Abstract
We used a new approach called panhandle polymerase chain reaction (PCR) to clone an MLL genomic translocation breakpoint in a case of acute lymphoblastic leukemia of infancy in which karyotype analysis was technically unsuccessful and did not show the translocation partner. Panhandle PCR amplified known MLL sequence 5′ of the breakpoint and 3′ sequence from the unknown partner gene from a DNA template with an intrastrand loop schematically shaped like a pan with a handle. The 7-kb panhandle PCR product contained the translocation breakpoint in MLL intron 8. The partner DNA included unique nonrepetitive sequences, Alu and mammalian apparent LTR-retrotransposon (MaLR) repetitive sequences, and a region of homology to expressed sequence tags. MaLR sequences have not been found before near leukemia-associated translocation breakpoints. The nonrepetitive sequences were not homologous to known partner genes of MLL. Screening of somatic cell hybrid and radiation hybrid lines by PCR and fluorescence in situ hybridization analysis of normal metaphase chromosomes mapped the partner DNA to chromosome band 4q21. Reverse transcriptase-PCR identified an MLL-AF-4 chimeric mRNA, indicating that panhandle PCR identified a fusion of MLL with a previously uncharacterized AF-4 intronic sequence. Panhandle PCR facilitates cloning translocation breakpoints and identifying unknown partner genes.
TRANSLOCATIONS OF THE MLL gene at chromosome band 11q23 are the most common chromosomal aberrations in de novo leukemias of infants and leukemias related to chemotherapy with DNA topoisomerase II inhibitors.1-7 Exogenous damage to the MLL gene may be the common feature in the epidemiology of de novo and treatment-related cases.8-11 An understanding of the pathogenesis requires molecular cloning of MLL genomic breakpoints. However, MLL has approximately 30 different translocation partners and several breakpoints involve partner genes that have not yet been cloned.2 Of cases with MLL gene rearrangement at the level of the Southern blot and successful karyotypes, the karyotypes in one third of cases do not show structural abnormalities of chromosome band 11q23 but, rather, show more proximal abnormalities of 11q, abnormalities of 11p, other structural abnormalities, numerical abnormalities, or no abnormality at all.4,7,12 The karyotypes in cases with partial duplication of several exons of the MLL gene may be normal or they may show trisomy of chromosome 11.13 In other cases with MLL gene rearrangement at the level of the Southern blot, there may be a lack of metaphase cells for karyotype analysis. Thus, MLL genomic breakpoints are a molecular cloning challenge because known 5′ sequence from MLL, but 3′ sequence from unknown partner genes, comprise many of the translocations.
Panhandle polymerase chain reaction (PCR) amplifies genomic DNA with known 5′ sequence and unknown 3′ sequence from template DNA with an intrastrand loop schematically shaped like a pan with a handle.14 We adapted panhandle PCR to clone the translocation breakpoint in a case of de novo acute lymphoblastic leukemia (ALL) of infancy. Southern blot analysis of DNA from the diagnostic marrow showed a germline band and two MLL gene rearrangements, suggesting that one MLL allele had been split by chromosomal translocation.12 The translocation partner was unknown because there were no mitoses on karyotype analysis.12 By adding sequence to the unknown 3′ partner gene that was complementary to a known 5′ MLL sequence, we were able to generate the genomic template with an intrastrand loop for panhandle PCR. This first application of panhandle PCR to amplify a genomic translocation breakpoint suggests that panhandle PCR may be the prototypic approach to identify other breakpoints where the partner gene is undetermined.
MATERIALS AND METHODS
The Institutional Review Board at the Children's Hospital of Philadelphia approved the use of patient and parental specimens as described below. The 3-month-old girl, previously designated patient 38,12 presented with a white blood cell count of 399 × 109/L and large extramedullary tumor burden typical of infant ALL. Consistent with early B-lineage ALL, the bone marrow was replaced by lymphoblasts of French-American-British L1 morphology that expressed tdt, CD34, HLA DR, and CD19, but not CD10, CD20, or myeloid antigens. We previously reported on detection and restriction mapping of the MLL gene rearrangement by Southern blot analysis.12
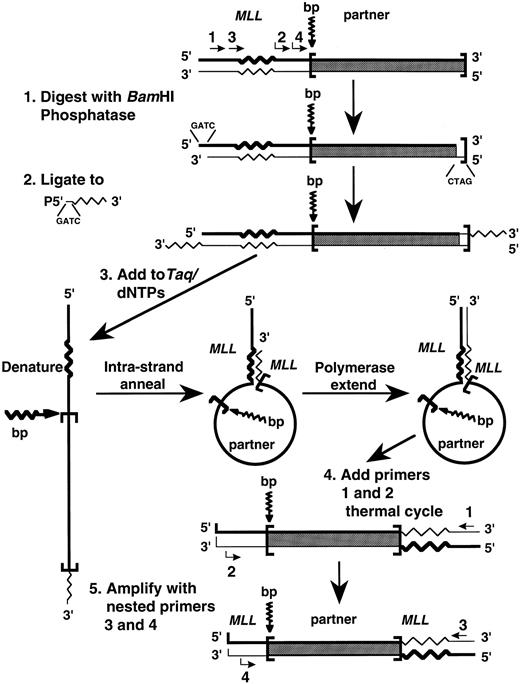
Panhandle PCR amplification of MLL genomic translocation breakpoint. We adapted the panhandle PCR strategy to amplify the genomic breakpoint from the der(11) chromosome with known 5′ MLL sequence juxtaposed to 3′ unknown partner DNA (Fig 1). Genomic DNAs were isolated from the leukemic marrow cells at the time of diagnosis and from control maternal peripheral blood mononuclear cells using 4 mol/L GITC-5.7 mol/L CsCl gradients. Five micrograms of genomic DNA was digested to completion with 40 U BamHI (New England Biolabs, Beverly, MA) to create a 5′ overhang, treated with 0.05 U calf intestinal alkaline phosphatase (Boehringer Mannheim Biochemicals, Indianapolis, IN) at 37°C for 30 minutes, and purified using a GENECLEAN II kit (BIO 101, Inc, La Jolla, CA).
Schematic of panhandle PCR strategy to amplify MLL genomic breakpoint on der(11) chromosome.
Schematic of panhandle PCR strategy to amplify MLL genomic breakpoint on der(11) chromosome.
A single-stranded 5′ phosphorylated oligonucleotide was ligated to the 3′ ends. The 32-nucleotide 3′ end of the oligonucleotide was complementary to nucleotide positions 92-123 in MLL exon 5 in the 5′ portion of BamHI fragment that defines the breakpoint cluster region (bcr).15 The 4-base 5′ end of the oligonucleotide was complementary to the 5′ overhang of BamHI-digested DNA, but would not reconstitute the BamHI site upon ligation. The sequence of the 5′ phosphorylated oligonucleotide was 5′-GAT CGA AGC TGG AGT GGT GGC CTG TTT GGA TTC AGG-3′. The 50 μL ligation reaction mixtures contained 2.5 μg DNA, a 50-fold molar excess of the 5′ phosphorylated oligonucleotide, 1 Weiss Unit T4 DNA ligase (Boehringer Mannheim Biochemicals), and 1 × ligase buffer (Boehringer Mannheim Biochemicals). Ligations were performed overnight at 4°C. The DNA again was purified using a GENECLEAN II kit (BIO 101, Inc).
The sense strand generated the genomic template with an intrastrand loop for panhandle PCR. A 200-ng aliquot of the digested, ligated DNA was added to 2.5 U Taq/Pwo DNA polymerase mix, 385 μmol/L of each dNTP, and PCR reaction buffer at 1.1× final concentration in 45 μL reaction mixtures (Expand Long Template PCR System; Boehringer Mannheim Biochemicals). Preheating reaction mixtures to 80°C before the addition of the DNA prevented nonspecific annealing and polymerization.16 Reaction mixtures then were heated at 94°C for 1 minute to make the template single-stranded.16 Intrastrand annealing of the ligated oligonucleotide to the complementary sequence in MLL and polymerase extension of the recessed 3′ end completed formation of the handle during a 2-minute ramp to 72°C and incubation at 72°C for 30 seconds.16 Intrastrand annealing brought the translocation breakpoint and unknown partner DNA within an intrastrand loop or pan-like structure.16
The next step was to add MLL primers and thermal cycle. The primers were sense with respect to MLL exon 5. MLL primer 1 (5′-TCC TCC ACG AAA GCC CGT CGA G-3′) was upstream to the MLL sequence that was complementary to the ligated oligonucleotide. MLL primer 2 (5′-TCA AGC AGG TCT CCC AGC CAG CAC-3′) was between the MLL sequence that was complementary to the ligated oligonucleotide and the breakpoint junction. The addition of 12.5 pmol of each primer in 2.5 μL brought concentrations in 50 μL final reaction volumes to 350 μmol/L of each dNTP and 1× PCR reaction buffer. After initial denaturation at 94°C for 1 minute, 10 cycles at 94°C for 10 seconds and 68°C for 7 minutes and 20 cycles at 94°C for 10 seconds and 68°C for 7 minutes (increment of 20 seconds/cycle) were used, followed by a final elongation at 68°C for 7 minutes. A nested PCR reaction using a 1-μL aliquot of the first reaction as template DNA enhanced the yield. Sequences of the nested MLL primers, also from MLL exon 5, were 5′-A GCT GGA TCC GGA AAA GAG TGA AGA AGG GAA TGT CTC GG-3′ and 5′-A GCT GGA TCC GTG GTC ATC CCG CCT CAG CCA C-3′. Conditions for nested PCR were the same as in the initial panhandle PCR reaction.
Subcloning and sequencing of the products of panhandle PCR. The panhandle PCR product from the der(11) chromosome in the leukemia DNA was agarose gel-isolated and subcloned into the BamHI site of pBluescript SK II (Stratagene, Inc, La Jolla, CA) using standard methodology. Automated sequencing of three entire individual genomic subclones identified the MLL genomic breakpoint and characterized the 3′ unknown partner DNA. Repeat regions in the partner gene were identified and masked using the Repeat Masker program available through the Washington University Human Genome Center (http://ftp.genome.washington.edu/cgi-bin/mrs/mrs_reg). Masked sequence was submitted for BLAST searches against the nonrepetitive nucleotide database using the server at the Japanese Genome Center at Kyoto (http://www.genome.ad.jp/SIT/BLAST.html).
Direct sequencing of MLL genomic translocation breakpoint. We amplified fresh aliquots of genomic DNA from the leukemia with primers encompassing the translocation breakpoint, which were designed from sequences of the subcloned products of panhandle PCR. Forward and reverse primers derived from MLL and from the partner DNA were 5′-GGG ACT TTC TGT TGG TGG AA-3′ and 5′-GAA ACA CCA GCA AAC CAA CC-3′, respectively, or 5′-ATA CAT GTT GGG TGG CAG G-3′ and 5′-GTC AAG GAA AGG TGG TAT ATC TCA-3′, respectively, and would yield products 450-bp or 411-bp long. The 50-μL PCR reaction mixtures contained 200 ng genomic DNA, 0.5 U Taq Gold DNA polymerase, 250 μmol/L of each dNTP, PCR reaction buffer at 1× final concentration (Perkin Elmer, Norwalk, CT), and 5 pmol of each primer. After the initial denaturation and Taq Gold activation at 95°C for 10 minutes, 35 cycles at 94°C for 15 seconds, 55°C for 15 seconds, and 72°C for 1 minute were used, followed by a final elongation at 72°C for 10 minutes. Products of reactions performed in duplicate for each primer set were isolated from a 1.5% agarose gel using a GENECLEAN II kit (Bio 101, Inc). Approximately 100 ng of pooled purified PCR products was used for each sequencing reaction. Sequencing was performed in both directions by automated methods to verify the translocation breakpoint by an independent method.
Chromosomal localization of the partner DNA. Panels of somatic cell hybrid DNAs and radiation hybrid DNAs were screened by PCR to localize the partner DNA. For the somatic hybrid screen, the 50-μL PCR reaction mixtures contained 500 ng somatic cell hybrid DNA (Bios Laboratories, New Haven, CT), 1.25 U AmpliTaq DNA polymerase, 200 μmol/L of each dNTP, PCR reaction buffer at 1× final concentration (Perkin Elmer), and 12.5 pmol of each primer. Forward and reverse primer sequences were 5′-CCT ACA CCC AGC CAA ACT GT-3′ and 5′-ATG GTA CCA GAA CAG GGC AG-3′, respectively, and would yield a product 267 bp in length. After initial denaturation at 94°C for 9 minutes, 35 cycles at 94°C for 1 minute, 55°C for 1 minute, and 72°C for 2 minutes were used, followed by a final elongation at 72°C for 7 minutes. Human and hamster genomic DNAs were controls. Twenty-microliter aliquots of PCR reaction mixtures were electrophoresed in 4% Nusieve agarose gels (FMC Corp, Rockland, ME). Reactions mixtures yielding products were compared with the known human chromosome complement of the somatic hybrid panel to determine the location of the partner gene.
For the radiation hybrid screen, the primers were the same. The 20 μL PCR reaction mixtures contained 25 ng DNA from the Stanford G3 radiation hybrid panel (Research Genetics, Huntsville, AL), 0.5 U Taq Gold DNA polymerase, 250 μmol/L of each dNTP, PCR reaction buffer at 1× final concentration (Perkin Elmer), and 5 pmol of each primer. After initial denaturation and Taq Gold activation at 95°C for 10 minutes, we used a two-phase touchdown protocol for annealing and extension. The first phase included 16 cycles at 95°C for 45 seconds and 70°C for 1 minute (decrease 0.7°C/cycle) to reach a final combined annealing and extension temperature of 59°C. The second phase included 26 cycles at 95°C for 45 seconds, 55°C for 30 seconds, and 72°C for 1 minute and was followed by a final elongation at 72°C for 5 minutes. PCR reaction mixtures were electrophoresed in 4% Nusieve agarose gels (FMC Corp). Reactions yielding products and reactions without products were scored as 1 and 0, respectively. Results were submitted to the radiation hybrid server of the Stanford Human Genome Center (http://www-shgc.stanford.edu/rhserver2/rhserver_form.html) to determine the location of the partner DNA.
For further verification of the location of the partner gene, the 7-kb subclone 34-1 containing the genomic breakpoint junction was used as probe in fluorescence in situ hybridization (FISH) analysis. The probe was labeled with biotin-16-dUITP and FISH analysis was performed on metaphases from peripheral blood lymphocytes of a normal male using standard methods.17
Reverse transcriptase-PCR (RT-PCR) analysis. RT-PCR analysis was performed to evaluate whether the translocation fused MLL with the AF-4 gene. We used the Superscript Preamplification System and random hexamers for synthesis of cDNA from 4 μg total RNA according to the manufacturer's directions (GIBCO BRL, Gaithersburg, MD). The 100 μL initial RT-PCR reaction mixtures contained 2 μL random hexamer-primed cDNA, 2.5 U AmpliTaq DNA polymerase, 200 μmol/L of each dNTP, PCR reaction buffer at 1× final concentration (Perkin Elmer), and 100 pmol of each primer. The forward primer from MLL exon 6, MLLEx6S, and the reverse primer from the AF-4 gene, LTG4AS2, have been described.18 After initial denaturation at 95°C for 2 minutes, 35 cycles at 95°C for 1 minute, 62°C for 2 minutes, and 72°C for 1 minute were used, followed by a final elongation at 72°C for 10 minutes. Second round RT-PCR reactions were performed using 2-μL aliquots of the initial RT-PCR reactions as the templates. Primers and conditions were the same as for initial RT-PCR, except that the annealing temperature was 65°C. The cell line RS4:11, which is known to have an MLL genomic breakpoint in intron 7 and to yield a 627-bp product with these primers, was the positive control.18
RT-PCR reaction mixtures were electrophoresed in VisiGel Separation Matrix (Stratagene, Inc) to visualize the products. Products of four second round reactions were electrophoresed in 1% agarose and gel-purified for sequencing using a GENECLEAN III kit (Bio 101, Inc). Seventy nanograms of purified RT-PCR products (∼10 ng/100 bases) was used for direct automated sequencing with the same forward and reverse primers as those used for RT-PCR.
Automated sequencing of expressed sequence tag (EST) H73415. EST H73415 (Genome Systems, St Louis, MO) derived from the Soares human fetal liver spleen cDNA library (dbEST Id:375797) was obtained as a bacterial stab in the vector pT7T3D-Pac and isolated as individual colonies from an LB/agar plate containing 100 μg/mL ampicillin. The entire EST was sequenced in forward and reverse directions with a T3 sequencing primer and sequencing primers used to characterize the partner DNA.
RESULTS
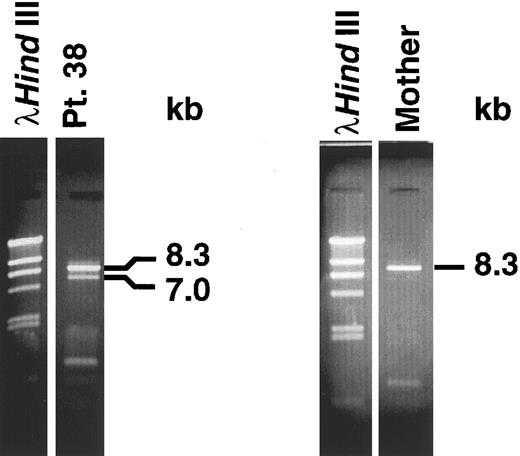
Panhandle PCR identifies genomic breakpoint in MLL intron 8. We obtained the predicted 8.3-kb panhandle PCR product from the normal MLL genes in control maternal DNA. We obtained both a 7-kb product from the der(11) chromosome and an 8.3-kb product from the normal MLL allele in the leukemia (Fig 2). We subcloned the 7-kb product from the der(11) chromosome and sequenced three individual genomic subclones.
Panhandle PCR products. A 7.0-kb product from der(11) chromosome and 8.3-kb product from the normal MLL allele were obtained from leukemic marrow DNA of patient 38 (left). An 8.3-kb product from normal MLL alleles was obtained from maternal peripheral blood lymphocyte DNA (right).
Panhandle PCR products. A 7.0-kb product from der(11) chromosome and 8.3-kb product from the normal MLL allele were obtained from leukemic marrow DNA of patient 38 (left). An 8.3-kb product from normal MLL alleles was obtained from maternal peripheral blood lymphocyte DNA (right).
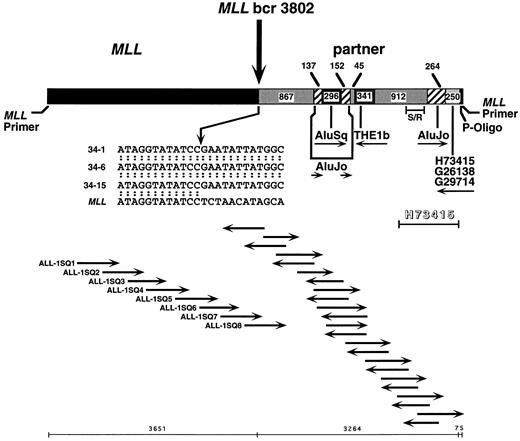
Previous Southern blot analysis had localized the translocation breakpoint to within the first 4,464 bp of the 8.3-kb MLL genomic bcr.12 Automated sequencing of the 5′ bcr in subclone 34-1 from panhandle PCR identified the MLL genomic breakpoint at nucleotide 3802 in intron 8 and partial sequence of the partner DNA (Fig 3). The sequencing strategy is shown in Fig 3. Sequencing of two additional subclones from panhandle PCR, 34-6 and 34-15, with primers ALL-1-SQ7 and ALL-1-SQ8 that identified the breakpoint in subclone 34-1, verified the MLL genomic breakpoint at nucleotide 3802 (Fig 3).
Sequence of der(11) in individual subclones, 34-1, 34-6, and 34-15, from panhandle PCR. The 5′ 3,651 bp include MLL forward nested primer and MLL bcr sequence. The 3,224 bp of 3′ sequence are partner DNA. The most 3′ 75 bp of sequence extend from ligated phosphorylated oligonucleotide (P-Oligo) through reverse nested primer. Sequencing strategy and orientation of sequencing primers are shown at bottom. Primers ALL-1-SQ7 and ALL-1-SQ8 identified the breakpoint in subclone 34-1 and verified the breakpoint in subclones 34-6 and 34-15. Comparison with normal MLL genomic sequence localized the breakpoint to nucleotide 3802. The insert at middle is the breakpoint junction sequence in subclones 34-1, 34-6, and 34-15, where the arrow shows the breakpoint. The partner DNA includes two unique nonrepetitive sequences 867 bp and 912 bp in size, Alu and THE1b MaLR repeats, and a 3′ region of homology to 255 bp of existing sequences of known ESTs. Each subclone ended at the 3′ side with the ligated oligonucleotide and MLL reverse nested primer sequence. Sequences were the same in all three subclones. Further sequencing of EST H73415 in entirety in forward and reverse directions showed homology at 1,033 of 1,034 bases in the indicated region (shadowed). S/R indicates a 267-bp region amplified in somatic cell hybrid and radiation hybrid screens.
Sequence of der(11) in individual subclones, 34-1, 34-6, and 34-15, from panhandle PCR. The 5′ 3,651 bp include MLL forward nested primer and MLL bcr sequence. The 3,224 bp of 3′ sequence are partner DNA. The most 3′ 75 bp of sequence extend from ligated phosphorylated oligonucleotide (P-Oligo) through reverse nested primer. Sequencing strategy and orientation of sequencing primers are shown at bottom. Primers ALL-1-SQ7 and ALL-1-SQ8 identified the breakpoint in subclone 34-1 and verified the breakpoint in subclones 34-6 and 34-15. Comparison with normal MLL genomic sequence localized the breakpoint to nucleotide 3802. The insert at middle is the breakpoint junction sequence in subclones 34-1, 34-6, and 34-15, where the arrow shows the breakpoint. The partner DNA includes two unique nonrepetitive sequences 867 bp and 912 bp in size, Alu and THE1b MaLR repeats, and a 3′ region of homology to 255 bp of existing sequences of known ESTs. Each subclone ended at the 3′ side with the ligated oligonucleotide and MLL reverse nested primer sequence. Sequences were the same in all three subclones. Further sequencing of EST H73415 in entirety in forward and reverse directions showed homology at 1,033 of 1,034 bases in the indicated region (shadowed). S/R indicates a 267-bp region amplified in somatic cell hybrid and radiation hybrid screens.
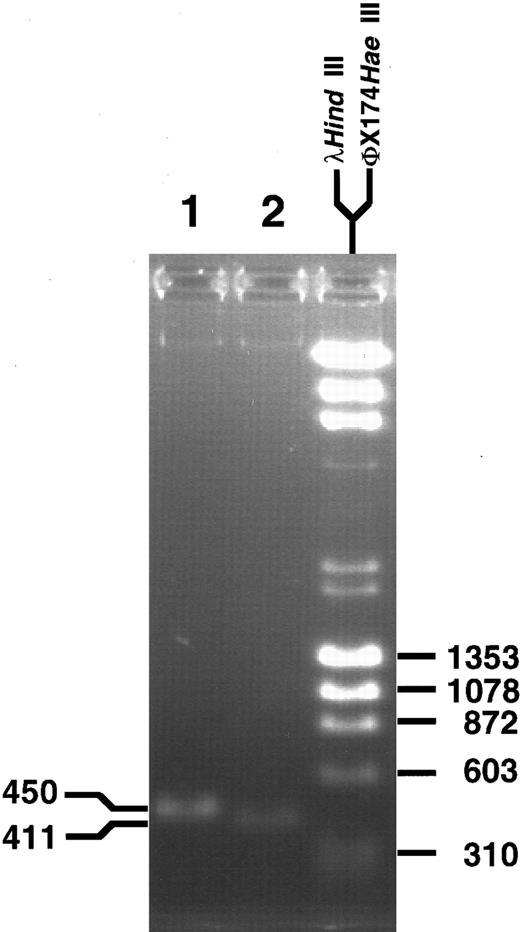
For further confirmation of the breakpoint sequence, fresh aliquots of genomic DNA from the leukemia were amplified in conventional PCR reactions with two different primer sets encompassing the translocation breakpoint. The PCR products were 450 and 411 bp long, as expected from locations of the primers in MLL and partner DNA (Fig 4). Direct sequencing of the PCR products in both directions identified the translocation breakpoint at nucleotide 3802 in MLL and showed fidelity of the sequence in the three subclones of products of panhandle PCR.
PCR products obtained with primer sets from MLL and partner DNA encompassing the genomic breakpoint. Direct sequencing in both directions confirmed the translocation breakpoint at nucleotide 3802 in the MLL genomic bcr.
PCR products obtained with primer sets from MLL and partner DNA encompassing the genomic breakpoint. Direct sequencing in both directions confirmed the translocation breakpoint at nucleotide 3802 in the MLL genomic bcr.
The partner DNA is not homologous to known partner genes of MLL. Complete forward and reverse sequences of the partner DNA in subclones 34-1, 34-6, and 34-15 were then determined. Figure 3 summarizes the results. The first 867 bp 3′ of the breakpoint were a unique nonrepetitive sequence. An AluJo interrupted by an AluSq was immediately downstream, followed by a THE1b member of the mammalian apparent LTR-retrotransposon (MaLR) family of repeats, 912 bp of unique nonrepetitive sequence, and another AluJo . The most 3′ 250 bp of partner DNA in each subclone were identical to existing sequences of ESTs H73415, G26138, and G29714. Sequences of the phosphorylated MLL oligonucleotide used to generate the template for panhandle PCR and the reverse nested primer followed last. The unique sequences of the partner DNA were not homologous to known partner genes of MLL.
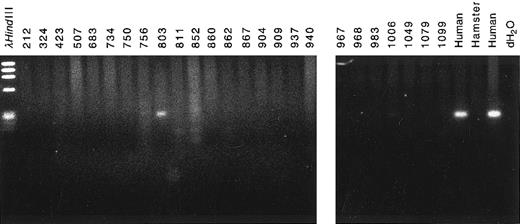
The partner DNA originated from chromosome band 4q21. To determine the chromosomal location of the partner DNA, we screened panels of somatic cell hybrid DNAs and radiation hybrid DNAs by PCR. Amplification of a PCR product from cell line 803 in the somatic hybrid panel (Bios Laboratories, New Haven, CT) indicated that the partner DNA was from human chromosome 4 (Fig 5). PCR amplification of radiation hybrid lines in the Stanford G3 radiation hybrid panel showed that the partner DNA was in the same bin as the framework marker D4S1542 at chromosome band 4q21 (data not shown). The PCR primers used to screen the panels of somatic hybrid DNAs and radiation hybrid DNAs were from a more 5′ region of the partner DNA than the 250-bp region of homology to existing sequences of ESTs H73415, G26138, and G29714 (Fig 3). Thus, the chromosome band 4q21 location of the ESTs independently corroborated the location of the partner DNA.
Chromosomal localization of partner DNA by screening panel of somatic cell hybrid DNAs by PCR. Cell line 803 contained the complement of human chromosome 4 (Bios Laboratories). Human and hamster genomic DNAs were positive and negative controls, respectively.
Chromosomal localization of partner DNA by screening panel of somatic cell hybrid DNAs by PCR. Cell line 803 contained the complement of human chromosome 4 (Bios Laboratories). Human and hamster genomic DNAs were positive and negative controls, respectively.
For further verification of the location of the partner DNA, subclone 34-1 containing the genomic breakpoint junction was used as probe in FISH analysis. The probe consisted of 3,651 bp of MLL sequence extending from the nested forward primer to the translocation breakpoint, 3,264 bp of sequence from the partner gene, and an additional 75 bp of MLL sequence extending from the ligated phosphorylated oligonucleotide through the reverse nested primer used for PCR (Fig 3). We examined 20 metaphases from human peripheral blood lymphocytes of a normal male. Signal was detected on at least one chromosome 11 in 9 of 20 cells. Signal was detected at proximal 4q in 5 of 20 cells (data not shown). Because of the small size of the probe, signal was not detected in every cell. However, more importantly, there was no significant hybridization elsewhere in the genome. These data are consistent with a location of the partner DNA at chromosome band 4q21 and indicate that panhandle PCR amplified a genomic translocation breakpoint involving MLL and partner DNA from chromosome band 4q21.
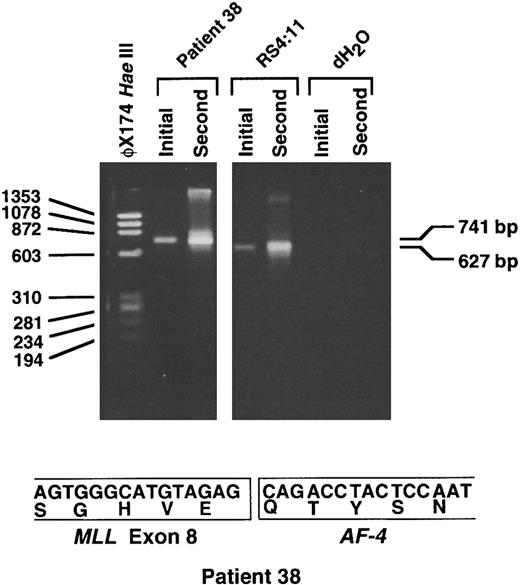
RT-PCR analysis shows MLL-AF-4 chimeric mRNA. Because the partner DNA originated from chromosome band 4q21, we performed RT-PCR analysis on randomly primed cDNA from the leukemic cells of patient 38 to evaluate whether the translocation joined MLL to AF-4. Initial and second round RT-PCR reactions with sense and antisense primers from MLL and AF-4, respectively, showed the predicted 627-bp product in the positive control cell line RS4:11.18 In the leukemia of patient 38, initial and second round reactions gave a single 741-bp product (Fig 6). Direct sequencing of the products of four separate second round reactions showed an in-frame fusion of MLL exon 8 to the AF-4 gene at position 1459 of the AF-4 cDNA.19 These data indicate that the unknown partner DNA that panhandle PCR had amplified was from a previously uncharacterized region of the AF-4 gene.
RT-PCR analysis showing MLL-AF-4 chimeric mRNA. Initial and second round RT-PCR reactions were performed using random hexamer-primed cDNA prepared from total RNA from the leukemic cells of patient 38. Sequences of sense MLL primer from exon 6 and LTG4AS2 antisense primer from AF-4 have been described.18 Four separate initial and second round RT-PCR reactions each gave a single 741-bp product, as shown by example in the figure (top). Direct sequencing of the products of each second round reaction showed an in-frame fusion of MLL exon 8 to AF-4 at position 1459 in AF-4 cDNA19 (bottom). The predicted 627-bp product was obtained in positive control cell line RS4:11 (top). RNA negative controls (dH2O) for initial and second round reactions also are included (top).
RT-PCR analysis showing MLL-AF-4 chimeric mRNA. Initial and second round RT-PCR reactions were performed using random hexamer-primed cDNA prepared from total RNA from the leukemic cells of patient 38. Sequences of sense MLL primer from exon 6 and LTG4AS2 antisense primer from AF-4 have been described.18 Four separate initial and second round RT-PCR reactions each gave a single 741-bp product, as shown by example in the figure (top). Direct sequencing of the products of each second round reaction showed an in-frame fusion of MLL exon 8 to AF-4 at position 1459 in AF-4 cDNA19 (bottom). The predicted 627-bp product was obtained in positive control cell line RS4:11 (top). RNA negative controls (dH2O) for initial and second round reactions also are included (top).
The partner DNA is identical to EST H73415. The most 3′ 250 bp of partner DNA in subclones 34-1, 34-6, and 34-15 from panhandle PCR were identical to existing sequences of ESTs H73415, G26138, and G29714 (Fig 3). We obtained and sequenced the entire EST H73415 (Genome Systems) from the Soares human fetal liver and spleen cDNA library (dbEST Id:375797), in forward and reverse directions. The EST was 1,034 bp in length. The sequence of the EST was homologous to the 3′ 1,034 bp of the partner DNA in subclones 34-1, 34-6, and 34-15 (Fig 3). The homology was in 1,033 of 1,034 bp and extended 5′ through the AluJo into the 912-bp region of unique nonrepetitive sequence, where the EST subclone ended (Fig 3). Neither the sequence of the partner DNA in the full-length products of panhandle PCR nor the region of homology with EST H73415 contained intron-exon boundaries or shared homology with the full-length AF-4 cDNA.19 These results suggest that the partner DNA in the panhandle PCR products was a previously uncharacterized intronic region of the AF-4 gene.
DISCUSSION
In the present study, we implemented panhandle PCR14,16,20 as a new approach to MLL genomic breakpoint cloning. We designed the panhandle PCR strategy to amplify the breakpoint from the der(11) chromosome with known 5′ MLL sequence and 3′ sequence from an unknown partner gene in the case of infant ALL, but the same methodology and primers should lend easily to the amplification of additional MLL genomic breakpoints within the bcr. Because rearrangement sizes on Southern blot analysis with the B859-BamHI probe-restriction enzyme combination in the case of infant ALL were 2 kb and 7 kb,12 the 7-kb panhandle PCR product was consistent with the Southern blot and, moreover, indicated that the 7-kb rearrangement on Southern blot analysis was from the der(11) chromosome. Consistent with restriction mapping,12 the MLL genomic breakpoint in the panhandle PCR products was at position 3802 in intron 8 in the 5′ bcr. Confirmation of the breakpoint by the independent method of direct genomic sequencing validated this result. Thus, with a newer DNA polymerase and proof reading enzyme, panhandle PCR successfully amplified 7-kb and 8.3-kb products from the der(11) chromosome and the normal MLL allele in the case of infant ALL and an 8.3-kb product from the normal MLL alleles in control maternal DNA.
One objective was to identify the partner gene of MLL in the case of infant ALL in which karyotype analysis did not give information about the translocation partner. At the time of this work, 10 partner genes of MLL already had been cloned.19 21-27 The partner DNA did not share homology with known partner genes of MLL, but the most 3′ 250 bp of partner DNA were homologous to three identical ESTs. The known location of the ESTs was chromosome band 4q21. By sequencing EST H73415, we further determined that the 3′ 1,034 bp of partner DNA matched the sequence of the full-length EST. In addition, three independent methods including screening of somatic cell hybrid and radiation hybrid lines by PCR with primers upstream from the region of homology with the ESTs and FISH analysis of normal metaphase chromosomes mapped the partner DNA to chromosome band 4q21.
Screening of radiation hybrid DNAs previously indicated that the nearest linked marker to the AF-4 gene, the most common partner gene of MLL in infant ALL, was also D4S1542 at chromosome band 4q21.28 Nakamura et al19 have published on the full-length cDNA sequence of the AF-4 gene, but there is only limited intronic sequence information on AF-4. Neither the nonrepetitive sequences in the partner DNA nor the ESTs were homologous to known sequences of AF-4. Nonetheless, RT-PCR proved definitively that the partner gene of MLL in the case of infant ALL was AF-4. Because the sequence of the partner DNA in the panhandle PCR products did not show intron-exon boundaries or regions of homology with the full-length AF-4 cDNA, these results suggest that the partner DNA amplified by panhandle PCR was a new intronic sequence of the AF-4 gene.
The intronic region of the AF-4 gene included unique nonrepetitive sequences and both Alu and MaLR repeats. Alu and MaLR are distinct families of interspersed repetitive sequences that occur within the genome after retroposition through RNA intermediates.29,30 Alu sequences, the most prevalent human repeats, previously have been found in the MLL genomic bcr and ascribed a role in MLL gene translocations.31 MaLR sequences such as the THE1b, which comprise 0.5% to 2% of the human genome,29 have not been found before near leukemia-associated translocation breakpoints.
These results constitute a novel application of panhandle PCR: amplification of a genomic translocation breakpoint in which the partner gene was undetermined. The unknown partner DNA amplified by panhandle PCR was a previously uncharacterized intronic region of the AF-4 gene at chromosome band 4q21. Panhandle PCR should facilitate discovering additional partner genes of MLL and may be the prototypic approach for cloning translocation breakpoints besides those involving MLL in which the partner gene is undetermined.
ACKNOWLEDGMENT
The authors thank Terence Williams for help with figure preparation.
C.A.F. is supported by National Institutes of Health Grant No. 1R29CA66140-02, American Cancer Society Grant No. DHP143, a Leukemia Society of America Scholar Award (1996-2001), the National Childhood Cancer Foundation, a National Leukemia Research Association Grant in Memory of Maria Bernabe Garcia, and the Children's Hospital of Philadelphia High Risk High Impact Grant. C.S.K. is supported by National Institutes of Health Grant No. 5R25CA4009. P.C.N. is supported by National Institutes of Health Grant No. CA42232.
Address reprint requests to Carolyn A. Felix, MD, Division of Oncology, Leonard and Madlyn Abramson Pediatric Research Center, Room 902B, Children's Hospital of Philadelphia, 324 S 34th St, Philadelphia, PA 19104-4318.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal