Abstract
Oncostatin M (OSM) and leukemia inhibitory factor (LIF ) are members of the interleukin-6 (IL-6) subfamily of cytokines that use a common signal transducer gp130. Human OSM (hOSM) and LIF share a functional high-affinity receptor that is composed of gp130 and LIF receptor β subunit (LIFRβ). A second high-affinity receptor for hOSM was recently found to be formed by gp130 and the hOSM receptor β subunit. However, the nature of murine OSM (mOSM) and its receptors has remained unknown. Using the recently cloned mOSM cDNA, we produced recombinant mOSM and studied its biological activity and receptor structure. Murine hematopoietic cell lines M1 and DA1.a, an embryonic stem cell line CCE, and Ba/F3 transfectants expressing gp130 and LIFRβ responded to murine LIF (mLIF ) and hOSM equally well, while these cells responded to mOSM only at a 30-fold to 100-fold higher concentration than those of mLIF and hOSM. In contrast, NIH3T3 cells responded to mOSM, but not to mLIF and hOSM. Scatchard plot analyses showed that mOSM bound to gp130 with low-affinity (kd = 2.8 to 4.2 nmol/L) and that the binding affinity did not increase in the presence of LIFRβ. However, mOSM bound to NIH3T3 cells with high-affinity (kd = 660 pmol/L), whereas mLIF did not bind to NIH3T3 cells at all. These results indicate that unlike hOSM, mOSM and mLIF do not share the same functional receptor, and mOSM delivers signals only through its specific receptor complex. Further studies in mice will define the physiological roles of OSM.
MULTIPLE CYTOKINES regulate cell growth and differentiation in a wide variety of biological systems including hematopoiesis, osteogenesis, and neurogenesis. Cytokines also play a crucial role in the immune and inflammatory responses.1 It has been known that more than two different cytokines often induce the same biological response in the same target cells, while each cytokine exhibits its specific biological activity.2 Functional overlap among a subset of cytokines is now well explained by the findings that receptors for a subset of cytokines with similar functions share a signaling subunit, eg, the common β subunit for the interleukin-3 (IL-3)/granulocyte-macrophage colony-stimulating factor (GM-CSF )/IL-5 receptors; the common γ subunit for IL-2, IL-4, IL-7, IL-9, and IL-15 receptors; and gp130 for the IL-6, IL-11, oncostatinM (OSM), leukemia inhibitory factor (LIF ), ciliary neurotrophic factor, and cardiotrophin-1 receptors.3-5 On the other hand, cytokine specificity is mainly due to the cell-type specific expression of cytokine receptors, eg, the erythropoietin (EPO) receptor is predominantly expressed in erythroid cells6 as is the IL-5 receptor α subunit in eosinophils.7
Among the IL-6/LIF family of cytokines, OSM and LIF are the most closely related.8,9 They are not only structurally related, but, additionally, their genes are tightly linked in human chromosome 22q12 and mouse chromosome 11, suggesting that they arose by gene duplication.10-12 In humans these two cytokines are known to exhibit extensive overlapping biological activities, eg, induction of differentiation in M1 myeloid leukemic cells,13 inhibition of differentiation of embryonic stem (ES) cells,14 and induction of acute phase protein expression in primary hepatocytes.15,16 On the other hand, OSM exhibits unique activities that are not shared with LIF, eg, growth inhibition of A375 human melanoma cells,17,18 autocrine growth stimulation of Kaposi sarcoma,19 induction of tissue inhibitor of metalloproteinase-1 (TIMP-1) and other early response genes on human fibroblasts and hepatoma cells,20,21 and the stimulation of DNA synthesis in rabbit vascular smooth muscle cells.22
Binding studies using human OSM and LIF have established the presence of two distinct hOSM receptors.23-25 The type I hOSM receptor is identical to the high-affinity LIF receptor, which is composed of the LIF receptor β subunit (LIFRβ) and the IL-6 signal transducer gp130. LIF binds to LIFRβ with low-affinity, and this binding affinity is increased by gp130, which does not bind LIF by itself.23 In contrast, hOSM binds to gp130 with low-affinity and the binding affinity is increased by LIFRβ, which does not bind OSM by itself.23,26 As LIF and OSM bind to the same receptor, they compete for high-affinity binding with each other. Thus, the common functions between LIF and OSM are mediated by a common receptor consisting of gp130 and LIFRβ (type I OSM receptor). Furthermore, there are several cell lines that bind OSM, but not LIF with high-affinity, and the OSM binding is not competed by LIF in those cells,24 suggesting the presence of a second OSM specific receptor (type II OSM receptor). This possibility was recently proven by the isolation of the OSM receptor specific β subunit (OSMRβ) cDNA.25 The second high-affinity OSMR is composed of gp130 and OSMRβ, and the distribution of gp130/LIFRβ (type I OSMR) and gp130/OSMRβ (type II OSMR) partly overlaps. Thus, OSM function is believed to be manifested through two types of functional receptors. However, as the murine OSM had not been identified until very recently, characterization of OSM and its receptors was entirely based on studies using hOSM.
During the screening of cytokine-inducible genes, we recently cloned a murine OSM (mOSM) cDNA as an immediate early response gene induced by IL-2, IL-3, and EPO in hematopoietic cells.27 This gene induction is mediated by the activation of STAT5 (signal transducer and activator of transcription-5). Murine OSM is produced most abundantly in bone marrow cells, and the level of OSM expression in bone marrow is much higher than that of murine LIF (mLIF )27 (M.I. and A.M., unpublished data). Using mOSM cDNA, we have produced recombinant OSM and characterized its biological activities, as well as its functional receptors. In this study, we present evidence that, in contrast to human receptors, mOSM and mLIF do not share the same functional high-affinity receptor.
MATERIALS AND METHODS
Cells and cytokines.DA1.a and Ba/F3 cells were maintained in a RPMI 1640 medium supplemented with 10% fetal calf serum (FCS) in the presence of murine IL-3 (100 U/mL). M1 cells were cultured in RPMI 1640 containing 10% FCS. NIH3T3 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf serum. CCE cells were maintained in DMEM supplemented with 15% FCS in the presence of mLIF (10 ng/mL).
Human and murine LIF, hOSM, human IL-6, and soluble IL-6 receptors were purchased from R&D Systems (Minneapolis, MN). Murine IL-3 was produced in silkworms as previously described.28
Production of recombinant mOSM.Mature mOSM (Fig 1) cDNA lacking the native leader sequence was amplified by polymerase chain reaction (PCR) with a set of primers [5′-ATATGCGGCCGCGCCTGGCTGCTCCAAC-3′ (sense) and 5′-GACATAGAATTCCTATCTCCGGCTGCGTGTGGAGCC-3′ (antisense)]. After endonuclease digestion with Not I and EcoRI, the PCR product was cloned into pSRαCHFX vector (kindly provided by S. Albright and F. Lee, DNAX, Palo Alto, CA), which contains SRα promoter29 followed by sequences encoding CD8α signal peptide linked with Flag epitope tag (Eastman Kodak, New Haven, CT) and eight histidine residues to generate an epitope-tagged mOSM (Fig 1A). The expression plasmid was transfected into COS7 cells by the diethyl aminoethyl (DEAE)-dextran method as described previously.30 Four days after transfection, the culture medium was collected and the epitope-tagged mOSM was purified by column chromatography using Ni2+-nitrilo–triacetic acid resin (Qiagen, Chatsworth, CA) and anti-Flag M2 affinity resin (Eastman Kodak) according to the manufacturers' instructions. Fractions exhibiting the growth inhibitory activity for NIH3T3 cells were pooled, concentrated by ultrafiltration, and quantitated by BCA Microassay kit (Pierce, Rockford, IL). The purity of mOSM was verified by 8% to 16% polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate (SDS) followed by staining with express Coomassie stain (Diversified Biotech, Boston, MA).
Production of the recombinant mOSM. (A) Schematic representation of structure of the native mOSM and the epitope-tagged recombinant mOSM. (B) The recombinant mOSM (rmOSM) protein expressed in COS7 cells. Culture supernatant from COS7 cells transfected with the epitope-tagged mOSM cDNA was used to purify mOSM to homogeneity and the purified protein was analyzed by 8% to 16% polyacrylamide gel electrophoresis in the presence of SDS. Position of molecular weight standards are indicated on right by kilodalton (kD).
Production of the recombinant mOSM. (A) Schematic representation of structure of the native mOSM and the epitope-tagged recombinant mOSM. (B) The recombinant mOSM (rmOSM) protein expressed in COS7 cells. Culture supernatant from COS7 cells transfected with the epitope-tagged mOSM cDNA was used to purify mOSM to homogeneity and the purified protein was analyzed by 8% to 16% polyacrylamide gel electrophoresis in the presence of SDS. Position of molecular weight standards are indicated on right by kilodalton (kD).
Transfection into cells.Murine gp130 cDNA (kindly provided by Dr Taga, Tokyo Medical and Dental University, Tokyo, Japan) and mLIFRβ cDNA (kindly provided by Dr Tomida, Saitama Cancer Center) were placed under the SRα promoter of the expression vector pME18S (K. Maruyama and A.M., unpublished data). The drug selectable gene fragments conferring resistance to G418 or hygromycin B was introduced into the plasmid constructs. Ba/F3 cells were transfected with the linealized plasmids by electroporation, and transfectants were selected with G418 (1 mg/mL) or hygromycin B (1 mg/mL) as previously described.30
Radioiodination and binding assay.The purified recombinant mOSM and mLIF were radioiodinated using the Bolton-Hunter reagent kit (ICN, Costa Mesa, CA) according to the manufacturer's instructions. The specific radioactivity was determined by self-displacement analysis as previously described.31 The specific radioactivities of [125I]-labeled mOSM and mLIF were 1.08 × 105 and 3.39 × 105 cpm/ng, respectively. Maximal binding capacity for the iodinated mOSM and mLIF was 0.66 and 0.64, respectively. Scatchard plot analysis was performed as described previously.30 In brief, cells (1 to 2 × 106) were incubated in 100 μL binding buffer (DMEM containing bovine serum albumin [1 mg/mL] and 20 mmol/L HEPES [pH 7.4]) containing various concentrations of [125I]-labeled mOSM or mLIF in the presence or absence of a 100-fold excess amount of unlabeled ligands at 4°C for 2 hours. Cell-bound radioactivity was separated from the free radioactivity by centrifugation through an oil layer as described previously. The data of the binding assays were analyzed by the Ligand program.32
Proliferation and growth inhibition assays.Colorimetric assay using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma) described by Mosmann33 was performed to measure the cell proliferation or the growth inhibition of Ba/F3 transfectants, DA1.a cells, and NIH3T3 cells. In brief, 2 × 104 of Ba/F3 transfectants and DA1.a cells or 2 × 103 of NIH3T3 cells were incubated in 100 μL medium with various concentrations of cytokines in 96-well multiwell plates. After 2 or 4 days (Ba/F3 and DA1.a or NIH3T3, respectively), 10 μL of MTT (5 mg/mL in phosphate-buffered saline [PBS]) was added to each well and further incubated for 4 hours before the colorimetry.
Inhibition of M1 cell proliferation was measured by using [3H] thymidine as described elsewhere.10 In brief, M1 cells (2.5 × 104) were incubated in 100 μL of medium containing various concentrations of mOSM or mLIF for 3 days in 96-well multiwell plates. Cells were then incubated with [3H] thymidine for 4 hours, and radioactivity incorporated in the cells was measured.
RESULTS
Preparation of recombinant mOSM.OSM is produced as a precursor form carrying the signal peptide and the C-terminal tandem repeats of charged amino acid residues, which are removed by proteolytic cleavage in the cells (Fig 1A). There are four such repeats in the C-terminal region of the mOSM precursor, whereas there are only two in hOSM.27 Linsley et al34 reported that the proteolytic cleavage of the C-terminal portion makes hOSM sevenfold to 10-fold more active than the unprocessed form. Therefore, to obtain recombinant mOSM with higher specific activity, the C-terminal portion was removed and the modified open reading frame was inserted downstream of the Flag epitope and histidine tag sequences for detection and purification (Fig 1A). Addition of such epitope tags to mOSM did not affect the biological activity in a growth inhibition assay of NIH3T3 cells (see below). After transient transfection into COS7 cells, the supernatants were collected and subjected to the purification process as described in Materials and Methods. Figure 1B depicts the purified recombinant mOSM as a single band of 36 kD.
Biological activities of mOSM.We first evaluated biological activities of mOSM on LIF-responsive cell lines. M1 is a murine myeloid cell line that ceases proliferation and differentiates to macrophages in response to murine IL-6, mLIF, or hOSM.10 Like these cytokines, mOSM also inhibited the proliferation of M1 cells and induced their differentiation (Fig 2A and data not shown). However, more than 30-fold higher concentration of mOSM was required for the full growth inhibition of M1 when compared with mLIF or hOSM (Fig 2A and data not shown). We next examined the response of DA1.a, an IL-3–dependent murine lymphoid subline that also proliferates in response to LIF.35 While mLIF stimulated its proliferation, mOSM did not stimulate cell growth (Fig 2B). One of the most prominent activities of mLIF is to inhibit the differentiation of mouse ES cells and to maintain their totipotency. We thus examined if mOSM can inhibit differentiation of CCE, an ES cell line whose differentiation is inhibited by either mLIF or hOSM.36 As shown in Fig 3, mOSM was again approximately 100-fold less active than hOSM in this assay.
Comparison of biological activities between mOSM and mLIF. (A) Inhibition of [3H] thymidine uptake in mouse M1 myeloid leukemia cells by mOSM (•) and mLIF (○). (B through D) Growth stimulation of mouse myeloid DA1a cells (B), a Ba/F3 transfectant expressing gp130 and LIFRβ (C), and a Ba/F3 expressing gp130 alone (D). As a control in (D), various concentrations of human IL-6 were added in the presence of 100 ng/mL of soluble human IL-6 receptor α subunit (open triangles with broken line). Data are averages of duplicate measurements.
Comparison of biological activities between mOSM and mLIF. (A) Inhibition of [3H] thymidine uptake in mouse M1 myeloid leukemia cells by mOSM (•) and mLIF (○). (B through D) Growth stimulation of mouse myeloid DA1a cells (B), a Ba/F3 transfectant expressing gp130 and LIFRβ (C), and a Ba/F3 expressing gp130 alone (D). As a control in (D), various concentrations of human IL-6 were added in the presence of 100 ng/mL of soluble human IL-6 receptor α subunit (open triangles with broken line). Data are averages of duplicate measurements.
Inhibition of differentiation of ES cells. Supernatants of COS cells transfected with mock, mOSM, and hOSM cDNAs were concentrated 20-fold by ultrafiltration. CCE (ES cell line) cells (3.5 × 102) were incubated in 24-well multiwell plates with a serial dilution of the supernatants as indicated for 5 days. Cells were stained with Giemsa solution after methanol fixation. Dispersed and compact stain dots in each well represent differentiated and undifferentiated ES cell colonies, respectively.
Inhibition of differentiation of ES cells. Supernatants of COS cells transfected with mock, mOSM, and hOSM cDNAs were concentrated 20-fold by ultrafiltration. CCE (ES cell line) cells (3.5 × 102) were incubated in 24-well multiwell plates with a serial dilution of the supernatants as indicated for 5 days. Cells were stained with Giemsa solution after methanol fixation. Dispersed and compact stain dots in each well represent differentiated and undifferentiated ES cell colonies, respectively.
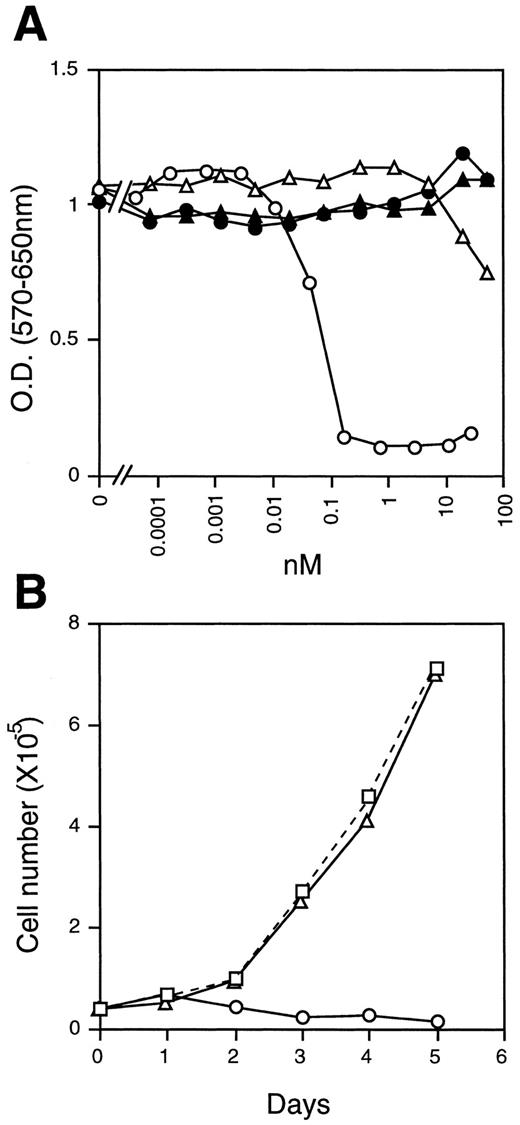
These results led us to suspect that specific activity of the recombinant mOSM might be very low for some reason. However, this was not the case. We found that the proliferation of NIH3T3 cells was severely suppressed in the presence of mOSM (Fig 4A and B). Interestingly, these cells were completely insensitive to mLIF, hLIF, and hOSM (Fig 4A and B). To confirm that the difference between mOSM and hOSM we observed was not due to the modification of mOSM, we expressed the mOSM cDNA encoding the mature form without a tag in COS7 cells and compared its activity with the tagged-mOSM. Untagged mOSM also showed a poor activity on DA1a cells, whereas it inhibited the growth of NIH3T3 cells as efficiently as the tagged mOSM (data not shown). These results suggested that mOSM does not transmit signals through the functional LIF receptor.
Growth inhibition of NIH3T3 cells. (A) NIH3T3 cells (2 × 103/well) were cultured in the presence of various concentrations of mOSM (○), hOSM (▵), mLIF (•), and hLIF (▴) for 4 days in 96-well multiwell plates. Cell growth was monitored by the MTT assay. (B) Time course of cell growth in the presence of mOSM. NIH3T3 cells (4 × 104/well) were incubated in the absence (□) or presence of mOSM (○) or hOSM (▵) in 24-well multiwell plates. Viable cell numbers were counted daily by the trypan-blue exclusion method. Data are averages of duplicate measurements.
Growth inhibition of NIH3T3 cells. (A) NIH3T3 cells (2 × 103/well) were cultured in the presence of various concentrations of mOSM (○), hOSM (▵), mLIF (•), and hLIF (▴) for 4 days in 96-well multiwell plates. Cell growth was monitored by the MTT assay. (B) Time course of cell growth in the presence of mOSM. NIH3T3 cells (4 × 104/well) were incubated in the absence (□) or presence of mOSM (○) or hOSM (▵) in 24-well multiwell plates. Viable cell numbers were counted daily by the trypan-blue exclusion method. Data are averages of duplicate measurements.
To verify the poor agonistic activity of mOSM on the gp130/LIFRβ complex, we established two Ba/F3 transfectants expressing either gp130 alone (Ba/F3-gp130) or both gp130 and LIFRβ (Ba/F3-gp130 + LIFRβ). Similar to the observation in the DA1.a assay, Ba/F3-gp130 + LIFRβ proliferated well in response to mLIF, but not to mOSM (Fig 2C). In contrast, Ba/F3-gp130, as well as Ba/F3 parental cells, did not respond to either of mLIF or mOSM, although the addition of human IL-6 and soluble IL-6 receptor conferred a proliferative response of Ba/F3-gp130 cells (Fig 2D). These results confirmed that mOSM does not deliver signals through the gp130/LIFRβ complex.
Binding of mOSM to the receptors.The poor agonistic activity of mOSM on LIF-sensitive cell lines could be due to its inability to bind tightly to the gp130 + LIFRβ complex. To examine this possibility, we performed binding studies using iodinated mOSM and mLIF. As shown in Fig 5A and 5B, Scatchard plot analysis demonstrated that only NIH3T3 cells expressed high-affinity receptors for mOSM (kd = 660 pmol/L), whereas M1, Ba/F3-gp130, Ba/F3-gp130 + LIFRβ exhibited only low-affinity binding sites (kd = 2.8 to 4.2 nmol/L). As control experiments, we demonstrated that mLIF binds to M1 and Ba/F3-gp130 + LIFRβ with high-affinity (kd = 139 pmol/L and 80 pmol/L, respectively), but does not bind to Ba/F3-gp130 and NIH3T3 cells (Fig 5C). The numbers and affinities of the binding sites on the cell lines we analyzed are summarized in Table 1. These results indicate that mOSM binds to gp130 with low-affinity and that unlike the type I hOSM receptor, the presence of LIFRβ does not change its binding affinity. These results are consistent with the observation that mOSM is fully active only in NIH3T3 cells, but not in mLIF-responsive cell lines, whereas mLIF does not act on NIH3T3 cells (Figs 2-4). The results indicate that the high-affinity functional receptor for mOSM in NIH3T3 cells is not the gp130 + LIFRβ complex and is most likely the complex of gp130 and the OSM specific receptor β subunit.
Scatchard analyses of mOSM and mLIF binding. Binding of [125I]-labeled mOSM (A and B) or [125I]-labeled mLIF (C) to NIH3T3 cells (•), M1 cells (○), Ba/F3 cells (▵), Ba/F3 expressing gp130 (▪), and Ba/F3 expressing gp130 and LIFRβ (□). Each point represents an average of duplicate measurements. The dissociation constants and receptor numbers of mOSM and mLIF for these cells are summarized in Table 1.
Scatchard analyses of mOSM and mLIF binding. Binding of [125I]-labeled mOSM (A and B) or [125I]-labeled mLIF (C) to NIH3T3 cells (•), M1 cells (○), Ba/F3 cells (▵), Ba/F3 expressing gp130 (▪), and Ba/F3 expressing gp130 and LIFRβ (□). Each point represents an average of duplicate measurements. The dissociation constants and receptor numbers of mOSM and mLIF for these cells are summarized in Table 1.
Binding Properties of mOSM and mLIF to Various Murine Cell Lines
| Cell Line . | 125I-ligand . | kd1* . | Sites/Cell . | kd2* . | Sites/Cell . |
|---|---|---|---|---|---|
| NIH3T3 | mOSM | 660 pmol/L | 6,710 | 1.52 nmol/L | 11,200 |
| mLIF | ND | ND | ND | ND | |
| M1 | mOSM | ND | ND | 4.25 nmol/L | 3,650 |
| mLIF | 139 pmol/L | 936 | ND | ND | |
| Ba/F3 | mOSM | ND | ND | ND | ND |
| mLIF | ND | ND | ND | ND | |
| Ba/F3-gp130 | mOSM | ND | ND | 2.79 nmol/L | 1,130 |
| mLIF | ND | ND | ND | ND | |
| Ba/F3-gp130 +LIFRβ | mOSM | ND | ND | 3.21 nmol/L | 448 |
| mLIF | 80.0 pmol/L | 443 | 1.87 nmol/L | 1,940 |
| Cell Line . | 125I-ligand . | kd1* . | Sites/Cell . | kd2* . | Sites/Cell . |
|---|---|---|---|---|---|
| NIH3T3 | mOSM | 660 pmol/L | 6,710 | 1.52 nmol/L | 11,200 |
| mLIF | ND | ND | ND | ND | |
| M1 | mOSM | ND | ND | 4.25 nmol/L | 3,650 |
| mLIF | 139 pmol/L | 936 | ND | ND | |
| Ba/F3 | mOSM | ND | ND | ND | ND |
| mLIF | ND | ND | ND | ND | |
| Ba/F3-gp130 | mOSM | ND | ND | 2.79 nmol/L | 1,130 |
| mLIF | ND | ND | ND | ND | |
| Ba/F3-gp130 +LIFRβ | mOSM | ND | ND | 3.21 nmol/L | 448 |
| mLIF | 80.0 pmol/L | 443 | 1.87 nmol/L | 1,940 |
Abbreviation: ND, not detected.
kd1 and kd2 are dissociation constants for high-affinity binding sites and low-affinity binding sites, respectively.
DISCUSSION
In this study, we have investigated the function and the receptor of mOSM by using the recombinant mOSM protein we produced, and we have reached the conclusion that unlike hOSM, mOSM does not use the gp130/LIFRβ complex as a functional receptor. This conclusion is supported by several observations. First, all of the mLIF responsive mouse cell lines we examined, including M1, DA1.a, CCE, and the Ba/F3 transfectant expressing gp130 and LIFRβ, did not respond to mOSM at a concentration comparable to that of mLIF or hOSM. Second, the proliferation of NIH3T3 cells was inhibited by mOSM, but not by LIF or hOSM. Finally, binding studies using mOSM have shown that mOSM binds to gp130 with low-affinity and that, unlike hOSM, LIFRβ does not increase the binding affinity of mOSM to the receptor. The binding study also shows that LIF-nonresponsive NIH3T3 cells express the high-affinity mOSM receptor, but not the LIF receptor. The difference between mouse and human receptors is somewhat reminiscent of the IL-3 receptors, ie, there are two IL-3 receptor β subunits in the mouse, one is a low-affinity IL-3 binding protein and the other one is the common β subunit for the IL-3/GM-CSF/IL-5 receptors, while the human has only one common β subunit. Hence, each species appears to have acquired divergent cytokine responsiveness by generating an extra receptor chain or by altering the affinity of receptor subunits to cytokines during evolutional processes.
The fundamental difference between hOSM and mOSM appears to be the interaction between OSM and LIFRβ. Although the direct binding of OSM to LIFRβ has not been detected even in the human system, the active hOSM receptor is a tripartite complex formed between hOSM, gp130, and LIFRβ that induces the dimerization of the intracellular domains of gp130 and LIFRβ to trigger signaling. It is very likely that the interaction of mOSM with LIFRβ is so weak that mOSM cannot induce the formation of an active receptor complex between gp130 and LIFRβ. Thus, in the presence of an extremely high concentration of mOSM, LIF-responsive cell lines exhibit weak responses to mOSM (Fig 2A through C). A similar observation has been made in human GM-CSF (hGM-CSF ), ie, the low-affinity binding subunit of the hGM-CSF receptor (hGM-CSFRα) does not form a high-affinity hGM-CSF receptor with the mouse β subunit, but hGM-CSF induces weak signals at an extremely high concentration when both hGM-CSFRα and the mouse β subunit are coexpressed.37
The results shown in this study clearly demonstrate that OSM and LIF do not use the same receptor in mice. Thus, in contrast to human OSM and LIF, which exhibit extensive overlapping activities by sharing the same receptor, the functions of these two cytokines appear to be segregated in mice (Fig 6). It should be noted that functions of hOSM in mouse cell lines and presumably bovine OSM in the transgenic mice that have been previously reported38 are most likely mediated by the mLIF receptor, but not by the mOSM receptor. A recent report described the extrathymic development of double and single positive T cells in bovine OSM-transgenic mice and hOSM-injected mice.39 However, our results suggest that bovine and human OSM stimulate the T-cell development through the LIF receptor. In fact, a similar abnormal T-cell development was previously observed in LIF-transgenic mice.40 The role of OSM in the extrathymic T-cell development has to be verified by using mOSM in mice. Likewise, other biological activities of hOSM, which have been reported in several rodent models, must be reevaluated using mOSM.
A proposed model of the interaction of OSM to receptors in the human and mouse systems. A mouse OSMRβ subunit is a yet to be cloned putative molecule. Thick arrows represent high-affinity binding, while the thin arrows indicate low-affinity binding.
A proposed model of the interaction of OSM to receptors in the human and mouse systems. A mouse OSMRβ subunit is a yet to be cloned putative molecule. Thick arrows represent high-affinity binding, while the thin arrows indicate low-affinity binding.
Based on our data, as well as a recent report by Mosley et al,25 we propose the receptor structures of mouse and human OSM and LIF as shown in Fig 6. The structure of the high-affinity mOSM receptor, which is composed of gp130 and OSMRβ (type II OSM receptor in the human) needs to be proven by cloning an mOSM specific receptor, and there is still the possibility that the mOSM receptor is completely distinct from the human type II OSM receptor. However, the model we propose is most probable because (1) NIH3T3 cells express gp130 and the addition of human IL-6 and soluble IL-6 receptor suppressed cell growth as seen in the case of mOSM (M.I., T.H., and A.M., unpublished data), and (2) proliferation of NIH3T3 cells was inhibited by hOSM at a very high concentration (Fig 4A). The type II OSM receptor seems to be species-specific because hOSM does not induce TIMP-1 gene expression or growth suppression in rodent fibroblasts and mOSM does not exhibit these activities in human fibroblasts (C.R. Richards, C. Kerr, D. Pennica, T.H., A.M., F. Botelho, and C. Langdon, submitted).
Important questions still remain to be addressed: what are the functional differences between OSM and LIF, and what is the molecular basis for the differences? Because both LIF and OSM receptors use gp130 as a signal transducer, they may mediate similar signals. Thus, a simple explanation is that LIFRβ and OSMRβ are differentially expressed in different types of cells. However, this is not always the case because some human tumor cell lines express both receptors and respond differently to OSM and LIF.24 It is, therefore, more likely that LIFRβ and OSMRβ are not only binding subunits for their ligands, but they also actively participate in signaling. If this is the case, the two receptor systems may deliver different signals even if they are expressed in the same cells. Supporting this idea, hOSM induced MAP kinase activity through the hOSM specific receptor much more potently than LIF and triggered tyrosine phosphorylation of several proteins that were not induced by LIF.24 We have addressed this question by examining whether mLIF and mOSM exhibit the same growth inhibitory activity in NIH3T3 cells when both OSM and LIF receptors are coexpressed. Proliferation of NIH3T3 transfectant expressing the LIFRβ cDNA was inhibited by mLIF. However, the inhibitory effect was much weaker than that of mOSM, even though the transfectants highly expressed the mLIFRβ (M.I., T.H., and A.M., unpublished results). Similarly, the induction of TIMP-1 gene expression by mOSM in NIH3T3 cells was much higher than that by IL-6 or hOSM (C.R. Richards, C. Kerr, D. Pennica, T.H., A.M., F. Botelho, and C. Langdon, submitted). To further characterize the OSM receptor, a putative OSMRβ gene needs to be isolated. Finally, as the functional receptors for OSM and LIF are distinct in mice, further studies on mOSM and mLIF will define their physiological roles.
ACKNOWLEDGMENT
We thank Drs T. Taga (Tokyo Medical and Dental University) and M. Tomida (Saitama Cancer Center Research Institute) for providing us with the murine gp130 cDNA and LIFRβ cDNA, respectively. We are grateful to C. Mawson for critical reading of this manuscript.
Supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan, a grant from New Energy and Industrial Technology Development Organization, and a research grant from The Toray Research Foundation.
Address reprint requests to Atsushi Miyajima, PhD, Institute of Molecular and Cellular Biosciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113, Japan.



![Fig. 2. Comparison of biological activities between mOSM and mLIF. (A) Inhibition of [3H] thymidine uptake in mouse M1 myeloid leukemia cells by mOSM (•) and mLIF (○). (B through D) Growth stimulation of mouse myeloid DA1a cells (B), a Ba/F3 transfectant expressing gp130 and LIFRβ (C), and a Ba/F3 expressing gp130 alone (D). As a control in (D), various concentrations of human IL-6 were added in the presence of 100 ng/mL of soluble human IL-6 receptor α subunit (open triangles with broken line). Data are averages of duplicate measurements.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.165/5/m_bl_0006f2.jpeg?Expires=1767727027&Signature=AiN-AaLZsNAjvgpRi2chOvQFsxqEkDoFmLc9pl9OeWDXDwss9Wibr69S-Msk9K24FpcK-tV~4KodnLX3qgl8vFV2sEjVgNq86B2D-wEpUVPzBdOyi2z~A9Jl1yT6BsHEO6F10Uk5s1DWCWqB6FPy1DRqJ0DwNkRA6Sf-sThza1wOoOa8za5rPN-7lh6DSjxpDNaxjSV9QCsWVNMH1rFiQGFnzTp95-USswRzjxQAxZ1qWh8qaaLkHnWoUDqvl~tSroA~XwVF13Yx2OporPLhl1Wqt7VFT0oi6fQERbmRKjlBS89lOUbdcsLEaMHmpqOba9H8I4tqP3qm9F2qf7fE6w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 5. Scatchard analyses of mOSM and mLIF binding. Binding of [125I]-labeled mOSM (A and B) or [125I]-labeled mLIF (C) to NIH3T3 cells (•), M1 cells (○), Ba/F3 cells (▵), Ba/F3 expressing gp130 (▪), and Ba/F3 expressing gp130 and LIFRβ (□). Each point represents an average of duplicate measurements. The dissociation constants and receptor numbers of mOSM and mLIF for these cells are summarized in Table 1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.165/5/m_bl_0006f5.jpeg?Expires=1767727027&Signature=ayIB7zsEdciOwWbgELC6LXNDjkOZCeAuwfvTweI1BGDWtsPafMrBIquphdrx9Ue3XpHTXFXq--qye65d5Uc-bFYaA~Yu8jBrPj2FDDj82hOfPpdrtxfFniXxw7CpQeHah0smURVRa4za49SWoOuVNk9We5xexXvL1hHxjtUqE6q6O6JxV4q32AiZ83r2wW9cUdi5j68KOTlebKtXNI0sDzR8Fge4na6IjUz8Uc~-Fo2Ba~qLGVnhg2LiA7xak6WziNSb9FNLtEyOfucMj~k7rL8~jGBFj5BBY4-4LB9dCbSl9r~FYTLFa8301dwk17ktzLqy4Qxwx4mJURjpirDobA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal