Abstract
We searched for immediate early cytokine responsive genes and isolated a novel gene, CIS (Cytokine Inducible SH2 containing protein) that is induced in hematopoietic cells by a subset of cytokines including interleukin-2 (IL-2), IL-3, and erythropoietin (EPO). The mutant IL-2 receptor that fails to activate STAT5 could not induce CIS, suggesting that STAT5 is involved in the cytokine-inducible expression of CIS. We cloned the 5′-flanking region of the CIS gene and found that about 200 bases upstream of the transcription-initiation site contain four potential STAT5 binding sites (MGF boxes). Luciferase reporter assays showed that these MGF boxes were essential for EPO-dependent promoter activity. Expression of STAT5 and the EPO receptor in HEK293 cells conferred EPO-dependent activation of the CIS promoter. These data indicate that CIS is a target of the JAK-STAT5 pathway of cytokine receptors. CIS contains an SH2 domain and binds to tyrosine-phosphorylated EPO and IL-3 receptors. In HEK293 cells expressing STAT5 and the EPO receptor, EPO-dependent tyrosine phosphorylation of STAT5, as well as EPO-dependent CIS-promoter activation, was suppressed when CIS was coexpressed. Moreover, the induction of oncostatin M, another STAT5 target, as well as the tyrosine-phosphorylation of STAT5, were partially suppressed by CIS expression in Ba/F3 cells. Thus, CIS is a feedback modulator of STAT5; its expression is induced by STAT5 and it negatively modulates STAT5 activation.
GROWTH, DIFFERENTIATION, and functions of immune and hematopoietic cells are controlled by multiple cytokines, including interleukins (ILs) and colony-stimulating factors (CSFs).1 These cytokines exert their biological functions through specific receptors expressed on their target cells. A novel subfamily of protein tyrosine kinases, known as Janus kinases (JAKs), plays an important role in cytokine signaling. This pathway, originally found in interferon (IFN) signaling, is now known to be shared by various cytokines.2,3 This signaling system is unique for its direct link of the receptor-ligand interaction on the cell surface to gene expression in the nucleus. JAKs associate with cytokine receptors and are stimulated when cytokines bind to their cognate receptors. The activated JAKs, in turn, convert latent cytoplasmic transcription factors, known as STATs (signal transducer and activator of transcription), into active forms by tyrosine phosphorylation. The tyrosine phosphorylated STATs form homo or heterodimers and translocate into the nucleus where they bind to their specific target sequences, most of which are related to gamma interferon (IFN-γ) activated sites (GAS), a key regulatory element in the promoter of IFN-γ–inducible genes.4
The STAT family presently includes six members, each of which functions in a specific cytokine system.4 STAT5, which was originally identified as mammary gland factor (MGF ) regulated by prolactin,5 is activated by multiple cytokines such as IL-2, IL-3, IL-5, granulocyte macrophage colony-stimulating factor (GM-CSF ), erythropoietin (EPO) and thrombopoietin (TPO).6-11 Although IL-2 and the other cytokines activate different JAKs, they all activate STAT5. The consensus STAT5 recognition sequence TTCNNNGAA (MGF box) overlaps STAT1 and STAT3 binding motifs. However, in contrast to STAT1 and STAT3, the target genes of STAT5 in tissues other than mammary gland remain largely unknown.
We searched for immediate early genes induced by multiple cytokines, using IL-3 and EPO-dependent hematopoietic cells and identified a novel immediate early gene, CIS.12 It encodes an SH2 domain, which stably associates with tyrosine-phosphorylated IL-3 and EPO receptors. Because CIS is induced by IL-2, IL-3, GM-CSF, and EPO, but not by IL-6, granulocyte colony-stimulating factor (G-CSF ) and stem cell factor (SCF ), it seems to be a target of STAT5. We analyzed the promoter region of the murine CIS gene and found that MGF boxes are essential for induction by EPO. Moreover, CIS expression inhibited the EPO-dependent activation of STAT5, indicating that CIS is a kind of feedback modulator of STAT5.
MATERIALS AND METHODS
Cells.Murine IL-3–dependent lymphoid Ba/F3 cells and their transformants were maintained in RPMI medium supplemented with 10% fetal calf serum (FCS) and 10% conditioned medium from the WEHI-3B cell line as a source of IL-3.13 Ba/F3 transformants expressing Dexamethasone (Dex)-inducible CIS, as well as the EPO receptor (BF-CIS-ER),12 the IL-2 receptor β chain (F-7), and the C-terminal truncated IL-2 receptor β chain (H-4) were described previously.9 L cell transformants stably expressing the EPO receptor (L-ER) were obtained by transfection with pEF-EPOR14 and selection with G418. L, L-ER, and HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% FCS.
Cloning of the 5′-flanking region of the CIS gene.The 5′-flanking region of the CIS gene was cloned by means of cassette-ligation mediated polymerase chain reaction (PCR)15 using a Takara in vitro PCR cloning kit (Takara, Japan). Briefly, mouse genomic DNA was digested with EcoRI, then ligated with an EcoRI cassette oligonucleotide. PCR was performed using a sense cassette primer and the antisense CIS cDNA primer, AAGGAGAAAGGAGCCAATGGGTCCAGGGGT according to the manufacturer's instructions. The PCR product was cloned into the pCRII vector (Invitrogen, San Diego, CA). The full-length cloned promoter region, BglII/Xho I fragment and mutated fragments were subcloned into the upstream region of the luciferase gene in pGV (Nippon Gene, Tokyo, Japan). In vitro mutagenesis of the MGF boxes proceeded as described.16 The following primers were designed to replace consecutively two MGF boxes (TTCNNNGAA) with GAATTC (EcoRI site): A (TCCAGATCTCACAGCAC), B (TTAGAATTCCGGGCGGGAGCTCCTAGA), C (ATCGAATTCATCTGTCAAAGGTGTTT), D (ATAGAATTCCCGCGGGCTTTGCGGCG), E (TTAGAATTCGGGCTGGGACGCAGCGG), F (AAGCTTGAGAAAGGAGCCAATGGG). The PCR fragment produced by A and B primers (AB) and that by C and F primers (CF ) were joined using EcoRI sites, then ligated into the pGV basic vector (construct B). Similarly, fragments produced by AD and EF primers were ligated into pGV (construct C). Construct D lacking all MGF boxes were produced by replacing the Sac I-HindIII fragment of construct B with that of construct C. Reporter gene containing wild-type promoter region is designated construct A.
Luciferase assay.Plasmids (10 μg/transfection) of various CIS promoter-luciferase constructs in pGV were introduced into L-ER cells using a calcium-phosphate method as described.17 After 24 hours, the cells were factor depleted for 5 hours, then stimulated with 5 ng/mL IL-3, 10 U/mL EPO, or 10 ng/mL IL-2 for 2 hours. Cell extracts were prepared and luciferase activity was measured as described.5,6 HEK293 cells were also transfected with pGV-promoter-reporter gene constructs in combination with the expression vector (10 μg/transfection) carrying the EPO receptor (pXM-EPOR), STAT5 (pXM-MGF ), and CIS (pME-CIS). These plasmids have been described previously.6,7,12 16
Northern hybridization.The BF-CIS-ER cells were factor-depleted for 6 hours in RPMI medium containing 1% bovine serum albumin (BSA) in the presence of absence of 500 nmol/L Dex, then stimulated with 10 ng/mL IL-3 or 10 U/mL EPO. For Northern blotting, total RNA (5 μg) was separated on 1.0% agarose gels containing 2.4% formaldehyde, then transferred to positively charged nylon membranes. After fixation under calibrated ultraviolet (UV) irradiation, the membranes were hybridized with DIG-labeled riboprobes prepared using a DIG-RNA labeling kit (Boehringer, Tokyo, Japan). The blot was visualized using alkaline-phosphatase labeled anti-digoxigenin (DIG) antibody and a chemiluminescent substrate according to the manufacturer's instructions.
Detection of tyrosine-phosphorylation of STAT5.BF-CIS-ER or transfected HEK293 cells (5 × 107/sample) were incubated in the medium containing 0.5% FCS and 0.2 mmol/L sodium vanadate for 15 minutes at 37°C, then stimulated with 10 ng/mL IL-3 or 10 U/mL EPO at 25°C for 10 minutes. Cells were then solubilized with 1 mL of 1.0% Triton X-100 in phosphate-buffered saline (PBS) containing 50 mmol/L NaF, 10 mmol/L sodium pyrophosphate, 1 mmol/L sodium vanadate, and 1% aprotinin. After centrifugation, cleared lysates were incubated with 4 μL of anti-STAT55 for 1 hour at 4°C. The immune complexes adsorbed with protein A-Sepharose were resolved on 8.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), immunoblotted with monoclonal antiphosphotyrosine antibody (4G10) or anti-STAT5, and detected as described.14
RESULTS
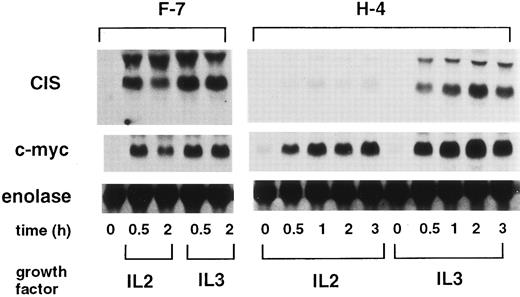
Mutant IL-2 receptor β chain that fails to activate STAT5 cannot induce CIS.To determine the STAT5 requirement for CIS-induction, we used a mutant IL-2 receptor β chain, which does not activate STAT5, but supports cell proliferation. Fujii et al9 showed that the H mutant of the IL-2 receptor β chain lacking the 147 carboxyl-terminal amino acid activates JAK1 and JAK3 and supports cell growth, but fails to activate STAT5. As shown in Fig 1, both IL-2 and IL-3 induced CIS in Ba/F3 cells expressing the wild-type IL-2 receptor β chain (F-7). However, in H-4 cells expressing the H mutant receptor, IL-2 failed to induce CIS. Because IL-2 still induced c-myc, the H-mutant receptor was actually activated by IL-2. Failure of CIS-induction by IL-2 was not due to the alteration of the CIS gene itself, as IL-3 still induced CIS in H-4 cells. These data suggest that STAT5 plays an important role in CIS-induction.
CIS-induction by IL-2 in F-7 and H-4 cells. Ba/F3 cells expressing the wild-type IL-2 receptor β chain (F-7) or cells expressing the H-mutant (H-4) were factor-depleted for 4 hours, and stimulated with either IL-2 or IL-3 for the indicated periods (h). Total RNA (5 μg/lane) was separated from the cells and blotted with CIS, c-myc, and α-enolase probes.
CIS-induction by IL-2 in F-7 and H-4 cells. Ba/F3 cells expressing the wild-type IL-2 receptor β chain (F-7) or cells expressing the H-mutant (H-4) were factor-depleted for 4 hours, and stimulated with either IL-2 or IL-3 for the indicated periods (h). Total RNA (5 μg/lane) was separated from the cells and blotted with CIS, c-myc, and α-enolase probes.
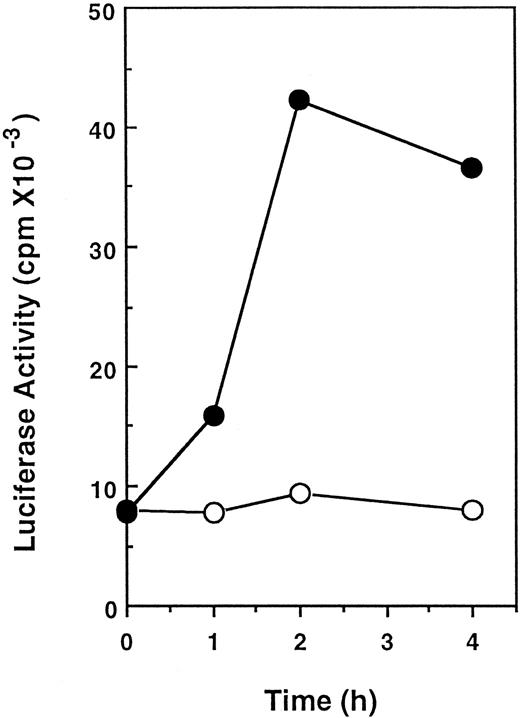
Isolation of the 5′-flanking region of the CIS gene and its promoter activity.To explore the involvement of STAT5 on CIS-induction, we cloned the 5′-flanking region of the CIS gene. Approximately 0.8 kb of the 5′-flanking region of the CIS gene obtained by cassette-ligation mediated PCR is shown in Fig 2. The 5′-end of mRNA was determined by means of rapid amplification of the cDNA ends (RACE)-PCR as described previously.12 The major transcriptional-initiation site (the bold arrow in Fig 2) determined by this procedure was located 45 bases upstream from the translation start codon (the broken arrow), and a minor transcriptional start site was found 130 bases upstream (not shown). Although there was no apparent TATA box, there was a GC-rich region near the transcription-initiation site. Four MGF boxes (TTCNNNGAA) were located close to the transcription-initiation site. One of these sequences was identical to that of β-casein (TTCTTGGAA). Two consecutive MGF boxes (boxes 1, 2 and boxes 3, 4) were separated by an insertion of approximately 80 bp. To examine the role of these elements in the activation by cytokines, this promoter region was linked to the luciferase reporter gene and transfected into Ba/F3 cells. As shown in Fig 3, stimulation with IL-3 increased the luciferase activity in Ba/F3 cells transformed with the reporter gene fourfold to fivefold. This indicates that a 5′-flanking region of approximately 0.8 kb is sufficient to confer IL-3–dependent promoter activity.
The nucleotide sequence of the 5′-flanking region of murine CIS gene. The 5′-untranslated flanking sequence obtained by PCR is numbered as minus from the major transcription-initiation site. The bold arrow indicates a major transcription-initiation site and the broken arrow indicates the first ATG codon. Four MGF boxes are underlined.
The nucleotide sequence of the 5′-flanking region of murine CIS gene. The 5′-untranslated flanking sequence obtained by PCR is numbered as minus from the major transcription-initiation site. The bold arrow indicates a major transcription-initiation site and the broken arrow indicates the first ATG codon. Four MGF boxes are underlined.
IL-3–induced activation of the reporter gene in Ba/F3 cells. Cells were transfected with 50 μg of the reporter gene carrying 0.8-kb promoter region linked to luciferase gene and 5 μg of pSV2neo by electroporation. After culture in the presence of 1 mg/mL G418 for 2 weeks, a pool of G418-resistant cells was starved for 16 hours in the absence of growth factor and then stimulated with (•) or without (○) 10 ng/mL IL-3 for the indicated periods. Cell extracts were prepared and luciferase activity was measured.
IL-3–induced activation of the reporter gene in Ba/F3 cells. Cells were transfected with 50 μg of the reporter gene carrying 0.8-kb promoter region linked to luciferase gene and 5 μg of pSV2neo by electroporation. After culture in the presence of 1 mg/mL G418 for 2 weeks, a pool of G418-resistant cells was starved for 16 hours in the absence of growth factor and then stimulated with (•) or without (○) 10 ng/mL IL-3 for the indicated periods. Cell extracts were prepared and luciferase activity was measured.
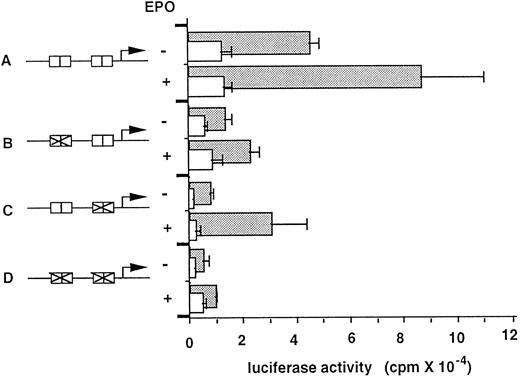
For a convenient reporter gene assay, we used L cell fibroblast cell line stably expressing the EPO receptor (L-ER). In this transformant, EPO induced tyrosine phosphorylation of JAK2, as well as the EPO receptor, and EPO also induced expression of endogenous CIS and c-myc (data not shown). Approximately 540 bases of the upstream region of the CIS gene (A construct; wild-type) was sufficient to confer EPO-inducible promoter activity (Fig 4). Promoter activity was not EPO-dependently increased in parental L cells lacking the EPO receptor. To determine the effect of MGF boxes on promoter activity, each or both of the consecutive MGF boxes were deleted (B, C, and D constructs). Deletion of one of the two consecutive MGF boxes decreased promoter activity, but still retained EPO inducibility (constructs B and C). However, disruption of all four MGF boxes severely abolished basal promoter activity and EPO dependence (construct D). These data indicate that MGF boxes were essential for EPO-dependent promoter activity, and the presence of all four MGF boxes was required for full promoter activity.
EPO stimulation of the CIS promoter-luciferase reporter gene in L-ER cells. Schematic representations of each reporter construct are shown in the lower portion (A-D). Boxes indicate MGF boxes, and their disruptions are shown in B, C, and D. The plasmids (10 μg) were introduced into wild-type L cells (open bars) or transformants expressing the EPO receptor (closed bars) using a calcium phosphate method for 12 hours. After transfection, cells were incubated in the presence or absence of EPO for 24 hours and luciferase activity was measured. Luciferase activity (arbitrary units) from triplicate experiments is shown.
EPO stimulation of the CIS promoter-luciferase reporter gene in L-ER cells. Schematic representations of each reporter construct are shown in the lower portion (A-D). Boxes indicate MGF boxes, and their disruptions are shown in B, C, and D. The plasmids (10 μg) were introduced into wild-type L cells (open bars) or transformants expressing the EPO receptor (closed bars) using a calcium phosphate method for 12 hours. After transfection, cells were incubated in the presence or absence of EPO for 24 hours and luciferase activity was measured. Luciferase activity (arbitrary units) from triplicate experiments is shown.
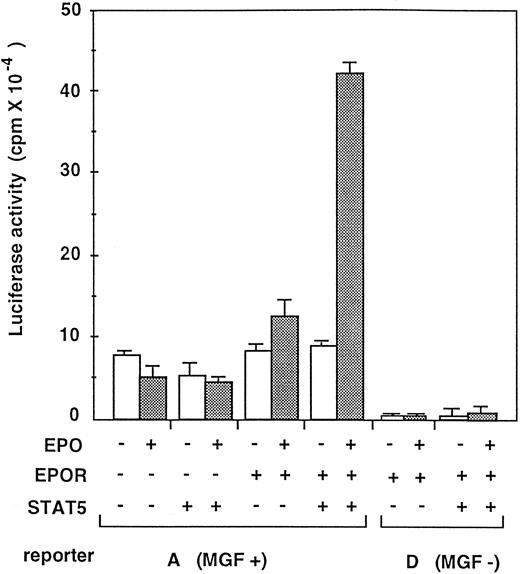
To determine whether or not the CIS promoter can really be activated by STAT5, the wild-type reporter construct A (MGF+) and the mutant reporter construct D, which lacks all four MGF boxes (MGF−) were transfected in HEK293 cells in combination with the EPO receptor and STAT5 (Fig 5). HEK293 cells have been used for high efficiency of transient transfection, and we found that they do not overexpress the EPO receptor and STAT5 compared with the COS cells that we used previously (data not shown). As shown in Fig 5, the A promoter construct was activated in an EPO-dependent manner when both the EPO receptor and STAT5 were coexpressed. Disruption of the MGF boxes abolished the EPO-dependent activation of CIS promoter (D, MGF−). These data strongly support the notion that CIS is a direct target of STAT5.
Reconstitution of CIS promoter activation by STAT5 and the EPO receptor in HEK293 cells. HEK293 cells were transfected with CIS promoter-luciferase reporter constructs in combination with the EPO receptor and/or STAT5 cDNAs in pXM expression vector. MGF+ means A construct carrying wild-type promoter and MGF− means D construct containing mutant promoter lacking all four MGF boxes, as shown in Fig 4. In minus (−), same amount of pXM plasmid without insert cDNA was added. Twenty-four hours after transfection, cells were stimulated with or without EPO (10 U/mL) and cell extracts were analyzed for luciferase activity.
Reconstitution of CIS promoter activation by STAT5 and the EPO receptor in HEK293 cells. HEK293 cells were transfected with CIS promoter-luciferase reporter constructs in combination with the EPO receptor and/or STAT5 cDNAs in pXM expression vector. MGF+ means A construct carrying wild-type promoter and MGF− means D construct containing mutant promoter lacking all four MGF boxes, as shown in Fig 4. In minus (−), same amount of pXM plasmid without insert cDNA was added. Twenty-four hours after transfection, cells were stimulated with or without EPO (10 U/mL) and cell extracts were analyzed for luciferase activity.
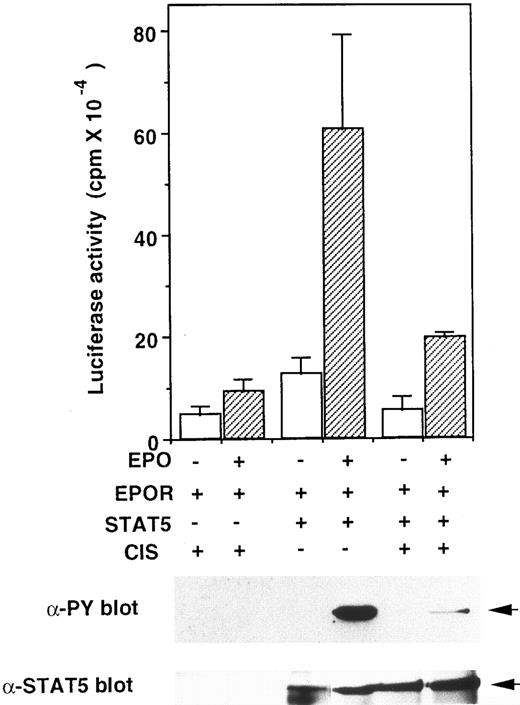
Forced expression of CIS suppressed STAT5 activation.We reported that CIS binds to the tyrosine phosphorylated EPO and IL-3 receptors and partially suppresses cell proliferation.12 Because CIS seems to modulate signal transduction from the receptor, we examined the effect of CIS expression on STAT5 activation. The EPO receptor, STAT5, CIS, and reporter A constructs were transiently expressed in HEK293 cells. As shown in Fig 6, upper graph, the EPO-induced luciferase activity decreased to about one third when CIS was coexpressed with STAT5. To examine STAT5 activation, STAT5 protein was precipitated with anti-STAT5 antibody and immunoblotted with antiphosphotyrosine antibody (Fig 6, lower panel). Tyrosine phosphorylation of STAT5 was decreased when CIS was coexpressed. These data suggest that CIS inhibits STAT5 trans-activation activity by suppressing its tyrosine-phosphorylation.
CIS inhibits STAT5 activity in a reconstituted system in HEK293 cells. Upper graph, plasmids for the EPO receptor, STAT5, and luciferase-reporter gene (construct A) were introduced into HEK293 cells as described in the legend to Fig 5, in the presence of an expression vector carrying the CIS gene (pME-CIS)(+) or unrelated protein (pME-CCF18)(−). After a 24-hour incubation in the presence or absence of EPO (10 U/mL), luciferase activity in each transfection was measured. Lower gels, after transfection, cells were cultured for 24 hours in DMEM containing 10% FCS without EPO, then starved for 12 hours in the medium containing 0.5% FCS. Cells were stimulated with EPO (10 U/mL) for 10 minutes. Cell extracts were immunoprecipitated with anti-STAT5 antibody, then resolved by SDS-PAGE and probed with antiphosphotyrosine (anti-PY) or anti-STAT5 antibodies.
CIS inhibits STAT5 activity in a reconstituted system in HEK293 cells. Upper graph, plasmids for the EPO receptor, STAT5, and luciferase-reporter gene (construct A) were introduced into HEK293 cells as described in the legend to Fig 5, in the presence of an expression vector carrying the CIS gene (pME-CIS)(+) or unrelated protein (pME-CCF18)(−). After a 24-hour incubation in the presence or absence of EPO (10 U/mL), luciferase activity in each transfection was measured. Lower gels, after transfection, cells were cultured for 24 hours in DMEM containing 10% FCS without EPO, then starved for 12 hours in the medium containing 0.5% FCS. Cells were stimulated with EPO (10 U/mL) for 10 minutes. Cell extracts were immunoprecipitated with anti-STAT5 antibody, then resolved by SDS-PAGE and probed with antiphosphotyrosine (anti-PY) or anti-STAT5 antibodies.
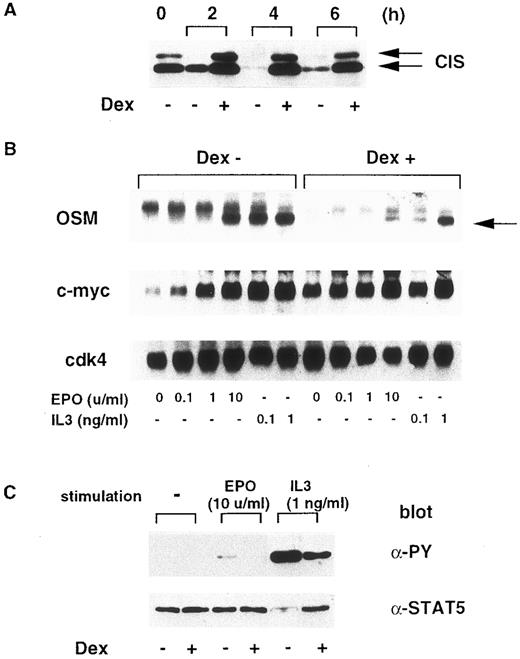
To determine the effect of CIS on cytokine-induced STAT5 activation in hematopoietic cells, we used BF-CIS-ER cells, which were transformed with Dex-inducible CIS and the EPO-receptor genes.12 As shown in Fig 7A, in the absence of Dex, the CIS protein level decreased rapidly after cytokines were removed. However, Dex maintained the CIS level even in the absence of growth factor up to 6 hours. We compared EPO or IL-3–induced oncostatin M (OSM) expression under these conditions. OSM is another target of the JAK-STAT5 pathway.16 As shown in Fig 7B, Dex suppressed EPO and IL-3–dependent OSM induction, but not c-myc. This suggests that CIS expression inhibits STAT5 activation. Next, we examined STAT5 tyrosine phosphorylation (Fig 7C). Although under our conditions, STAT5 was phosphorylated more efficiently by IL-3 than by EPO, Dex suppressed EPO and IL-3–dependent STAT5 phosphorylation. Tyrosine-phosphorylation of JAK2 was not affected by CIS expression (data not shown). These data indicate that forced CIS expression suppressed the cytokine-induced activation of STAT5.
Induction of CIS inhibits STAT5 activity in Ba/F3 cells. (A) After BF-CIS-ER cells were cultured in the absence of growth factor for 0, 2, 4, and 6 hours in the presence or absence of 500 nmol/L Dex, cell extracts were prepared and CIS content was measured by immunoblotting. (B) After 6 hours starvation in the presence or absence of Dex, cells were stimulated with the indicated concentrations of EPO or IL-3 for 20 minutes. Expression of OSM, c-myc, and cdk4 was measured by Northern blotting. (C) After starvation for 6 hours, cells were stimulated with 10 U/mL EPO or 10 ng/mL IL-3, and STAT5 was immunoprecipitated, resolved by SDS-PAGE, and tyrosine-phosphorylation was detected by immunoblotting with anti-PY. The STAT5 content was checked by reprobing the membrane with anti-STAT5.
Induction of CIS inhibits STAT5 activity in Ba/F3 cells. (A) After BF-CIS-ER cells were cultured in the absence of growth factor for 0, 2, 4, and 6 hours in the presence or absence of 500 nmol/L Dex, cell extracts were prepared and CIS content was measured by immunoblotting. (B) After 6 hours starvation in the presence or absence of Dex, cells were stimulated with the indicated concentrations of EPO or IL-3 for 20 minutes. Expression of OSM, c-myc, and cdk4 was measured by Northern blotting. (C) After starvation for 6 hours, cells were stimulated with 10 U/mL EPO or 10 ng/mL IL-3, and STAT5 was immunoprecipitated, resolved by SDS-PAGE, and tyrosine-phosphorylation was detected by immunoblotting with anti-PY. The STAT5 content was checked by reprobing the membrane with anti-STAT5.
DISCUSSION
We have searched for immediate early genes induced by multiple cytokines and identified the novel immediate early gene, CIS, which encodes an SH2 domain.12 We also isolated another immediate early gene, OSM, which is induced by a similar subset of cytokines. We characterized the promoter of the OSM gene and confirmed that OSM is a direct target of STAT5 in hematopoietic cells.16 Here we isolated and characterized the promoter region of the CIS gene. Several lines of evidence indicate that CIS is another target of STAT5. The promoter region of the CIS gene contained MGF boxes that are recognized by STAT5. The EPO-dependent promoter activity was abolished when MGF boxes in the promoter were deleted. Moreover, mutant IL-2 receptor β chain, which cannot activate STAT5, did not induce CIS. Recently, Mui et al18 reported that a dominant negative STAT5 (dnSTAT5), which lacks C-terminal trans-activation domain, suppresses CIS, as well as OSM, gene induction. These data indicate that CIS is a direct target of STAT5.
Mui et al18 also found that induction of pim-1 and Id-1 by IL-3 was suppressed by dnSTAT5 expression. However, in contrast to CIS and OSM gene promoters, there was no MGF boxes in the 5′-flanking region close to the transcription-initiation site in the murine pim-1 gene.19 Id-related protein has also been isolated as a growth factor-inducible gene.20 However, an MGF box was not present within the reported promoter region. Because GAS or MGF boxes are usually present within 200 to 300 bp upstream of initiation sites of the responsive genes, pim-1 and Id-1 may not be direct targets of STAT5. These genes may be regulated indirectly by STAT5. Analysis of promoters of pim-1 and Id-1 gene may provide a clue for STAT5-dependent, but indirect transactivation.
Although GAS or MGF boxes are present in cytokine-responsive genes, the extent to which they are used depends on many other considerations. For example, CIS was not induced by cytokines like IL-6 and G-CSF that activate STAT3,12 although MGF boxes in the CIS promoter were also recognized by STAT3. The potential for serine/threonine phosphorylation and interaction with other transcription factors will modify the potential ability of a STAT complex.21-23 The mechanism of this selection in hematopoietic cells is under investigation.
Although CIS is a target of STAT5, its physiological function remains unclear. We showed that CIS binds to tyrosine phosphorylated EPO and IL-3 receptors. Because forced expression of CIS partially inhibits cell proliferation, CIS modifies the function of cytokine receptors. CIS contains an SH2 domain, but no other motifs or enzymatic activities are found. Thus, CIS may mask phosphotyrosine residues of activated receptors and thereby downmodulate the signaling involved in SH2 domains. In support of this notion, CIS overexpression reduced the level of tyrosine phosphorylation of STAT5 (see Figs 6 and 7). Our preliminary data indicate that CIS binds to phosphorylated Y401 of the EPO receptor. Y401 is also one of the two major tyrosine residues that are essential for STAT5 activation.24-27 Thus, CIS binding to the receptor may reduce interaction between STAT5 and the receptor, resulting in the suppression of STAT5 activation. We found that the effect of CIS expression on STAT5 activation was partial. This may be because CIS cannot inhibit activation through other tyrosine residues like Y343, which are also involved in the activation of STAT5. Thus, CIS may be a modulator of STAT5, but not a complete inhibitor. One can argue that CIS directly binds to STAT5 and inhibits its DNA binding. However, such possibility is small, as tyrosine-phosphorylated IL-3 or EPO receptor, but not STAT5 were coimmunoprecipitated with CIS in Ba/F3 and FDC-P1 cells.12
Probably, CIS can compete with other SH2 proteins interacting with the cytoplasmic domain of the EPO receptor such as Syp, a tyrosine-phosphatase, which binds to phosphorylated Y401 and positively regulates Ras pathway and EPO-dependent proliferation.28 In support of this idea, the overexpression of CIS reduced the activation of MAK kinase (ERK2) in Dex-treated BF-CIS cells to about 50% of untreated cells (A. Yoshimura, unpublished data, August 1996). Although the role of STAT5 on proliferation is still controversial, the negative effect of CIS on proliferation may be explained by the competitive inhibition of SH2-containing signal transducers, including STAT5. Further study is under way to define the influence of CIS on the activation of other SH2 proteins.
ACKNOWLEDGMENT
We thank H. Ohgusu for excellent technical assistance, Drs T. Taniguchi and H. Fujii for F-7 and H-4 cells, Dr H. Wakao for anti-STAT5 antibody and technical support.
Supported in part by grants from the Ministry of Science, Education, and Culture of Japan, Kato Memorial Foundation (Tokyo, Japan), Japanese Foundation for Multidisciplinary Treatment of Cancer (Tokyo, Japan), Haraguchi Memorial Foundation (Tokyo, Japan), Uehara Memorial Foundation (Tokyo, Japan), Naito Memorial Foundation (Tokyo, Japan), and Kowa Life Science Foundation (Nagoya, Japan).
The nucleotide sequence reported in this paper will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases with the accession number D89613.
Address reprint requests to Akihiko Yoshimura, PhD, Institute of Life Science, Kurume University, Aikawamachi 2432-3 Kurume 839, Japan.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal