in the dot plot represent the windows used to collect G0 , G1 , and S/G2 + M cells, respectively. R4 designates the area in such a dot plot where dead cells are located.
Staining with PKH2 and PKH26.When needed, sorted G0CD34+ cells were stained with PKH2 (Sigma ImmunoChemicals) before their use in short-term culture as per manufacturer's instructions and as previously described.12 Briefly, cells were suspended in 1 mL of diluent and immediately transferred into a polypropylene tube containing 1 mL of 4 × 10−6 mol/L PKH2 in diluent at room temperature. After 5 minutes of incubation with frequent agitation, 2 mL of FBS were added to the suspension for 1 minute. The total volume was brought up to 8 mL with complete medium (Iscove's modified Dulbecco's medium [IMDM] supplemented with 10% FBS, L-glutamine, and antibiotics [penicillin, 100 U/mL and streptomycin, 100 μg/mL]), and the cells were washed three times in complete medium. All of the complete medium ingredients (except for FBS) were obtained from BioWhitaker, Walkersville, MD. After the first wash, cells were transferred to a new tube. After the third wash, cells were resuspended in complete medium and seeded in short-term culture as described next. A sample of PKH2-stained CD34+ cells was fixed in 1% paraformaldehyde immediately after staining to be used 7 or 14 days later for the determination of day 0 green fluorescence intensity. Staining of cultured (PKH2-stained cells) with PKH26 was performed as described previously for PKH2, substituting the former for the latter. A sample of PKH26-stained cells was fixed in 1% paraformaldehyde immediately after staining to be used 7 days later for the determination of green and red fluorescence intensities corresponding to day 7.
Limiting dilution analysis (LDA).Determination of the frequency of long-term hematopoietic culture-initiating cell (LTHC-IC) by stromal cell-free LDA was performed as previously described.12 Briefly, populations of freshly isolated CD34+ cells, or subsets of CD34+ cells were seeded at 64, 32, 16, and 8 cells in 100 μL of complete medium supplemented with 200 ng/mL stem cell factor (SCF), 25 ng/mL interleukin-3 (IL-3), 25 ng/mL IL-6, 10 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF ), and 2 U/mL erythropoietin into the wells of flat-bottomed 96-well plates in replicates of 48 wells per cell dose. Plates were incubated at 37°C in 100% humidified atmosphere containing 5% CO2 and fed with another 100 μL of complete medium and cytokines on day 7. On day 14, a total of 120 μL of medium were removed from each well followed by the addition of 150 μL of a mixture designed to deliver at a final concentration, 30% FBS, 1.1% methylcellulose, 5 × 10−5 mol/L 2-mercaptoethanol, 100 ng/mL SCF, 25 ng/mL IL-3, 25 ng/mL IL-6, 25 ng/mL GM-CSF, and 2 U/mL erythropoietin. Cells were mixed into the semisolid medium by gentle vortexing, and the plates were incubated as described previously. On day 28, the plates were scored under an inverted microscope for the presence of burst-forming unit-erythroid (BFU-E), colony forming unit-granulocyte-macrophage (CFU-GM) or colony forming unit-mixed (CFU-GEMM) in every well. Wells were considered positive, indicating the presence of at least one LTHC-IC in the original cell inoculum deposited into the well, if one or more hematopoietic colonies per well were detected. The number of negative wells per cell dose input was then calculated and the data used in a χ-minimalization assay13 to calculate a frequency of LTHC-IC.
Long-term hematopoietic cell cultures.Long-term hematopoietic cultures (LTHC) were initiated with 2 × 103 to 2 × 104 sorted CD34+, G0CD34+, G1CD34+, or CD34+ HLA-DR− CD15− cells. Cultures were established in 1 mL of complete medium in flat-bottomed 48-well plates as previously described.14 At initiation, and every 48 hours thereafter, cultures were supplemented with 200 ng/mL SCF, 100 ng/mL IL-3, and 100 ng/mL IL-6. At weekly intervals, cultures were demidepopulated by removal of half the cells followed by the addition of fresh medium and cytokines. Harvested cells were counted and assayed for their clonogenic progenitor cell content as described next.
Hematopoietic progenitor cell assays.Cells (1 × 103 freshly sorted cells or between 2 × 103 to 5 × 104 cultured cells/mL) were suspended in duplicate in plastic 35-mm tissue culture dishes containing 1 mL 30% FBS, 5 × 10−5 mol/L 2-mercaptoethanol, 1.1% methylcellulose, 100 ng/mL SCF, 25 ng/mL IL-3, 25 ng/mL IL-6, 25 ng/mL GM-CSF, and 2 U/mL erythropoietin in IMDM. Cultures were incubated in 100% humidified 5% CO2 in air at 37°C. After 14 days BFU-E, CFU-GM, and CFU-GEMM were enumerated using an inverted microscope.
Cell cycle analysis.Cell fractions were stained for cell cycle analysis as previously described.12 Briefly, cells were stained with equal volumes of staining buffer (0.1 mg/mL propidium iodide + 0.6% Nonidet P-40 in PBS) and 2 mg/mL RNAse, agitated, and incubated on ice for 30 minutes in the dark. Samples were analyzed on a FACScan flow cytometer calibrated with chicken erythrocytes. The percentage of cells in G0 /G1 phases of cell cycle was analyzed using Modfit software (Verity Software House, Topsham, ME).
Two dimensional cell tracking in vitro.Activation and proliferation of G0CD34+ cells in response to sequential exposure to single or multiple cytokines was analyzed by two-dimensional cell tracking. Sorted G0CD34+ cells were stained on day 0 with PKH2 as described previously. Individual wells of a flat-bottomed 48-well plate (Falcon #3072, BDIS) were seeded with 2 × 104 up to 4 × 104 PKH2-stained G0CD34+ cells in 1 mL of complete medium. Individual wells were supplemented with either 200 ng/mL SCF, 20 ng/mL Flt-3 ligand, 100 ng/mL IL-3, or 100 ng/mL IL-6. All cytokines used in these studies, except Flt-3 ligand, were a kind gift from Amgen (Thousand Oaks, CA). Flt-3 ligand was a kind gift from Immunex (Seattle, WA). The plates were incubated in 100% humidified 5% CO2 in air at 37°C, and these cytokines were replenished every 48 hours until day 7. On day 7, cells from each well were collected separately, washed three times, and each group of cells was stained with PKH26. Each group of cells was then split into four or more new wells and each well received the same or a new single or multiple cytokine combination as indicated in Results. Cells were incubated and supplemented with appropriate cytokines for an additional 7 days under the same conditions. On day 14, all the cells were harvested and analyzed on a FACScan for residual PKH2 and PKH26 using samples fixed on day 0 and day 7 to establish original fluorescence intensities. In some experiments, groups of cells identified by their proliferation history as assessed by retention of both membrane dyes (see Results for details) were sorted for further analysis.
Flow cytometric analysis of cyclin E and p27.Fractions of cultured cells were first isolated by flow cytometric cell sorting based on their proliferative history in the two-dimensional cell tracking experiments. Cells were then fixed and stained as previously described by Farahat et al.15 Briefly, cells were fixed for 10 minutes at room temperature in 2 mL of a 1:1 mixture of 4% paraformaldehyde in PBS and Becton Dickinson (BDIS) fluorescence-activated cell sorting lysing solution prepared in distilled water as per the manufacturer's recommendations. Cells were then centrifuged, washed in 0.5% Tween 20 in PBS, and incubated for 20 minutes at room temperature with the primary anticyclin E or anti-p27 monoclonal antibodies (Pharmingen, San Diego, CA). The cells were then washed twice and developed with a PE-Cy5–conjugated goat antimouse IgG1 monoclonal antibody (Caltag Laboratories, South San Francisco, CA). Control samples were stained with irrelevant mouse IgG1 .
RESULTS
Isolation of CD34+ cells residing in G0 . Exit of cells from G0 and entry into G1 is associated with transcription of RNA, which can be measured by RNA-specific dyes such as PY.10 When PY was used in conjunction with Hst, it was possible to generate a dot plot in which G0 cells were identified as those with 2n DNA and minimal RNA content (Fig 1). This allowed for the identification of a group of cells constituting approximately 5% to 10% of the total cells analyzed, which were isolated as G0CD34+ cells. To confirm the cell-cycle status of isolated G0CD34+ cells, these cells were stained with propidium iodide and analyzed. An excess of 99% of G0CD34+ cells were detected within the G0 /G1 phases of cell cycle (Fig 2). Similarly, cells isolated as G1 or as S/G2 + M cells contained the appropriate amount of DNA corresponding to their respective phases of cell cycle (Fig 2). In addition, we examined CD34+ cells isolated in different stages of the cell cycle for their expression of the nuclear antigen Ki67.16 17 Expression of this nuclear antigen was least among cells isolated as G0 and highest among those sorted as S/G2 + M with G1 cells displaying an intermediate Ki67 expression (data not shown).
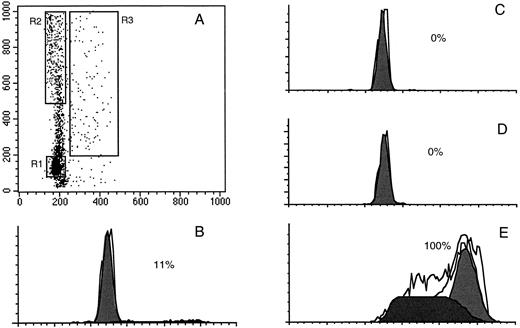
Dot plot display of a typical Hoechst 33342 (X axis) and Pyronin Y (Y axis) staining of fresh bone marrow CD34+ cells (A). The three sort windows R1, R2, and R3 were constructed to collect G0 , G1 , and S/G2 + M cells, respectively. After sorting, G0 , G1 , and S/G2 + M cells, along with unsorted total CD34+ cells were stained with propidium iodide and analyzed for their cell-cycle distribution as described in Materials and Methods. B depicts the cell cycle distribution of total freshly isolated BM CD34+ cells with only 11% being in S/G2 + M. C, D, and E illustrate the DNA content of sorted G0 , G1 , and S/G2 + M cells, respectively. As can be seen, no cells were detected with greater than 2n DNA among G0 and G1 cells (C and D, respectively), whereas all sorted S/G2 + M cells displayed a DNA content characteristic of their position in the cell cycle (E).
Dot plot display of a typical Hoechst 33342 (X axis) and Pyronin Y (Y axis) staining of fresh bone marrow CD34+ cells (A). The three sort windows R1, R2, and R3 were constructed to collect G0 , G1 , and S/G2 + M cells, respectively. After sorting, G0 , G1 , and S/G2 + M cells, along with unsorted total CD34+ cells were stained with propidium iodide and analyzed for their cell-cycle distribution as described in Materials and Methods. B depicts the cell cycle distribution of total freshly isolated BM CD34+ cells with only 11% being in S/G2 + M. C, D, and E illustrate the DNA content of sorted G0 , G1 , and S/G2 + M cells, respectively. As can be seen, no cells were detected with greater than 2n DNA among G0 and G1 cells (C and D, respectively), whereas all sorted S/G2 + M cells displayed a DNA content characteristic of their position in the cell cycle (E).
Because of its reported toxicity,9 different PY concentrations were evaluated before adopting a final working concentration. Only at concentrations lower than 5 μg/mL was viability consistently maintained above 95%. Therefore, a final working concentration of 1 μg/mL was chosen. It has been reported that at concentrations lower than 5 μmol/L (1.51 μg/mL), PY preferentially stains mitochondrial membranes in lieu of RNA. To confirm that when used at 1 μg/mL, PY will stain RNA and not just mitochondrial membranes, cells treated with RNase then stained with PY were compared with cells stained with PY without RNase treatment. PY fluorescence intensity of CD34+ cells treated with RNase was significantly lower than that of control cells, thus, verifying the RNA-specificity of PY staining performed under these conditions (data not shown).
Activation and progression of G0CD34+ cells into active phases of cell cycle.We hypothesized that G0CD34+ cells may be less responsive to cytokine stimulation and may require a longer period of time to traverse into active phases of cell cycle than mitotically heterogeneous populations of primitive HPCs, such as CD34+ HLA-DR− CD15− cells. Both groups of cells were plated in suspension cultures supplemented with SCF, IL-3, and IL-6 and were monitored periodically for their cell-cycle status. Whereas CD34+ HLA-DR− CD15− cells responded within 16 to 18 hours to cytokine stimulation and began to exit G0 /G1 , more than 95% of G0CD34+ cells remained nonresponsive to cytokine stimulation for at least 24 hours, suggesting that G0CD34+ cells are more quiescent than CD34+ HLA-DR− CD15− cells and less responsive to cytokine stimulation.
We recently reported on a group of quiescent HPCs termed cytokine nonresponsive (CNR) cells, which, in vitro, appear to resist cytokine stimulation and remain dormant for several days.12 We examined whether the frequency of CNR cells is higher in cultures initiated with G0CD34+ cells than what was previously established for cultures of total CD34+ cells.12,18 As can be seen in Fig 3, after 7 days of exposure to SCF, IL-3, and IL-6, 16% of cultured G0CD34+ cells tracked with PKH2 expressed the same level of PKH2 staining observed on day 0 confirming the cytokine nonresponsive status of these cells. We have previously shown, that under the same culture conditions, less than 5% of cultured total CD34+ cells remain as CNR cells.12 18 This percentage of CNR cells among G0CD34+ cells suggested once again the primitive and quiescent nature of these cells.
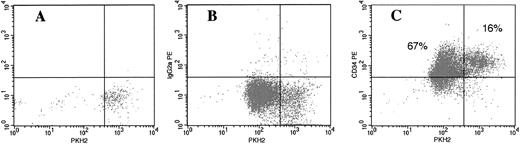
Cell tracking in vitro of cultured bone marrow G0CD34+ cells using the membrane dye PKH2. G0CD34+ cells were isolated, stained with PKH2, and cultured in IMDM containing 10% FCS in the presence of SCF, IL-3, and IL-6 for 7 days. A sample fixed in 1% paraformaldehyde on day 0 (A) was used to identify the fluorescence intensity corresponding to freshly stained cells. On day 7, cells were harvested and stained with either a PE-conjugated isotype control monoclonal antibody (B) or PE-conjugated CD34 (C). Samples were analyzed for PKH2 (X axis) and PE (Y axis) fluorescence to estimate the fraction of CD34+ cells remaining as CNR cells. As can be seen in C, 83% of cultured cells remained CD34+, and 16% could be identified as CNR cells.
Cell tracking in vitro of cultured bone marrow G0CD34+ cells using the membrane dye PKH2. G0CD34+ cells were isolated, stained with PKH2, and cultured in IMDM containing 10% FCS in the presence of SCF, IL-3, and IL-6 for 7 days. A sample fixed in 1% paraformaldehyde on day 0 (A) was used to identify the fluorescence intensity corresponding to freshly stained cells. On day 7, cells were harvested and stained with either a PE-conjugated isotype control monoclonal antibody (B) or PE-conjugated CD34 (C). Samples were analyzed for PKH2 (X axis) and PE (Y axis) fluorescence to estimate the fraction of CD34+ cells remaining as CNR cells. As can be seen in C, 83% of cultured cells remained CD34+, and 16% could be identified as CNR cells.
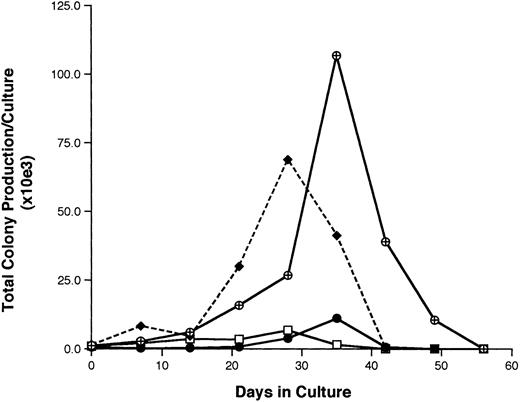
Functional properties of G0CD34+ cells.The hematopoietic potential of G0CD34+ cells was assessed in two different assays. In the first, our previously described stromal cell-free–limiting dilution analysis assay12 was used to calculate the frequency of LTHC-IC among CD34+ and G0CD34+ cells. In eight separate experiments, the mean LTHC-IC frequency among CD34+ cells was 2.3 ± 1.4, a value similar to what we previously reported for BM CD34+ cells.12 18 However, a significantly higher (P < .01) statistical mean of LTHC-IC was documented for G0CD34+ cells (4.0 ± 2.5), indicating that this group of cells is enriched for primitive HPCs. In the second approach, the ability of CD34+, G0CD34+, G1CD34+, and CD34+ HLA-DR− CD15− cells to sustain in vitro hematopoiesis was evaluated in LTHC.14,19CD34+ cells maintained the production of assayable progenitors for up to 4 weeks (Fig 4), whereas G1CD34+ cells sustained progenitor cell production for 1 additional week. On the other hand, a 10.2-fold higher level of production of progenitor cells was detected by week 4 in cultures initiated with CD34+ HLA-DR− CD15− cells compared with those initiated with CD34+ cells (Fig 4). However, peak production of assayable progenitors in G0CD34+ cultures did not occur until week 5 when a 95-fold increase in the number of input clonogenic cells was detected. Production of progenitor cells in these cultures was still detectable by week 7 (Fig 4).
A representative experiment depicting the total production of assayable progenitor cells (BFU-E plus CFU-GM, plus CFU-GEMM) in long-term bone marrow cultures initiated with freshly isolated total CD34+ (□) G0CD34+ (⊕), G1CD34+ (•), and CD34+ HLA-DR− CD15− (♦) BM cells. Cultures were maintained by the addition every 48 hours of SCF, IL-3, and IL-6 as described in Materials and Methods. Every week, half of the cultured cells were removed and assayed for clonogenic progenitor cells as described in Materials and Methods. The total production of clonogenic progenitors at every time point was calculated using the formula X = (number of clonogenic cells per culture) ÷ ()n, where “X” is the number of total colonies in culture and “n” is equal to the number of previous demidepopulations. All results were normalized to reflect those obtained from cultures initiated with 104 sorted cells. Identical results were obtained in four other experiments.
A representative experiment depicting the total production of assayable progenitor cells (BFU-E plus CFU-GM, plus CFU-GEMM) in long-term bone marrow cultures initiated with freshly isolated total CD34+ (□) G0CD34+ (⊕), G1CD34+ (•), and CD34+ HLA-DR− CD15− (♦) BM cells. Cultures were maintained by the addition every 48 hours of SCF, IL-3, and IL-6 as described in Materials and Methods. Every week, half of the cultured cells were removed and assayed for clonogenic progenitor cells as described in Materials and Methods. The total production of clonogenic progenitors at every time point was calculated using the formula X = (number of clonogenic cells per culture) ÷ ()n, where “X” is the number of total colonies in culture and “n” is equal to the number of previous demidepopulations. All results were normalized to reflect those obtained from cultures initiated with 104 sorted cells. Identical results were obtained in four other experiments.
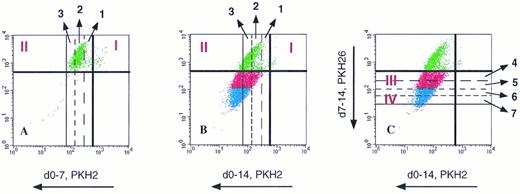
Estimation of the number of divisions attained by G0CD34+ cells in a two-dimensional tracking experiment. G0CD34+ cells were stained with PKH2 on day 0 and cultured for 14 days in the presence of IL-3. On day 7, cells were harvested and stained with PKH26 and analyzed (A). The dark vertical and horizontal lines in all three dot plots represent the lower limits of the fluorescence intensities of PKH2 and PKH26, respectively, achieved directly after staining. Using the relative PKH2 fluorescence loss (50% loss with every division along the X axis), it was possible to identify the position of these cells after every division. Thus, cells moving into phase II (upper left hand corner of dot plot) had been through one, two, or three divisions by day 7. Panel B, which was generated on day 14, depicts the same cells previously shown in A after they were allowed to proliferate for an additional week. The relative positions of the vertical lines designating one, two, and three divisions are still in place. Notice how dividing cells “track” along a 45° diagonal direction because both green and red dyes are lost proportionally as the cells divide, such that cells proliferating beyond three divisions now appear below the dark horizontal line representing the lower limit of PKH26 fluorescence. Panel C, which is composed of the same dot plot shown in B, illustrates how divisions 4, 5, 6, and 7 achieved between days 8 and 14 can be calculated along the Y axis using loss of PKH26 for measurement. Phase III cells were identified as those having gone through four or five divisions, and phase IV cells were defined as those having divided in excess of six times. Proliferation of cells compromising phases II, III, and IV was defined as depicted in Fig 6.
Estimation of the number of divisions attained by G0CD34+ cells in a two-dimensional tracking experiment. G0CD34+ cells were stained with PKH2 on day 0 and cultured for 14 days in the presence of IL-3. On day 7, cells were harvested and stained with PKH26 and analyzed (A). The dark vertical and horizontal lines in all three dot plots represent the lower limits of the fluorescence intensities of PKH2 and PKH26, respectively, achieved directly after staining. Using the relative PKH2 fluorescence loss (50% loss with every division along the X axis), it was possible to identify the position of these cells after every division. Thus, cells moving into phase II (upper left hand corner of dot plot) had been through one, two, or three divisions by day 7. Panel B, which was generated on day 14, depicts the same cells previously shown in A after they were allowed to proliferate for an additional week. The relative positions of the vertical lines designating one, two, and three divisions are still in place. Notice how dividing cells “track” along a 45° diagonal direction because both green and red dyes are lost proportionally as the cells divide, such that cells proliferating beyond three divisions now appear below the dark horizontal line representing the lower limit of PKH26 fluorescence. Panel C, which is composed of the same dot plot shown in B, illustrates how divisions 4, 5, 6, and 7 achieved between days 8 and 14 can be calculated along the Y axis using loss of PKH26 for measurement. Phase III cells were identified as those having gone through four or five divisions, and phase IV cells were defined as those having divided in excess of six times. Proliferation of cells compromising phases II, III, and IV was defined as depicted in Fig 6.
Definition of phases II, III, and IV of G0CD34+ cells in two-dimensional tracking experiments using sequential cytokine stimulation. Sorted cells were stained with PKH2 on day 0 and maintained with either FL or SCF for the first 7 days of culture. On day 7, cells were washed and stained with PKH26 (two dot plots on the left). After that, cells were split and cultured with different cytokines as indicated to the right. Cells traversing to the upper left hand corner were identified as phase II cells (see Fig 5). Cells receiving SCF as the secondary stimulus (middle column) were capable of few additional divisions only and, therefore, defined phase III cells. For ease of distinction, phase III cells were colored red. When cultured cells received IL-3 for a secondary stimulus (right two dot plots), few additional divisions beyond phase III were demonstrable. Cells capable of proliferating six or more times (see Fig 5) defined phase IV cells. For ease of distinction, phase IV cells were colored in blue. Dot plots used for this illustration were from the same experiment depicted in Fig 7.
Definition of phases II, III, and IV of G0CD34+ cells in two-dimensional tracking experiments using sequential cytokine stimulation. Sorted cells were stained with PKH2 on day 0 and maintained with either FL or SCF for the first 7 days of culture. On day 7, cells were washed and stained with PKH26 (two dot plots on the left). After that, cells were split and cultured with different cytokines as indicated to the right. Cells traversing to the upper left hand corner were identified as phase II cells (see Fig 5). Cells receiving SCF as the secondary stimulus (middle column) were capable of few additional divisions only and, therefore, defined phase III cells. For ease of distinction, phase III cells were colored red. When cultured cells received IL-3 for a secondary stimulus (right two dot plots), few additional divisions beyond phase III were demonstrable. Cells capable of proliferating six or more times (see Fig 5) defined phase IV cells. For ease of distinction, phase IV cells were colored in blue. Dot plots used for this illustration were from the same experiment depicted in Fig 7.
Sequential cytokine stimulation.Given the dormant nature of G0CD34+ cells (Fig 3), we speculated that these cells may be excellent targets for the examination of cell cycle activation and proliferation of primitive HPCs in response to sequential cytokine stimulation. A two-step assay system was employed in which proliferation during the primary stimulation period was tracked with PKH2, and continued proliferation or arrest of cultured cells was tracked in the secondary stimulation period with PKH26. Because 50% of either membrane dye is lost to daughter cells with every cell division, the relative fluorescence of stained cells could be used to infer the number of divisions that the cells had been through (Fig 5).
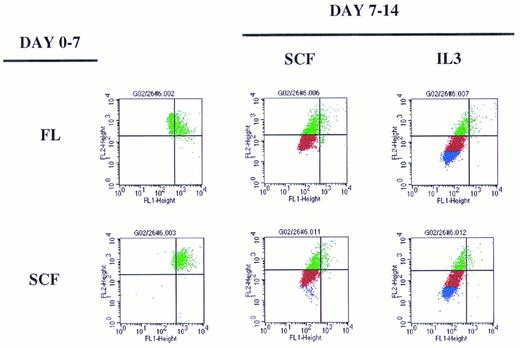
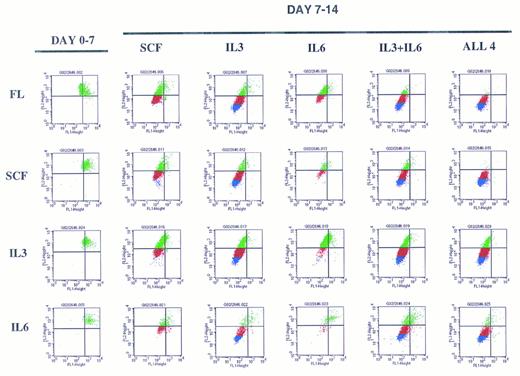
A representative experiment depicting sequential cytokine stimulation of G0CD34+ cells in a two-dimensional tracking experiment. Sorted G0CD34+ cells were stained with PKH2 on day 0 and cultured for 7 days (day 0-7, on the left) in the presence of one primary cytokine. On day 7, cells were harvested, washed, stained with PKH26, and cultured for an additional 7 days (day 7-14) in the presence of the secondary cytokine/cytokine combinations indicated to the right. Definition of phase III and IV cells, depicted here as red and blue events, respectively, was accomplished according to criteria described in Fig 6. A total of nine such experiments were performed with similar results obtained in all. However, because of the relatively small number of G0CD34+ cells obtained from a normal donor, FL was used in four experiments only, and IL-6 was used in only three experiments. SCF and IL-3 were used in all nine experiments.
A representative experiment depicting sequential cytokine stimulation of G0CD34+ cells in a two-dimensional tracking experiment. Sorted G0CD34+ cells were stained with PKH2 on day 0 and cultured for 7 days (day 0-7, on the left) in the presence of one primary cytokine. On day 7, cells were harvested, washed, stained with PKH26, and cultured for an additional 7 days (day 7-14) in the presence of the secondary cytokine/cytokine combinations indicated to the right. Definition of phase III and IV cells, depicted here as red and blue events, respectively, was accomplished according to criteria described in Fig 6. A total of nine such experiments were performed with similar results obtained in all. However, because of the relatively small number of G0CD34+ cells obtained from a normal donor, FL was used in four experiments only, and IL-6 was used in only three experiments. SCF and IL-3 were used in all nine experiments.
Overall examination of proliferation patterns of these cells for 2 weeks showed that cells could progress into four phases of proliferation. Phase I contained cytokine nonresponsive cells, which failed to proliferate (Fig 5). Phase II contained cells dividing up to three times within the first 7 days. Phases III and IV consisted of cells dividing between four and five divisions or greater than six divisions, respectively, by the end of the 14-day period (Fig 5). These patterns of divisions were established from examining the proliferation history of cells stimulated with sequential additions of single early/late or late/early5 cytokine combinations during the primary and secondary stimulation periods (Fig 6). The extent of proliferation after 7 days of primary stimulation defined phase II cells, which, as seen in Fig 6, would have divided up to three times. Cells exposed to SCF for the entire 14-day culture period underwent only two additional cell divisions (Fig 6), thus defining phase III cells as those dividing between four and five times (Fig 5). Cells stimulated with either FL or SCF during the first 7 days then exposed to IL-3 during the secondary stimulation period underwent maximal proliferation by day 14 (Fig 6) and, therefore, defined phase IV cells as those dividing up to or in excess of seven divisions (Fig 5).
Examination of the pattern of proliferation induced by different cytokine stimulation showed that regardless of the cytokine used for primary stimulation, G0CD34+ cells moved only to phase II by day 7, and a substantial percentage of cells remained in phase I (Fig 7) showing the cytokine nonresponsive nature of a fraction of these cells. Cells cultured in SCF for the entire 14-day period did not progress beyond phase III but proliferated into phase IV (with <20% of cells remaining in phases I and II) if IL-3, but not IL-6, was substituted for either cytokine on day 7 (Fig 7). Also, cells receiving FL as a primary stimulus followed by a secondary stimulus of IL-3 or IL-6 behaved in an analogus manner to those exposed to SCF between days 0 and 7. The same effects observed with IL-3 alone were observed if the combination of IL-3 plus IL-6 was substituted for FL or SCF on day 7, suggesting that these cytokines had distinct, and not additive or synergistic effects. G0CD34+ cells incubated with IL-3 for 14 days proliferated the most and progressed into phase IV; however, when SCF was substituted on day 7, cells failed to proliferate into phase IV. In fact, under all conditions examined, neither SCF nor IL-6 were capable of promoting proliferation of G0CD34+ cells into phase IV. Similarly, in experiments in which FL was used as a secondary stimulus, proliferation of G0CD34+ cells into phase IV was not demonstrable regardless of the nature of the primary stimulus (data not shown). Most intriguing was a group of cells detected in cultures initially stimulated with IL-3, which remained as a distinct population unable to progress out of phase II regardless of the nature of the secondary stimulus received on day 7. Phase II cells were also apparent in other cultures, but were most prominent in cultures maintained with IL-3 during the first 7 days. It is important to stress that the magnitude of proliferation of G0CD34+ cells during the first 7 days, especially in response to IL-3, was varied in as much as expected from inter- and intra-individual variation of primary cells. However, the general trend of proliferation patterns in all experiments, under all conditions tested, was consistent with that depicted in Fig 7.
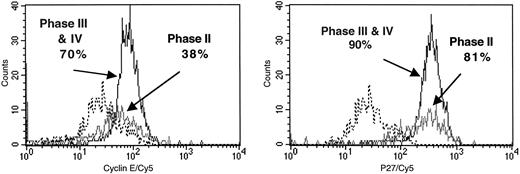
Proliferation arrest of phase II G0CD34+ cells.To better understand the nature of G0CD34+ cells arrested in phase II after 14 days in culture, their cell cycle status and continued expression of CD34 were examined. Furthermore, possible mechanisms responsible for the proliferation arrest of these cells were investigated. As can be seen in Table 1, a high percentage of phase II cells isolated from different cultures were residing in G0 /G1 , suggesting the inability of these cells to continue to proliferate after traversing into phase II. When IL-3 was included in the secondary stimulus of cells exposed to SCF or FL between days 0 and 7, a substantial fraction of phase III and IV cells were detected in S/G2 + M phases of cell cycle (Table 1). Almost one third (34%) of phase II cells still expressed CD34 (data not shown), suggesting that the proliferative potential of these cells was not exhausted. In contrast, only 8% of cells progressing into phases III and IV remained CD34+ by day 14 (data not shown). These findings prompted us to examine the role of cyclins and cyclin/cyclin-dependent kinase (cdk) inhibitors in the cell-cycle regulation of these cells. Figure 8 depicts cyclin E and p27 expression of phase II and phase III and IV cells isolated from a culture receiving IL-3 as the primary stimulus and IL-3 plus FL as the secondary stimulus. As can be seen in Fig 8, a similar percentage of phase II and phase III and IV cells expressed the cyclin-cdk inhibitor p27 (81% and 90%, respectively). In contrast, whereas 70% of phase III and IV cells expressed cyclin E, only 38% of phase II cells expressed this cyclin suggesting that proliferation of these cells may be arrested via a mechanism involving the downregulation of the expression of cyclin E rather than the upregulation or over-expression of p27.
Percentage of Phase II and Phase III Plus IV Cells Residing in G0/G1 After 14 Days of Sequential Cytokine Stimulation
| Phase . | Percentage of Cells in G0/G*1 . | ||||
|---|---|---|---|---|---|
| . | Cytokine Treatment: Day 0-7/Day 7-14 . | ||||
| . | SCF/SCF . | IL-3/FL . | IL-3/SCF . | FL/IL-3 . | SCF/IL-3 + SCF . |
| II | 92 | 93 | 92 | 88 | 81 |
| III and IV | 92 | 86 | 84 | 69 | 76 |
| Phase . | Percentage of Cells in G0/G*1 . | ||||
|---|---|---|---|---|---|
| . | Cytokine Treatment: Day 0-7/Day 7-14 . | ||||
| . | SCF/SCF . | IL-3/FL . | IL-3/SCF . | FL/IL-3 . | SCF/IL-3 + SCF . |
| II | 92 | 93 | 92 | 88 | 81 |
| III and IV | 92 | 86 | 84 | 69 | 76 |
Sorted G0 CD34+ cells were cultured under sequential cytokine stimulation as indicated for a period of 14 days. Phase II and phase III plus IV cells were sorted on day 14, stained with propidium iodide, and analyzed for cell cycle status. Results shown were derived from one representative experiment out of three. Similar results were obtained in the other two experiments. All experiments contained the same cytokine combinations shown in the first four columns.
Expression of cyclin E (left histogram) and p27 (right histogram) among phase II and phase III plus IV cells isolated from sequential cytokine stimulation of G0CD34+ cells in a two-dimensional tracking experiment. Cells were sorted based on their PKH2 and PKH26 fluorescence as defined in Fig 6 from cultures receiving IL-3 as a primary stimulus and SCF or FL as a secondary stimulus. Phase II cells were those shown in Figs 5, 6, and 7 as green events, and phases III plus IV were those shown as red and blue events, respectively. Cells belonging to Phases III and IV were collected together. Cells were stained for cyclin E and p27 as described in Materials and Methods. Each histogram is an overlay of three separate files derived from the analysis of isotype staining (- - - - -), phase II cells (─) and phase III plus IV cells (━). Positive values for every marker, obtained after background subtraction, are indicated. Similar data were obtained in two other experiments.
Expression of cyclin E (left histogram) and p27 (right histogram) among phase II and phase III plus IV cells isolated from sequential cytokine stimulation of G0CD34+ cells in a two-dimensional tracking experiment. Cells were sorted based on their PKH2 and PKH26 fluorescence as defined in Fig 6 from cultures receiving IL-3 as a primary stimulus and SCF or FL as a secondary stimulus. Phase II cells were those shown in Figs 5, 6, and 7 as green events, and phases III plus IV were those shown as red and blue events, respectively. Cells belonging to Phases III and IV were collected together. Cells were stained for cyclin E and p27 as described in Materials and Methods. Each histogram is an overlay of three separate files derived from the analysis of isotype staining (- - - - -), phase II cells (─) and phase III plus IV cells (━). Positive values for every marker, obtained after background subtraction, are indicated. Similar data were obtained in two other experiments.
DISCUSSION
In this communication, we described a new technique for the isolation of primitive, dormant HPCs from human BM and began to explore mechanisms and kinetics of proliferation of mitotically “synchronized” cells in response to individual, as well as sequential cytokine stimulation. Most, if not all previous studies attempting to examine cell-cycle progression of primitive human HPCs were hampered by the mitotic heterogeneity of test cells because the selection criteria of these cells had not been based on mitotic quiescence of isolated cells. Cells of phenotypically homogeneous populations of HPCs, such as CD34+ CD38−20,21CD34+/c-kitlow/CD38neg/low,22 or CD34+ HLA-DR−14,23,24 are usually defined as dormant only because the majority of these cells reside in G0 /G1 . Such cell cycle status determinations are usually made after the phenotypic selection of target cells using strategies that are neither suitable for the discrimination between G0 and G6,251 nor capable of maintaining cell viability.16,17 Recently, Goodell et al26 described an elegant approach for the isolation of primitive murine HPCs using different emission properties of Hst and went on to show that less than 2% of cells within this unique side population (SP) were in S/G2 + M. Because, in these studies, propidium iodide was used to examine the cell-cycle status of isolated SP cells, again no insight into the proportion of cells residing in G0 versus those in G1 could be gained.
In our studies, and probably for the first time, mitotically homogeneous cells residing in the most dormant phase of the cell cycle, namely G0 , were used for proliferation kinetics studies. It is important to point out that human or murine HPCs isolated by other methods, such as rhodamine 1236,27-30 or Hst and rhodamine 1237,8, may also be mitotically homogeneous. However, no solid proof to this effect was presented in these previous studies, and if so,25 no distinction between the G0 versus G1 status of rhodaminelo cells was attempted. The simplicity of the procedure we described in this study makes it possible to subfractionate any phenotypically defined population of cells, including subsets of CD34+ cells, into the different phases of cell cycle, thus making the investigations into cell cycle regulation of stem cells more accessible. Given that the only commonly used fluorescence channel occupied by this technique is the “red” (FL2) channel and that Hst is excited with UV light, any number of fluorochrome combinations could still be available for the isolation of phenotypically defined fractions of CD34+ cells before their restaining with Hst and PY.
Cells isolated with this technique were not only homogeneous as far as their cell cycle status is concerned, but were also highly enriched for dormant cells possessing primitive hematopoietic activity. A sizable fraction of G0CD34+ cells remained dormant over a period of 7 days persisting in culture as CNR cells. The fraction of CNR cells present among G0CD34+ cells was higher than we previously reported for total CD34+ cells12 and also higher than that surviving in vitro suicide as determined by Berardi et al.31 In addition, G0CD34+ cells were also enriched for LTHC-IC and for cells capable of initiating and sustaining in vitro hematopoiesis, two in vitro measurements that have long been recognized as indicators of primitive hematopoietic progenitor or stem cell activity.12,19 32-37
Proliferation is an integral component of differentiation and maturation of elements of the hematopoietic system. If responsiveness of HPCs to mediators of proliferation, such as cytokines and growth factors, is key to initiation of proliferation and progression through cell cycle, then it stands to reason that a better understanding of these events can be achieved if the effects of single cytokines on dormant, mitotically homogeneous cells are examined. We gain insight into initiation and maintenance of proliferation of G0CD34+ cells by the examination of sequential stimulatory effects of single cytokines. Using double tracking of cultured cells, we were able to identify two types of proliferation signals. The first, a proliferation progression signal, was capable of initiating proliferation and progression through cell cycle and sustaining proliferation through a limited number of cell divisions. All four cytokines examined delivered proliferation progression signals. The second, a proliferation competence signal, was required to sustain maximal proliferation and to propel cells into phase IV. Only IL-3 delivered proliferation competence signals. The degree of proliferation achieved during 14 days in response to different cytokines, and consequently, whether a given cytokine delivers proliferation competence signals cannot be explained by the length of cell cycle of G0CD34+ cells under the influence of various cytokine stimulation. Tanaka et al38 showed that SCF was more efficient than IL-3 in reducing the time required for HPCs to traverse through G1 , suggesting that, if capable, cells under the influence of SCF would undergo a higher number of cell divisions than those exposed to IL-3. Our studies do not support such an observation. Both SCF and IL-3, when present in culture for the entire 14-day period, were capable of inducing a similar number of division cycles allowing G0CD34+ cells to proceed through four or five divisions. However, only IL-3 sustained maximal proliferation of SCF or FL prestimulated G0CD34+ cells, an observation in line with that of Tanaka et al38 who showed that after a 5-day primary stimulation of murine HPCs with SCF and IL-11, “as a single factor, IL-3 was most effective in support of secondary colonies,” suggesting that SCF or FL alone did not provide a proliferation competence signal. Noteworthy in our observations, is that no additional proliferation potential capable of propelling dividing cells beyond phase IV was observed when cells were stimulated with all four cytokines between days 7 and 14 compared with that achieved with IL-3 alone, suggesting again that only IL-3 in this cytokine combination provided proliferation competence signals.
Our studies appear to confirm earlier reports regarding the effects of early cytokines such as FL and SCF on quiescent HPCs. Under our experimental conditions, all three cytokines examined, FL, SCF, and IL-3, were capable of initiating, and to different magnitudes, sustaining the proliferation of G0CD34+ cells. SCF alone sustained cycling and proliferation of human BM and cord blood CD34+ cells.16 Shah et al21 showed that FL induced substantial proliferation of early CD34+ CD38− BM progenitors. Similar results, using the same phenotype of HPC, CD34+ CD38− cells, were reported by Petzer et al.39 In addition, these investigators detected similar, albeit to a lesser degree, stimulatory activity by SCF.39 On the other hand, other investigators40 failed to detect any cycling of CD34+ CD38− cells when examined as a fraction of cultured total CD34+ cells. However, these observations were restricted to the first 48 hours of culture.40 Although unexpected, IL-3 profoundly stimulated the proliferation of G0CD34+ cells, an observation previously reported by Petzer et al.39 In fact, IL-3 was found to be superior to FL and SCF as a single agent in increasing the number of colony-forming cells among cultured CD34+ CD38− cells, an event that is directly associated with proliferation and increase in cell numbers.39 The novelty of our present studies lies in the fact that our experimental design allowed us to distinguish between the ability of a given cytokine to initiate proliferation and promote progression through cell cycle versus the capacity to maintain and sustain these activities. In addition, the technique of double tracking of dividing cells afforded a unique opportunity to investigate whether an orderly sequence of cell cycle progression and proliferation competence signals is required for support of maximal proliferation of primitive HPCs.
At first glance, it was possible to conclude that IL-3 was capable of sustaining maximal proliferation of G0CD34+ cells. However, closer examination of the pattern of proliferation of G0CD34+ cells maintained in IL-3 for 14 days showed that a sizable fraction of cells were unable to progress beyond phase II, a stage of proliferation in our experiments equivalent to up to 4 divisions. Phase II cells proceeded successfully into phases III and IV if the original proliferation stimulus resulted from exposure to SCF or FL. IL-6, as reported by several investigators21,39 was unable to promote sustained proliferation but appeared to be capable of delivering the necessary proliferation progression signals required for resumption of proliferation in response to competence signals. Examination of the mechanisms responsible for the proliferation arrest of phase II cells showed the possible involvement of cyclin-induced disruption of cell-cycle regulation.
Cyclins are integral components of cell-cycle regulation, without which regulation of cell-cycle progression by phosphorylation-dependent signaling pathways would be disrupted. Cyclins, which function as regulatory subunits of cdk are expressed in a cell-cycle stage-specific manner such that the cdk2/cyclin E complex is an essential element for progression of cells through the late G1 restriction point.4 Other components of cell-cycle regulation include different cdk inhibitors that are also cell-cycle stage- or cdk/cyclin-specific. p27 is a specific inhibitor of the activity of the cdk2/cyclin E complex. Because phase II cells appeared to be arrested in G0 /G1 , we investigated the expression, or lack thereof, of cyclin E and p27 among phase II cells. Our results suggest that unscheduled proliferation competence signals received by a subset of G0CD34+ cells result in the disruption of cell-cycle regulation possibly via a mechanism involving the downregulation of expression of cyclin E.
Activation and proliferation of G0CD34+ cells in response to different cytokines showed that, although these cells are mitotically homogeneous, they remain functionally heterogeneous. Studies in the murine system showed that phenotypically homogeneous HSC candidates isolated using different techniques25,26 consist of cells belonging to different phases of the cell cycle. These studies went on to establish that quiescent primitive HPCs (G0 /G1 ) provided a higher degree of radioprotection than their cycling (S/G2 + M) counterparts25 or to confirm that, regardless of their mitotic status, primitive HPCs possessed equivalent long-term engraftment potential.26 Our studies seem to take this controversy a step further by suggesting that even cells isolated from within a very specific phase of cell cycle are composed of subgroups of primitive HPCs with varying degrees of dormancy, cytokine responsiveness, and hematopoietic potential. One plausible explanation for this observation is that G0CD34+ cells most likely contain progeny of differentiating HPCs dividing in vivo, which, at the time of isolation, had traversed through mitosis and re-entered G0 . Such a group of cells, although mitotically identical to other cells within G0CD34+ cells, would be functionally distinct from more primitive HPCs.
In conclusion, we have described a novel approach for the isolation of mitotically homogeneous CD34+ cells enriched for primitive HPCs. This technique, which can easily distinguish between cells residing in different stages of cell cycle, may be a powerful tool for the examination of cell-cycle activation and regulation of hematopoietic cells and to investigate cell-cycle–associated events in these cells. Using this methodology, we examined initiation of proliferation and progression through cell cycle of CD34+ cells residing in G0 in response to sequential individual cytokine stimulation. Our results suggest that orderly activation of CD34+ cells residing in G0 and a programmed response to sequential cytokine stimulation may be part of a control mechanism required for maintenance of proliferation of primitive HPCs. These studies also suggest that unscheduled stimulation of primitive HPCs may result in disruption of cell cycle regulation, possibly through altered expression of cell-cycle stage-specific cyclins. Such findings may prove to be important in the areas of ex vivo expansion and retrovirus-mediated gene transfer.
This work was supported by National Institutes of Health Grant RO1 HL55716 and a research award from the Phi Beta Psi Sorority to E.F.S. A.G. is supported by a fellowship from the Fonds National de la Recherche Scientifique (FNRS, Brussels, Belgium) and by a travel grant from the Centre Anticancéreux prés l'ULg (University of Liège, Belgium).
Address reprint requests to Edward F. Srour, PhD, Indiana University School of Medicine, 975 West Walnut Street, IB 442, Indianapolis, IN 46202-5121.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal