Abstract
To better understand the molecular mechanism(s) by which growth and differentiation of the primitive hematopoietic stem cell is initiated, as well as the means by which the maturing cell can commit to development along a specific cell lineage, we elected to study the Id family of helix-loop-helix (HLH) transcriptional regulators. Some members of the HLH family are expressed in a stage-specific manner during hematopoietic development and can regulate the ability of immature hematopoietic cells to terminally differentiate. None of the four Id family genes were detected in the most primitive progenitors. Id-1 was widely expressed in proliferating bi- and unipotential progenitors, but its expression was downregulated in cells of increasing maturity; conversely, Id-2 and, to a limited extent, Id-3 gene expression increased as cells matured and lost proliferative capacity. Id-2 expression ran counter to that of Id-1 not only during maturation, but during periods of cell growth and arrest as well. This is quite distinct from the nonhematopoietic tissues, in which these two factors are coordinately expressed and suggests that Id-1 and Id-2 might be regulating very different events during hematopoiesis than they regulate in other cell types.
A POOL OF PRIMITIVE hematopoietic stem cells (PHSC) reside in the adult bone marrow as a reservoir of uncommitted and undifferentiated blood cell precursors. In order to maintain an appropriately balanced system, these cells must respond to cues from the extracellular environment by proliferation and subsequent differentiation into mature cell types. Each stage of this process, from the initial activation of the resting G0 stem cell through the critical event of lineage commitment to the final stages of differentiation is ultimately controlled by the amounts and types of transcriptional regulators expressed in the cell. Members of the helix-loop-helix (HLH),1 the homeobox (Hox),2-4 basic-zipper (bZ),5,6 LIM,7,8 and zinc finger9 10 transcription factor families have been detected in cells during some stage(s) of hematopoietic development. Thus, members of any of these groups could potentially play a role in the regulation of some or all of the key developmental switch points that must be negotiated in order for a normal blood cell to mature.
The HLH family of transcriptional regulators may play an important role in hematopoiesis. Members of this family have been shown to be key players in the regulation of neural development,11,12 sex determination,12 and myogenesis,13,14 as well as to contribute to regulation of development in the pancreas,15 bone,16,17 and fatty tissue.18 There is evidence to suggest that this family may control hematopoietic development as well.1,4,19-24 Numerous HLH regulators, including E47/E12,1,4,24 SCL,1,4,25 lyl-1,1,4,22 and most of the Id factors4 26-28 have been identified in at least a subset of normal hematopoietic tissues and cell lines. These regulators include both known transcriptional activators (eg, E12/E47 and SCL) as well as transdominant negative regulators (eg, the Id proteins).
The function of HLH transcriptional regulators is dictated by three structural elements: (1) the HLH protein dimerization interface, (2) a basic DNA binding region, and (3) an N-terminal transactivation domain. The HLH structure is a protein dimerization motif that is used to combine HLH monomer subunits into functionally important dimers.29 Upstream and extending from helix one there is usually a basic region, which is a DNA binding half-site. When these proteins dimerize they juxtapose the two half-sites into an active DNA-binding element that can then recognize and bind to specific sequence motifs called E-boxes. Finally, in some of these molecules, there is an N-terminal transactivation domain that affects transcriptional activation once the active dimer has been localized to its correct position in a promoter or enhancer regulatory unit.22,30,31 There are variants that function as negative regulators as well. In some instances, alternative splicing generates HLH monomers lacking the transactivation region; heterodimerization between this monomer and a full-length HLH activator would be expected to decrease or extinguish the activation potential of the resultant dimer.32 Alternately, there is a subclass of HLH forms that completely lack the basic DNA binding region. These proteins make up the Id (inhibitor of DNA binding) family, which can heterodimerize with HLH members to form dimers containing an incomplete DNA binding region, which then cannot bind to DNA.26,27,33 34 These Id family proteins act through sequestration of the HLH activators in nonfunctional complexes.
In addition to these structural classifications, HLH family regulators are subdivided according to their expression patterns. Some regulators are widely expressed, such as the universal E protein family (eg, E2A, E2-2, HEB; also called class I proteins); other class II factors are expressed in a tissue-restricted fashion (eg, MyoD and myogenin in muscle tissue; SCL and lyl-1 in hematopoietic cells).1,35 Class III proteins, such as the Id family of negative regulators, show an intermediate pattern of distribution.26,27,33,34 Most importantly, many of these regulators are highly expressed in normal hematopoietic tissues during the stage in which these cells must be able to respond to the external signaling that controls their ability to continue proliferation or commit to differentiate into a mature blood cell.1 4
Both class I and class II factors have been demonstrated to be essential for normal hematopoietic development. Mice lacking the class I gene E2A have a complete block in B cell development while other hematopoietic lineages remain largely normal.24,36 Blood development is profoundly limited in mice lacking the class II SCL gene; embryos suffer a complete block in early erythropoiesis with virtual absence of myeloid cells, suggesting that this gene is essential for normal growth and development of early myelo-erythroid progenitor cells.37 A working model for the role of HLH factors in differentiation proposes that the ability of a cell to differentiate may be controlled at the transcriptional level by a heterodimer between a class II tissue-specific HLH factor and a member of the class I E proteins.38 This heterodimer would then be able to activate expression of genes required for maturation. The action of this dimer is negatively regulated by the class III Id family, which can prevent differentiation by sequestration of class I E proteins in nonfunctional complexes.40 Since most tissue-specific class II proteins only function as heterodimeric complexes with the class I proteins,35 41-43 this prevents tissue-specific factors from initiating the maturation process.
In a previous study, we surveyed the expression patterns of several HLH transcriptional regulators (eg, E12/E47, SCL, ly1-1, and Id-1) in proliferating hematopoietic cells and cell lines.4 This work identified developmental stage-specific and cell cycle-related variations in expression of these genes, particularly Id-1. Since Id-1 belongs to a family of at least four similar transdominant negative regulators, we first needed to study the expression of Id-1 in the context of the other four family members in order to determine which Id family gene might be functionally important for regulating different aspects of hematopoietic growth and development. To do this, we first determined the range of expression of all four Id family genes in hematopoietic cells and tissues during growth and differentiation, then assayed whether the expression of some or all of these genes was specifically modulated in response to the defined environmental cues that stimulate normal hematopoietic growth and development.
MATERIALS AND METHODS
Cell Lines and Cell Culture
Cell lines were cultured in medium alone, medium containing an appropriate recombinant cytokine, or medium supplemented with conditioned medium, as suitable for the individual cell lines. P3X, Namalwa, EL-4, HL60, K562, MEL, WEHI 3, M1, and RAW 246.7 were grown in RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum (HI-FCS); 18-81 and 22D6 were grown in the same medium supplemented with 50 μmol/L 2mercaptoethanol (2ME). BW cells were grown in RPMI 1640 plus 20% FCS. T1165 were cultured in RPMI 1640/10% HI-FCS supplemented with 5,000 U/mL rIL-6 (R&D, Minneapolis, MN). HCD57 were grown in IMDM plus 30% HI-FCS, 20 μmol/L 2ME and 0.3 U/mL erythropoietin (Toyobo, New York, NY). M07E cells were cultured in DMEM/20% HI-FCS supplemented with 100 U/mL recombinant human GM-CSF (R&D). MC6 cells were cultured in McCoy's medium/10% FCS supplemented with amino acids, vitamins, and 10% WEHI 3-conditioned medium (WEHIcm). NFS60 and FDCP1 cells were routinely grown in RPMI 1640/10% HI-FCS supplemented with 10% or 25% WEHI3cm, respectively.
Cell-Cycle Synchronization
Cells growing actively in early- to mid-log phase were precultured for 36 hours in isoleucine-deficient medium (isoleucine-deficient DMEM supplemented with 10% dialyzed HI-FCS and 25% dialyzed WEHIcm) to arrest their growth. After that time, one half of the cells were resuspended in complete growth medium (DMEM plus 25% WEHIcm and 10% HI-FCS) while the control cells were resuspended in DMEM plus 10% HI-FCS only. Cells were grown for 48 hours, during which time samples from both cytokine replete and control populations were withdrawn for RNA preparation44 and samples from the cytokine-replete proliferating population removed for analysis of cell cycle progression by 3H-thymidine uptake.45 Additionally, cell growth and viability was monitored by cell counts and trypan blue exclusion.
Cytokine Starvation Experiments
FDCPI cells were cultured in RPMI 1640 supplemented with 25% WEHIcm and 10% FCS. For cytokine-response studies, these cells were starved of growth factors by culture in RPMI plus 0.5% HI-FCS for 24 hours before transfer into fresh medium supplemented recombinant murine (rm) IL-3 (Collaborative Research, Waltham, MA) or rmGM-CSF (Amgen, Thousand Oaks, CA) at 100 U/mL each. In each case, maximal cell growth stimulation was predetermined by a dose-response titration of the individual growth factor (not shown); a concentration of each factor that gave maximal growth response was selected for use in each of these experiments. Samples were removed for RNA analysis at multiple time points after cytokine stimulation.
RNA Isolation and Analysis
Ten micrograms of unselected cytoplasmic RNA44 from each time point was separated by electrophoresis on a formaldehyde gel before transfer to Hybond N membrane. Each filter was hybridized to 32P-random labeled Id gene probes. After a high stringency wash, filters were exposed to film and the specific bands later quantitated on a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). To normalize for loading efficiency each filter was stripped and rehybridized to a 28S ribosomal RNA probe46 or a GAPDH probe47 as indicated. Id-1 was quantitated after a 24-hour exposure; Id-2 quantitated after 3 to 5 days, Id-3 after 3 days; GAPDH after 3 hours; and 28S after 15 to 30 minutes.
Isolation, Culture, and Analysis of Normal Murine Bone Marrow and Fetal Liver Cells
Cells isolated from CBA/J bone marrow and day 12 fetal liver were used in this work.
Progenitor studies (PR1 through PR40).Bone marrow cells were plated in methylcellulose in the presence of IL-1, IL-3, SCF, and erythropoietin for 27 hours. One or two cells from four to eight cell colony starts were processed individually for RT-PCR. The remaining sibling cells were transferred into individual secondary methylcellulose cultures containing the same cytokine supplement. After 6 to 10 days, secondary colonies were picked, spread on glass slides, and scored for content of various lineage on the basis of May-Grunwald-Giemsa staining.
Hematopoietic mature (HMI through HM20).CBA/J marrow cells were grown under the conditions described above. Fifty to 150 cells were taken for RT-PCR analysis after 6 to 8 days' culture, while the remaining cells were analyzed morphologically on glass slides to confirm the lineage assignments. HM1 and HM2 were sampled from growing, nonhemoglobinized erythroid precursors at the BFU-E stage of maturation; HM3 and HM4 (erythroid) were taken from visibly hemoglobinized colonies; HM5 and HM6 (neutrophil) were propagated in IL-3; HM7, HM8, HM15, and HM16 (mast) were taken after 25 to 46 days bulk culture in IL-3; HM9 and HM10 (megakaryocytic) were amplified from single cells; HM11 and HM12 (day 12 fetal liver B220+) were from bulk cultures growing in IL-7; HM13 and HM14 were bulk cultures of splenic T cells growing in medium containing concanavalin A; HM17 and HM18 (macrophage) were cultured in IL-3 alone; and HM19 and HM20 (macrophage) were cultured in IL-3 plus M-CSF.
RT-PCR Analysis of Gene Expression in Single Cells and Small Populations
Individual cells picked from colony starts were analyzed by RT-PCR.4 For small pools of cells, RT-PCR was performed using primers and conditions as previously described.48 Five micrograms of each amplified product was separated by agarose gel electrophoresis and transferred to Hybond N membrane. Each blot was subsequently probed with cDNA probes of the individual Id family genes. Due to the high degree of sequence conservation between human and murine Id genes at both the DNA and protein levels (Id-1 mRNA, 83% identity at the DNA level; Id-2, 88%; Id-3, 86%), our murine Id probes were also effective for detection of human Id family mRNAs. The Id-1 probe was a gift of H. Weintraub. pE:Id(S) is a BamHI/EcoR1 fragment consisting of the total cDNA sequence from pMH18DR blunt-ended into pEMSV-scribe α2 then released by Sma I. The Id-2 probe was a gift of X-H. Sun and consisted of a BamHI/XhoI fragment consisting of the full-length Id-2 cDNA. Id-3 is a 0.94-kb EcoRI fragment originally cloned as h1h462 (ATCC#63120). The Id-4 probes were a gift of F. Sablitzky: an EcoRI fragment containing the “full” 1.7 kb cDNA sequence and “probe b,” an additional 0.7 kb of alternatively spliced Id-4, were used. Finally, a rat GAPDH probe was used for normalization of loading efficiency and RNA integrity.5 The isolated fragments were random labeled to 0.5 to 1 × 109 cpm/μg and hybridized to the filters at high stringency.48
RESULTS
The Id Family Genes Are Widely Expressed in Hematopoietic Cell Lines
To determine the range of Id family gene expression in hematopoietic cells, we initially analyzed a panel of 18 cell lines. These cells included both murine and human lymphoid (P3X and T1165 plasmacytoma, Namalwa mature B, 22D6 and 18-81 preB, and EL4 and BW T-cell lines), monocyte/macrophage (M1 and RAW 246.7), megakaryotic (MO7E), erythroleukemia (K562 and MEL), mastocytoma (MC6), and myeloid (WEHI-3, MC6, FDCP1, NFS60, HL60) cells.
Id-1 gene expression was detected in most of the cell lines examined (Fig 1A), although high level expression was restricted to cells of the myeloid lineages (WEHI-3, NFS60, and FDC-P1). Id-2 was more widely expressed than Id-1, including strong expression in lymphoid cell lines of both B- and T-cell origin, as well as in cells derived from the monocyte/macrophage lineages. The highest level of its expression was seen in the MC6 mastocytoma cell line. High level Id-3 gene expression was restricted to lymphoid cell lines, especially those immortalized at early stages of B-cell development (Fig 1C; 22D6 and 18-81 pre-B–cell lines). As the cell lines become increasingly mature (Namalwa is a mature B cell type derived from a Burkitt lymphoma, P3X is a myeloma, and T1165 is a hybridoma), Id-3 expression continues to drop. In P3X murine myeloma cells Id-3 is only expressed at 0.3% of its level in 18-81 murine pre-B cells. This inverse relationship between Id-3 gene expression and B-cell differentiation stage is similar to that reported for Id-1 in other cell types in which its expression declines during forced differentiation of cell lines in vitro.23,26 28 Id-4 gene expression was undetectable.
Id family genes are widely expressed in hematopoietic cell lines. Expression of the four Id family genes was assayed by Northern blot in total cellular RNA isolated from 18 murine and human hematopoietic cell lines. Signal intensity was measured by phosphorimager before the blots were normalized by rehybridization to a 28S RNA oligonucleotide. Values are comparable only within the group of samples hybridized to a single probe; different exposure times (Id-1 exposed for 24 hours, Id-2 exposed for 120 hours, Id-3 exposed for 72 hours, and 28S RNA exposed for 30 minutes) were used for each probe to maximize clarity of the phosphorimage. Values were plotted as the relative intensity (pixel number) of the specific bands above background for each probe.
Id family genes are widely expressed in hematopoietic cell lines. Expression of the four Id family genes was assayed by Northern blot in total cellular RNA isolated from 18 murine and human hematopoietic cell lines. Signal intensity was measured by phosphorimager before the blots were normalized by rehybridization to a 28S RNA oligonucleotide. Values are comparable only within the group of samples hybridized to a single probe; different exposure times (Id-1 exposed for 24 hours, Id-2 exposed for 120 hours, Id-3 exposed for 72 hours, and 28S RNA exposed for 30 minutes) were used for each probe to maximize clarity of the phosphorimage. Values were plotted as the relative intensity (pixel number) of the specific bands above background for each probe.
The Id Family Genes Are Differentially Expressed During the Early Stages of Normal Hematopoietic Development
While examination of Id family gene expression in cell lines provides an excellent foundation on which to base further study of these regulators, we also wished to examine earlier stages of hematopoietic development in normal cells. To do this, we used a unique culture system that allowed us to analyze gene expression in individual progenitor cells that have been extracted from primitive colony starts. As a comparison and a control, we also examined Id family gene expression in maturing cells derived from normal bone marrow.
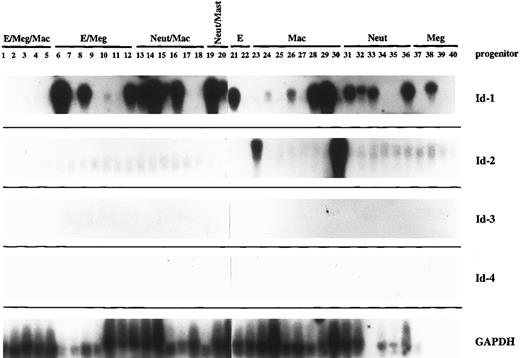
Hematopoietic progenitor cells.Murine bone marrow cells were plated in methylcellulose at low density in the presence of a cytokine cocktail (IL-1, IL-3, SCF, and erythropoietin) that has been shown to support growth of these cells.4 When individual colonies contained four to eight cells they were separated and one or sometimes two cells were removed for single-cell RT-PCR amplification of their mRNA. The remainder of the cells were separated in the dish and allowed to grow to maturity, when phenotype was assigned by May-Grunwald-Giemsa staining. PCR-amplified cDNA from 40 individual progenitor cells was assayed by southern hybridization to Id family probes (Fig 2). The same samples probed in parallel with GAPDH serve as an RNA integrity control for this experiment.
The Id family genes are differentially expressed during the early stages of normal hematopoietic development. Murine bone marrow was plated in methylcellulose at low density in the presence of IL-1, IL-3, SCF, and erythropoietin, then allowed to grow until individual colonies contained four to eight cells. At that time, colonies were separated and individual cells removed for single-cell RT-PCR analysis of their gene expression. Five micrograms of the amplified product from 40 such progenitor cells was Southern blotted and probed with high specific activity probes for each of the Id family genes and for GAPDH. Phenotype was assigned by May-Grunwald-Giemsa staining of the mature cells that developed from each colony start. E, erythroid; Meg, megakaryocytic; Mac, macrophage; Neut, neutrophil.
The Id family genes are differentially expressed during the early stages of normal hematopoietic development. Murine bone marrow was plated in methylcellulose at low density in the presence of IL-1, IL-3, SCF, and erythropoietin, then allowed to grow until individual colonies contained four to eight cells. At that time, colonies were separated and individual cells removed for single-cell RT-PCR analysis of their gene expression. Five micrograms of the amplified product from 40 such progenitor cells was Southern blotted and probed with high specific activity probes for each of the Id family genes and for GAPDH. Phenotype was assigned by May-Grunwald-Giemsa staining of the mature cells that developed from each colony start. E, erythroid; Meg, megakaryocytic; Mac, macrophage; Neut, neutrophil.
None of the Id family genes were highly expressed in the most primitive tripotential progenitors (PR1-PR5), which did express significant levels of GAPDH, a housekeeping gene. However, in the slightly more mature bipotential and unipotential precursors, Id-1 was widely expressed in a lineage-nonspecific manner (PR 6-40). While Id-2 may be expressed at extremely low levels in all of the progenitor cells in this study, high levels of its expression were limited to two unipotential progenitor cells, both of the macrophage lineage (PR #23 and PR #30). Thus regulation of Id-1 and Id-2 gene expression does not appear to be coordinate in the hematopoietic system as it has been reported for other tissue types.27,49,50 Id-3 was clearly not expressed in any of the 40 progenitor cells examined here. However, it is important to note that the culture conditions used for the generation of the colony starts (ie, IL-1, IL-3, SCF, and erythropoietin) favor development of the myeloid lineages at the expense of lymphoid development.51 Therefore, it is possible that expression of Id-3 would be upregulated during an earlier phase of development, perhaps corresponding to an early pre-B–cell precursor, if conditions promoting lymphoid development had been used in these cultures.
We also did not detect any expression of the Id-4 gene in these progenitor cells. However, analysis of its expression may be more complicated than the other three known Id family genes, since it appears to be posttranscriptionally regulated by alternative splicing.34 Two splice forms, 2.0 and 1.7 kb, predominate in the thymus, which is the only lymphohematopoietic organ in which the differential expression of Id-4 splice forms has been examined. We used two probes for this work: one that preferentially detects the 1.7 and 2.0-kb forms and one that hybridizes mainly to the 2.0-and 3.7-kb form (and not the 1.7-kb form). Since the 5′ and 3′ termini and all of the splice forms remain to be defined, it is difficult to say whether we have exhausted the possibility that one or more of the three Id-4 splice forms is actually expressed in hematopoietic cells during this phase of development.
Another notable finding in this work was the discordance in gene expression among individual cells at the same stage of maturation. For example, eight cells were withdrawn from bipotential colonies that later went on to produce neutrophils and macrophages (PR 13-20). Only six of the eight colonies produced high levels of Id-1 mRNA. We will show later that Id-1 mRNA levels vary widely during cell cycle, so perhaps this variation in gene expression merely reflects cell cycle position of the individual cells removed for analysis. Alternatively, it may merely mean that the progenitors in the colony are staggered with respect to maturation, with some cells lagging slightly behind and not upregulating Id-1 expression yet. However, we also saw variations in GAPDH and other housekeeping gene expression in these cells. There is some evidence to support the possibility that mRNA transcription from individual templates proceeds in an intermittent manner, so that the total level of mRNA accumulation in a cell population reflects the number of templates active at a given point in time rather than the sum activity of all templates, which are constantly active.48,52 53 If this were the case, then it would be possible to see all-or-none levels of gene expression in single cells drawn from colonies at the same stage of maturation. This interpretation is supported by the relative uniformity of gene expression among pools of lineage-defined cells, where variations in single cell gene expression should be averaged out over the whole population (Fig 3, below). Therefore, to interpret these data, we feel that is it most accurate and informative to assess gene expression at the “population” level, with gene expression within each group of like progenitors considered together, rather than on an individual cell basis.
Expression of Id family genes in mature hematopoietic cells. Single lineage colonies derived from terminally maturing cells were divided: 50 to 150 cells were sampled by RT-PCR analysis and the remaining cells were analyzed morphologically on glass slides to confirm the lineage assignments. After southern transfer, the blots were probed at high stringency for the expression of individual Id family genes and for GAPDH.
Expression of Id family genes in mature hematopoietic cells. Single lineage colonies derived from terminally maturing cells were divided: 50 to 150 cells were sampled by RT-PCR analysis and the remaining cells were analyzed morphologically on glass slides to confirm the lineage assignments. After southern transfer, the blots were probed at high stringency for the expression of individual Id family genes and for GAPDH.
Hematopoietic mature cells.Colonies or liquid cultures near maturity were assayed by RT-PCR and Southern blot (Fig 3). In these maturing cells, Id-1, Id-2, and Id-3 were all expressed, although again this expression was not coordinate. With the exception of the 2-day 12 fetal liver B220+ cultures (HM 11 and 12), there was no absolute correlation between Id-1 gene expression and cell lineage. However, when gene expression in the more primitive progenitor cells shown in Fig 2 was compared with these maturing colonies, there is a distinct and inverse correlation between expression of Id-1, Id-2, and cell maturation: with increasing maturity, levels of Id-1 expression declined and levels of Id-2 expression increased. High level Id-3 expression was restricted to the two fetal liver B220+ colonies (HM11 and 12), which mirrors its pattern of expression in the B-lymphoid cell lines shown in Fig 1. Again, Id-4 gene expression was not detected.
Differential Expression of Id Family Genes During Cell Cycle
Id-1 has been shown to regulate differentiation by sequestering class I HLH activators until they are required for differentiation to proceed. Certainly by preventing expression of lineage-specific genes, Id-1 can play a role in preventing cellular differentiation. However, regulation of hematopoietic cell differentiation is strongly affected by cell cycle, with cells being most receptive to differentiation signals during G1 .54 55 Therefore, we wished to test whether any/all of these Id family regulators might be involved in the control of hematopoietic cell cycle progression at the G1/S phase boundary.
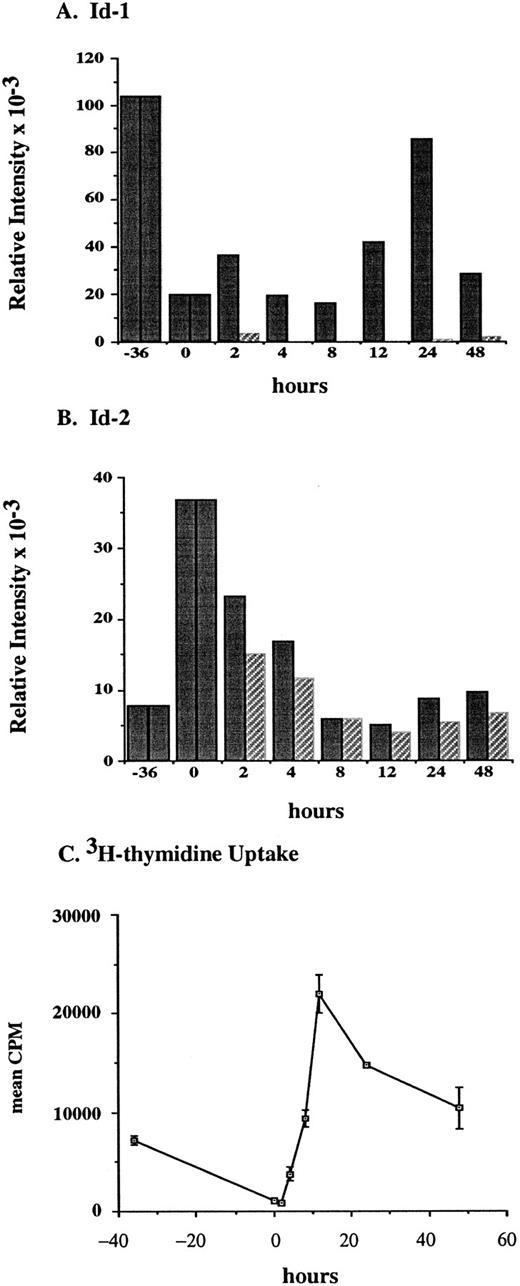
To determine whether Id genes are modulated during the G1 phase of the hematopoietic cell cycle, FDCP1 cells were synchronized by isoleucine (ILE) deprivation then followed through G1 and the S phase of one cell cycle. FDCP1 is a nontransformed, cytokine-dependent cell line isolated after long term culture of murine bone marrow56 that has retained growth and cell cycle characteristics of a normal diploid cell.57 After 36 hours of ILE deprivation in the presence of cytokines, the cells have cycled back to a point in mid-G1 phase where they arrested in synchrony. Then, half of the arrested cells were transferred into medium containing ILE plus cytokines while the other half were transferred into medium containing ILE but lacking cytokines. These cells were followed for 48 hours as indicated and Id family gene expression was measured by Northern blot and phosphorimage analysis. Synchronization was confirmed by following 3H-thymidine uptake in the cytokine-replete, responsive population during the course of the experiment.
Id-1 gene expression was high in the asynchronously proliferating log-phase cells, but it dropped off to less than 20% of the log-growing levels after 36 hours in medium lacking ILE but still containing the cytokines that would otherwise drive proliferation of these cells (Fig 4A and D). After refeeding the cells with complete medium containing WEHIcm, we saw a biphasic pattern of Id-1 gene expression: the first peak at 2 hours being approximately twofold over that seen in the growth-arrested cells; the second peak at 24 hours being a little over fourfold induction (solid bars). In the absence of exogenous cytokines, cells did not proliferate and Id-1 expression was not induced (striped bars). Since detailed cell cycle progression analysis of FDC-P1 shows that S phase begins between 12 and 16 hours after release from isoleucine block,45 Id-1 gene expression first peaked in mid G1 then again during S phase: a transient decline in Id-1 gene expression occurs during transit of the late G1 checkpoint that regulates whether a cell continues to cycle and whether it commits to differentiation.
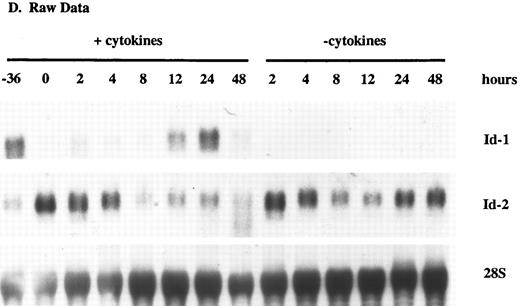
Id-1 and Id-2 are differentially expressed during cell cycle. Growth of FDCP1 cells was arrested in early G1 by isoleucine deprivation. After 36 hours, half the cells were transferred into normal growth medium (time = 0), with 25% WEHIcm as a complex source of cytokines (▪), while the other half were transferred into fresh medium without WEHIcm (▨). Cells were allowed to grow for 48 hours, during which time samples were removed for RNA isolation and northern hybridization to (A) Id-1 and (B) Id-2 probes. Signal intensity was quantitated by phosphorimage analysis before the blots were normalized by rehybridization to a 28S RNA oligonucleotide. Values were plotted as the relative intensity (pixel number) of the specific bands above background for each probe. Values are comparable only within the group of samples hybridized to a single probe; different exposure times were used for each probe to maximize clarity of the phosphorimage. (C) Cell cycle progression was monitored by 3H-thymidine uptake of the cytokine-replete, proliferating population.
(D) Raw data.
Id-1 and Id-2 are differentially expressed during cell cycle. Growth of FDCP1 cells was arrested in early G1 by isoleucine deprivation. After 36 hours, half the cells were transferred into normal growth medium (time = 0), with 25% WEHIcm as a complex source of cytokines (▪), while the other half were transferred into fresh medium without WEHIcm (▨). Cells were allowed to grow for 48 hours, during which time samples were removed for RNA isolation and northern hybridization to (A) Id-1 and (B) Id-2 probes. Signal intensity was quantitated by phosphorimage analysis before the blots were normalized by rehybridization to a 28S RNA oligonucleotide. Values were plotted as the relative intensity (pixel number) of the specific bands above background for each probe. Values are comparable only within the group of samples hybridized to a single probe; different exposure times were used for each probe to maximize clarity of the phosphorimage. (C) Cell cycle progression was monitored by 3H-thymidine uptake of the cytokine-replete, proliferating population.
(D) Raw data.
Since Id-1 and Id-2 have been reported to be coordinately regulated in several model systems, we were intrigued to find that Id-2 levels increased by a factor of five at the same time Id-1 levels declined (Fig 4B and D). After refeeding with ILE-replete medium Id-2 levels declined transiently whether cells were exposed to cytokines (solid bars) or not (striped bars), reaching log growth levels by 48 hours after release from ILE block.
We also stripped and reprobed these blots with Id-3 before normalizing with 28S rRNA but no Id-3 mRNA was detectable (not shown). Additional experiments with FDCP1 cells have failed to reveal detectable levels of Id-3 or Id-4 gene expression (Fig 1), even using RT-PCR analysis (data not shown).
Id-1 and Id-2 Are Differentially Regulated in Response to Cytokine Stimulation of FDCP1 Cells
In the previous experiment, we demonstrated a biphasic modulation of Id-1 levels as the cells traversed the G1 to S phase boundary. In the next set of experiments, we wishedto determine if these changes were cytokine-responsive and/or cytokine-specific. To do this, we again used FDCP1 cells which proliferate in response to IL-3 and GM-CSF. Cells starved of cytokines for 24 hours in medium (RPMI 1640/0.05% HI-FCS) were transferred into medium containing 100 U/mL rmIL-3, 100 U/mL rmGM-CSF, or no recombinant cytokines (medium alone). Cells were cultured in 0.5% HI-FCS plus/minus recombinant cytokines to minimize the contribution of exogenous growth factors contributed by the serum. This percentage of serum was determined by previous experimentation to be the minimum amount required to support viability of the unstimulated control cell population through the time course of this experiment (data not shown).
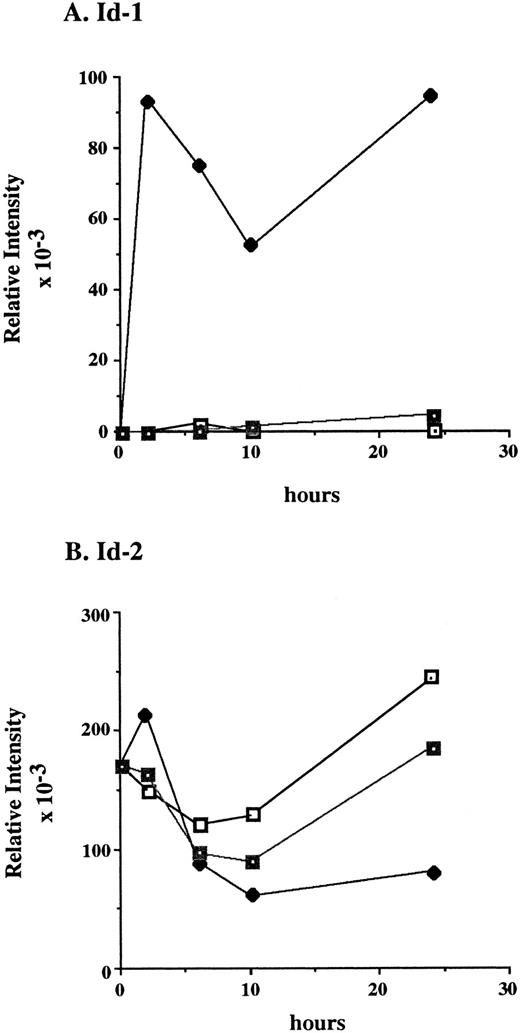
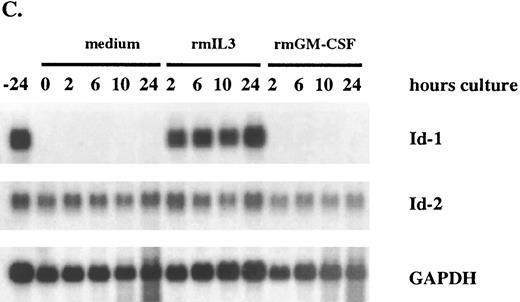
After a 24-hour incubation without cytokines, cell growth was arrested and Id-1 expression fell to undetectable levels. However, its expression increased swiftly and dramatically within 2 hours of supplementation with rmIL-3, remaining elevated over the time course of the experiment (Fig 5A and C). The biphasic pattern of Id-1 gene expression seen in this experiment is similar to that seen with ILE-synchronized cells (Fig 4A), which may reflect a partial synchronization of these cells by the cytokine withdrawal. In contrast, Id-1 gene expression was only slightly upregulated after treatment with rmGM-CSF. The overall difference in Id-1 gene expression between rmIL-3 and rmGM-CSF–treated FDCP1 cells may signify that rmIL-3 more effectively induces Id-1 gene expression and/or it may merely reflect the fact that FDCP1 cells respond more quickly to rmIL-3 than to rmGM-CSF.57
Differential response of Id-1 and Id-2 to cytokine stimulation in vitro. FDCP1 cells were starved of cytokines for 24 hours, then transferred into fresh medium (□) or medium containing 100 U/mL of either rIL-3 (♦) or rGM-CSF (▪) at time zero. The cells were allowed to grow for 24 hours, during which time samples were removed for RNA preparation and northern hybridization to (A) Id-1 and (B) Id-2 probes. Signal intensity was quantitated by phosphorimage analysis before the blots were normalized by rehybridization to GAPDH. Values were plotted as the relative intensity (pixel number) of the specific bands above background for each probe. Values are comparable only within the group of samples hybridized to a single probe; different exposure times were used for each probe to maximize clarity of the phosphorimage. (C) Raw data.
Differential response of Id-1 and Id-2 to cytokine stimulation in vitro. FDCP1 cells were starved of cytokines for 24 hours, then transferred into fresh medium (□) or medium containing 100 U/mL of either rIL-3 (♦) or rGM-CSF (▪) at time zero. The cells were allowed to grow for 24 hours, during which time samples were removed for RNA preparation and northern hybridization to (A) Id-1 and (B) Id-2 probes. Signal intensity was quantitated by phosphorimage analysis before the blots were normalized by rehybridization to GAPDH. Values were plotted as the relative intensity (pixel number) of the specific bands above background for each probe. Values are comparable only within the group of samples hybridized to a single probe; different exposure times were used for each probe to maximize clarity of the phosphorimage. (C) Raw data.
The pattern of Id-2 gene expression was somewhat different (Fig 5B and C). Id-2 levels fell transiently over the first 8 to 10 hours after refeeding at the same time Id-1 gene expression was significantly upregulated (Fig 5A and C). In cells stimulated with rmIL-3, the levels of Id-2 remained low as the cells proliferated over the 24-hours course of this experiment. In cells refed with medium plus 0.5% serum, the cells did not proliferate and the level of Id-2 gene expression continued to rise. The rmGM-CSF — treated cells, which responded with only minimal growth, showed an intermediate depression in Id-2 gene expression. A similar, reciprocal, pattern of Id-1 and Id-2 gene expression was also seen in factor-starved NFS60 cells when compared with cells proliferating in the presence of cytokines (Cooper, manuscript in preparation). Thus expression of Id-2 consistently runs counter to that of Id-1, during periods of cytokine-stimulated growth (Fig 5), cell cycle arrest (Figs 4 and 5), and during the differentiation process (Figs 2 and 3).
DISCUSSION
Although the PHSC is a dormant (G0 ) cell,58-60 it retains the capacity to respond to external signaling by entering cell cycle and continuing to proliferate until its progeny differentiate into mature blood cells. For this to happen, gene programs that maintain the quiescent state of the stem cell need to be extinguished or modified and genes that support proliferation must be induced. Later, the gene program that supports proliferation of these undifferentiated progenitor cells needs to be repressed and expression of a new set of differentiation-specific genes increased in turn. Candidate regulators of this process must meet certain criteria: they should be differentially expressed during a relevant period of development, respond to cytokine stimulation by appropriate up- or down-regulation of their expression, and participate in control of a developing cell's ability to initiate and/or maintain proliferation or to mediate terminal differentiation. The Id family regulators of the HLH transcription factor family are excellent candidates for regulators of this process. This is the first comprehensive study describing the expression of the four Id family genes during growth and differentiation in the hematopoietic system. Several important findings are revealed in this work: (1) Id-1 is not highly expressed until normal progenitors have matured to the bipotential and unipotential stage, after which time its expression decreases withmaturation; (2) Id-2 expression does not parallel that of Id-1 as it does in other tissue types; these two Ids are expressed reciprocally during growth and differentiation; and (3) Id-2 and, to a limited extent, Id-3 are upregulated as cells mature.
In this work, we have observed that Id-1 levels decline with differentiation (Figs 2 and 3) and with growth arrest of proliferating cells (Figs 4 and 5), suggesting that Id-1 plays a similar role during hematopoietic and nonhematopoietic development. Similar to its function during myogenesis, Id-1 controls the ability of erythroid, granulocytic, and B lymphocytes to differentiate.23,28,39 While Id-1 may inhibit differentiation by preventing activation of lineage-specific genes, it is also likely that expression of Id-1 promotes the G1 to S phase transition as it does in nonhematopoietic cells. Regulation of hematopoietic cell differentiation is strongly affected by cell cycle position, with cells being most receptive to differentiation signals during G1 .54,55 Cytokines that act to arrest cells in G1 or to prolong this phase generally promote differentiation54,55,61-63 while conditions that shorten G1 lower the probability that differentiation will occur.55 64 Thus, as Id-1 levels decline during normal development, the cell loses its capacity to progress, lingers in G1 , and is therefore more likely to be in the correct position to respond positively to differentiation signals from the bone marrow microenvironment.
In nonhematopoietic tissues, Id-2 is coordinately expressed with Id-1 and also promotes cellular proliferation. This is clearly not the case in hematopoietic cells, where expression of Id-2 seems to correlate with a state of quiescence rather than cell growth: Id-2 is highly expressed in CD34+ human progenitor cells65 and Hoechtslo/Rhodaminelo stem cells (not shown), and in FDCP1 cells after cytokine starvation (Fig 5) or after growth arrest secondary to ILE deprivation (Fig 4). Because many hematopoietic lineages undergo withdrawal from cell cycle at some point in the differentiation process, the increase in Id-2 levels during hematopoietic differentiation (Fig 3) may also reflect a reduced proliferative capacity in these populations.66,67 The correlation of Id-2 expression with quiescence was an unexpected finding, particularly since Id-2 has been demonstrated to interact with Rb/Rb-related proteins and promote proliferation of nonhematopoietic cells.68 It may still be that Id-2 drives proliferation of hematopoietic cells; the increased levels of its expression during experimentally induced growth arrest may reflect the cell's attempt to continue cycling. However, it is also possible that the increase in Id-2 levels will inhibit the expression and/or action of factors required for cell cycle progression of maturing hematopoietic cells and will thus mediate their withdrawal from active cell cycle.
There may be a direct connection between the inverse expression of Id-1 and Id-2 in hematopoietic cells. A functionally important E box in the Id-2 promoter has been shown to regulate its activity in neuroblastoma and glioma cell lines.69,70 It is possible that the differentiation-related down-regulation of Id-1 and subsequent release of any sequestered class I E proteins permits the formation of heterodimers that may, in turn, activate Id-2, and possibly also Id-3, gene expression through E-box motifs in their own promoters. If the various Id factors have similar specificity for dimerization with individual E proteins, then it may be the transient period after Id-1 extinction and before induction of Id-2 or Id-3, which allows formation of transactivating heterodimers that can mediate early stages of the differentiation process. Alternatively, it may be that Id-1 and Id-2/Id-3 possess differential capacity for regulating specific HLH heterodimers that have functional importance in undifferentiated and differentiating cells, respectively. Feedback loops from the Id-2 or Id-3 could also result in repression of these genes later in the differentiation process and could account for the mechanism by which Id-3 expression is extinguished in terminally differentiated B (plasma) cells (Fig 1).71
Multiple class I (E2A, E2-2, HEB) and class II (SCL/Tal-1, lyl-1) proteins are expressed during hematopoietic development. In most systems, it is a heterodimer between a class I and a tissue-specific class II gene that regulates a cell's ability to differentiate.19 Our earlier work has shown that the class II gene SCL/Ta1-1, a positive regulator of erythroid development as a heterodimer with the class I E2A proteins, is widely expressed in hematopoietic progenitors as is lyl-1.4 Many developmentally important genes have sequences homologous or closely related to the SCL/Tal-1 and lyl-1 heterodimer preferred binding motifs: c-myb and the serum response element of c-fos, the promoter of MHC class II Drα gene, pro-interleukin 1β promoter, γ-globin enhancer, the erythroid promoters of the carbonic anhydrase II, band 3 and GATA-1 genes, and the enhancer elements of the immunoglobulin locus.42,72,73 While HLH class I/class III proteins have been demonstrated to be critical for expression of immunoglobulin genes,74 the relative contribution of HLH heterodimers to the expression of most lineage-specific genes remains to be determined.
This study has extended our earlier analysis of SCL, lyl-1, E2A, and Id-1 to a detailed examination of the dynamics of Id family gene expression in hematopoietic cells at different stages of development. Our findings support the hypothesis that multiple members of the Id family will be important regulators of stem cell activation, progenitor growth, and the lineage differentiation process.
ACKNOWLEDGMENT
Thanks to Drs Christopher Roman, Ellen Kittler, and Gary Stein for their thoughtful comments on this manuscript. We also wish to acknowledge and thank Drs Harold Weintraub, Xiao-Hong Sun, and Fred Sablitzky for their gifts of Id-1, Id-2, and Id-4 cDNA clones, respectively.
Supported by Grant No. CA 27466 to P.J.Q. from the National Institutes of Health and by operating grants to N.N.I. from the National Cancer Institute of Canada and the Medical Research Council of Canada.
Address reprint requests to Cathleen L. Cooper, PhD, University of Massachusetts Cancer Center, 373 Plantation St, Suite 202, Worcester, MA 01605.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal