Abstract
There has been increasing interest in the involvement of mammalian homeobox (HOX) genes in hematopoietic regulation. The HOX genes are clustered in 4 chromosomes in mice and humans. In general, 5′ end HOX gene expression is predominant in hematopoietic stem cell populations, whereas 3′ end HOX gene expression are primarily found in committed progenitor cells. Furthermore, HOX genes of the A cluster are generally found in myelomonocytic cells, B cluster genes in erythropoietic cells, and C cluster genes in lymphoid cells. The results presented here concentrate on a single gene, namely HOX B6. Preliminary observations using whole mount in situ hybridization showed that both HOX B6 and erythropoietin (EPO) gene expression occurred in exactly the same areas of the 8.5-day mouse embryo. As a consequence, we studied the expression of HOX B6 and EPO gene expression from 6.5 to 19.5 days of gestation, in the neonate, and in the adult. It was found that the sequential transfer of erythropoiesis in different organs during development was followed by a similar transfer of HOX B6 and EPO gene expression. Between days 16.5 and 17.5, both HOX B6 and EPO gene expression decrease in the fetal liver, even though hepatic erythropoiesis continues to decline and is transferred to the fetal spleen. Precisely at this time point, HOX B6 and EPO gene expression are transferred to both the fetal spleen and fetal kidney. However, surprisingly, expression of both genes increases again in the fetal liver just before birth. HOX B6 is expressed in cells from in vitro erythropoietic colonies (colony-forming unit-erythroid and burst-forming unit-erythroid) and TER-119+ erythroid cells but not in hematopoietic or nonhematopoietic stem cell populations. When the latter two populations are allowed to differentiate into erythropoietic cells, HOX B6 and erythroid-relevant markers are expressed. The results indicate that HOX B6 is intimately involved in the regulation of the erythropoietic system and could be a marker for this lineage.
HOMEOBOX (HOX) GENES regulate fundamental mechanisms during embryologic development.1 Originally discovered in Drosophila melanogaster, they were described as selector and segmentation genes responsible for the correct sequence of segment production.2 Mutations in these regulator genes for development are called homeotic mutations, of which the first to be described was bithorax.3 Another homeotic mutation transforms the antennae on the head of Drosophila into legs. This mutation was mapped to the Antennapedia (Antp) gene and, after sequencing this gene and looking for cross-hybridization within the Ultrabithorax and fushi terazu genes,4 a common sequence was found that was called the homeobox.5 This evolutionary, highly conserved region of 180 bp encodes a 60 amino acid polypeptide segment that is called the homeodomain and was used to identify not only the homeobox gene cluster in Drosophila, but also in other animal species. Whereas the homeobox gene cluster in Drosophila is located on chromosome 36 in mice and humans, there are four gene clusters each on different chromosomes. Thus, the HOX (mammalian Antp complexes) gene clusters A, B, C, and D are present on chromosomes 6, 11, 15, and 2 in the mouse and 7, 17, 12, and 2 in the human, respectively.7 Since the tandem organization of the individual genes and their high level of homology, it is thought that the genes were derived by duplication and cluster expansion of a primordial HOX gene.8
The three-dimensional structure of the homeodomain is that of a helix-turn-helix DNA-binding protein.9,10 Developmental expression of HOX genes usually begins at the time mesoderm is formed or just after. Several remarkable observations have been noted with respect to the expression of homeobox genes during development. First, homeoboxes at the 3′ end of the mammalian HOX complexes are similar in nucleotide sequence to those at the 3′ end in Drosophila homeotic complexes, in particular the labial gene, whereas those at the 5′ end of the mammalian complex resemble the Abdominal-B gene in Drosophila.11 12 Second, with few exceptions, within each HOX complex, every mammalian gene is transcribed in the same direction. This is not the case in Drosophila.13 Finally, there is not only structural but also functional resemblance between HOX genes and Drosophila homeotic genes. Thus, the genomic organization of the HOX genes is colinear with their expression in the various body regions, so that 3′ end genes are expressed anteriorly, whereas those at the 5′ end are expressed primarily posteriorly.14 15
In the mammal, some HOX genes are only expressed for a certain time during development. One example is the HOX B8 (formerly 2.4; see the Results). Enforced, ectopic expression of HOX B8 has been shown to enhance the production of immature myeloid progenitors and progression to a fully malignant state.16,17 Some HOX genes are expressed almost ubiquitously throughout the body after development, whereas others appear to be expressed in particular organs and tissues. Lawrence and Largman18 have summarized the involvement of HOX genes in the hematopoietic system. Although the function of these genes is largely unknown, an interesting observation has evolved. In general, 5′ end HOX genes are largely expressed in the hematopoietic stem cell compartment, whereas 3′ end HOX genes are expressed in committed progenitor cells. Furthermore, the expression of specific HOX clusters appears to be restricted to specific hematopoietic lineages. Thus, genes of the HOX A cluster are found in myelomonocytic cells,19,20 genes of the HOX C cluster in lymphopoietic cells,21 and genes of the HOX B cluster in erythropoietic cells.22-24 Many studies regarding the involvement of HOX genes in the hematopoietic system have concentrated on cell lines.19,20,22-26 However, more recently, studies on separated human CD34+ cells,27 in peripheral blood from leukemic patients,20,28 and in undifferentiated mouse bone marrow cells27 have been reported.
After a report by Shen et al29 showing that overexpression of HOX B6 in the erythroleukemic cell line K562 led to a reduction of the erythroid phenotype with concomitant reduction in α- and δ-globin and heme synthesis, we decided to study the expression of this gene during the development of the erythropoietic system in the mouse. Starting from 6.5-day-old mouse embryos, we report here a remarkable correlation between HOX B6 expression, erythropoietin (EPO) gene expression, and the pathway of erythropoietic ontogeny. HOX B6 is not expressed in hematopoietic stem cells, undifferentiated embryonic stem cells (ES cells), or primordial germ cells (PGCs). However, upon differentiation into the EPO lineage, all three stem cell populations express HOX B6. The results show that HOX B6 is closely associated with the regulation of the erythropoietic system and could be a marker for this lineage.
MATERIALS AND METHODS
Animals, preparation of cell suspensions, and treatment.For mating, both CBA/Ulm and C57Bl/6J mice were used. Embryos and fetuses were produced as described previously.30 Mating was allowed from 8 pm to 8 am. The morning after mating was denoted as 0.5 days. Only female adult animals were used for preparation of bone marrow. All adult animals were 10 to 12 weeks of age and were given food and water ad libitum. Mouse embryo cell suspensions from different gestational ages, fetal liver, and adult bone marrow were prepared by standard methods described previously.31 In some cases, adult female mice were treated with 5-fluorouracil (5-FU; Sigma Chemicals, Munich, Germany) to kill proliferating cells, thereby allowing an early hematopoietic stem cell population, the high proliferative potential colony-forming cell (HPP-CFC), to be assayed (see below).31 Mice were injected intraperitoneally with 150 mg/kg body weight 5-FU dissolved in 0.14 mol/L sodium chloride solution and sterile filtered through a 0.22-μm filter (Millipore, Hamburg, Germany).
ES cell culture.The ES cell line D3 was obtained from the American Type Culture Collection (ATCC; No. 1934-CRL; Rockville, MD). Upon initial thawing and cultivation in supernatant from a recombinant leukemia inhibitory factor (LIF )-producing cell line (see below), it was found that both ES cells and embryoid bodies (EB) were present. To obtain only ES cells, the cell line was subcloned by limiting dilution in 96-well plates (Nunc, Wiesbaden, Germany) such that only single colonies were produced per well. These were removed and expanded and one clone was used for the experiments described here.
ES cells were maintained in the undifferentiated state by the addition of 2,000 U/mL LIF, which was obtained from a CHO cell line producing the recombinant mouse molecule (kindly provided by Dr S. Pentz, Department of Medical Genetics, University of Ulm, Ulm, Germany). The LIF activity was measured using an enzyme-linked immunosorbent assay (Amersham-Buchler, Braunschweig, Germany). The D3 cell line was passaged every 2 to 3 days in Iscove's modified Dulbecco's medium (IMDM) containing LIF, 10% heat-inactivated (30 minutes at 56°C) fetal bovine serum (FBS), and 1 × 10−4 mol/L α-thioglycerol (α-TG). Cells were removed from the culture vessel by treatment with trypsin/EDTA (Sigma Chemical, Munich, Germany) for 4 minutes at 37°C. The cells were incubated at 37°C in 5% CO2 under normal atmospheric oxygen tensions.
Differentiating ES cells were produced by culturing a single-cell suspension in the absence of LIF using the hanging-drop method.32 Cells were counted and 4 or 5 drops of 1 × 103 ES cells in 20 μL of medium (IMDM, FBS, and α-TG) were transferred to a Petri dish lid that was then turned upside down and incubated for 24 hours to produce embryoid bodies. Thereafter, 2 mL of culture medium was added and the EBs were collected.
Culture of PGCs.Between 40 and 50 embryos at 8.5 days of gestation were prepared, the visceral yolk sac was removed, and the allantois/primitive streak region30 was disaggregated for 5 minutes in trypsin/EDTA solution at 37°C in a water bath with shaking action. The resulting cell concentration was adjusted to 1 × 104 cells in IMDM containing 10% FBS, 1 × 10−4 mol/L α-TG, 10 ng/mL LIF, 10 ng/mL recombinant human stem cell factor (rhSCF; a kind gift from Amgen, Thousand Oaks, CA), and 50 ng/mL recombinant mouse interleukin-3 (rmIL-3; DPC Biermann, Bad Neuheim, Germany) and transferred to hydrophilic Petriperm dishes (Specialized Culture Technologies, Voehringen, Germany) that had been precoated with human collagen and fibronectin.30 Colonies of PGCs were produced within 5 days and detected using both alkaline phosphatase staining and fluorescence microscopy indirect immunostaining using a mouse monoclonal antibody to the stage-specific embryonic antigen (SSEA-1; Developmental Studies Hybridoma Bank, Department of Biological Sciences, University of Iowa, Iowa City, IA). The undifferentiated PGC colonies were subjected to reverse transcription-polymerase chain reaction (RT-PCR) or secondary culture in methyl cellulose for 7 days in the presence of 500 U/mL EPO and 10 ng/mL IL-3 was performed (see below). The resulting colonies were then individually removed from the methyl cellulose and subjected to RT-PCR.
Culture of hematopoietic stem and progenitor cells.Culture of HPP-CFC from mouse bone marrow 3 days after in vivo injection of 5-FU and early (burst-forming unit-erythroid [BFU-E]) and late (colony-forming unit-erythroid [CFU-E]) erythropoietic progenitor cells has been described previously.30,31,33 HPP-CFC were stimulated with 1 ng/mL recombinant human rhIL-1β (DPC Biermann), 50 ng/mL rmIL-3 (DPC Biermann), 50 ng/mL rhIL-6 (a gift from Dr A. Shimosaka, Kirin Brewery, Tokyo, Japan), 50 ng/mL rhSCF (a gift from Amgen), and 15% supernatant from L929 cells. BFU-E were stimulated with 50 ng/mL IL-3 and 500 U/mL EPO and CFU-E were stimulated with 0.195 mU/mL EPO.33 Cultures for HPP-CFC and BFU-E were supplemented with 10% FBS and 10% horse serum, whereas CFU-E cultures contained 30% FBS. All cultures contained 1 × 10−4 mol/L α-TG and 1 × 10−12 g/mL pure mouse transferrin. About 20 1-mL cultures were prepared to obtain sufficient mRNA for further analysis. Colonies derived from CFU-E were cultured for 48 hours in an atmosphere containing 5% CO2 and 3.5% O2 before collection and detection of HOX B6 gene expression. Colonies derived from BFU-E were cultured under the same conditions and collected after 10 days of incubation.
Separation of fetal liver and adult bone marrow erythroblasts.Erythroblasts from 13.5-day fetal livers and adult bone marrow were separated from other cells by using the TER-119 antibody and magnetic beads. Cell suspensions were washed and pelleted and 10 μL of undiluted TER-119 antibody (Pharmingen, San Diego, CA) was added, and the cells were resuspended and incubated for 30 minutes at 4°C. Thereafter, the cells were washed in phosphate-buffered saline containing 5 mmol/L EDTA and 0.5% bovine serum albumin and incubated for 15 minutes at 4°C with microbeads conjugated to mouse antirat-κ antibodies (Miltenyi Biotec Inc, Sunnyvale, CA). Cells binding the antibody complex were separated using MiniMacs columns (Miltenyi Biotec Inc) according to the manufacturer's instructions. Both positive and negative selected cell populations were assayed for colony formation as well as morphology and histochemistry by benzidine staining. The remaining cells were then subjected to mRNA preparation and RT-PCR (see below).
Oligonucleotide Primers Used for RT-PCR
| Gene . | GenEMBL Bank Accession No. . | Primer Pairs . | Length of PCR Product (bp) . |
|---|---|---|---|
| EPO | M12930 | L: | 339 |
| 5′-AAGGAGGCAGAAAATGTCACGATG-3′ | |||
| R: 5′-ACCTGCTCCACTCCGAACACTCAC-3′ | |||
| EPO-R | J04843 | L: | 497 |
| 5′-CAATGAAGTAGTGCTCCTGGAC-3′ | |||
| R: 5′-GAGATGAGGACCAGAATGAGAG-3′ | |||
| b-FGF-R | M28998 | L: | 236 |
| 5′-ATGTGCAGAGCATCAACTGG-3′ | |||
| R: 5′-TCGTCGTCGTCATCATCTTC-3′ | |||
| GATA-1 | X03221 | L: | 367 |
| 5′-TGTGGTCTACAGCCTCTCAGCAC-3′ | |||
| R: 5′-GTGTAGTTCAAGTGAGGCAGAG-3′ | |||
| GATA-2 | M77810 | L: | 528 |
| 5′-TCTCAATCACCTCGACTCG-3′ | |||
| R: 5′-GACGGAGAGCATGAAGATG-3′ | |||
| Flk-2/Flt-3 | M64689 | L: | 299 |
| 5′-CTTCTGGTCAAATGCTGTGC-3′ | |||
| R: 5′-GGGAGTTCCGAGAGAGAAC-3′ | |||
| GM-CSF | X03221 | L: | 367 |
| 5′-TGTGGTCTACAGCCTCTCAGCAC-3′ | |||
| R: 5′-GTGTAGTTCAAGTGAGGCAGAG-3′ | |||
| Hox B5 | M26283 | L: | 485 |
| 5′-ATTATGGCAGTGGCAGCTCT-3′ | |||
| R: 5′-TGTGAAGCTTCCTCATCCAG-3′ | |||
| Hox B6 | J30782 | L: | 469 |
| 5′-CTGGAGCTGGAGAAGGAATT-3′ | |||
| R: 5′-TCCGATTCTCAGTTCTCACC-3′ | |||
| Hox B7 | Y00436 | L: | 164 |
| 5′-AAGCGAG&CCGCCAGACCTACACG-3′ | |||
| R: 5′-CCACTTCATGCGCCGGTTCTGAA-3′ | |||
| Hox B8 | X13721 | L: | 489 |
| 5′-AATCCAGGAGTTCTACCACG-3′ | |||
| R: 5′-GCAACTTCTACGGCTACCAC-3′ | |||
| HPRT | J00423 | L: | 229 |
| 5′-CACAGGACTAGAACACCTGC-3′ | |||
| R: 5′-GCTGGTGAAAAGGACCTCT-3′ | |||
| LIF-R | D17444 | L: | 460 |
| 5′-TGACTTCTTCACCTCCTCCT-3′ | |||
| R: 5′-TACAGATGGATGAGAGGACG-3′ | |||
| SCF | M59915 | L: | 320 |
| 5′-GCTTGACTACTCTTCTGGAC-3′ | |||
| R: 5′-GTGACACTGACTCGTGAATC-3′ | |||
| SCF-R | Y00864 | L: | 490 |
| 5′-TGTGTCTACATCCGTGAACTCC-3′ | |||
| R: 5′-GGTATAAGTGCCTCCTTCTGTG-3′ | |||
| CD34 | M81104 | L: | 618 |
| 5′-GTTACCTCTGGGATCCCTTCAGGCTC-3′ | |||
| R: 5′-GMTMCGTAACCAGTGGAGACCTC-3′ |
| Gene . | GenEMBL Bank Accession No. . | Primer Pairs . | Length of PCR Product (bp) . |
|---|---|---|---|
| EPO | M12930 | L: | 339 |
| 5′-AAGGAGGCAGAAAATGTCACGATG-3′ | |||
| R: 5′-ACCTGCTCCACTCCGAACACTCAC-3′ | |||
| EPO-R | J04843 | L: | 497 |
| 5′-CAATGAAGTAGTGCTCCTGGAC-3′ | |||
| R: 5′-GAGATGAGGACCAGAATGAGAG-3′ | |||
| b-FGF-R | M28998 | L: | 236 |
| 5′-ATGTGCAGAGCATCAACTGG-3′ | |||
| R: 5′-TCGTCGTCGTCATCATCTTC-3′ | |||
| GATA-1 | X03221 | L: | 367 |
| 5′-TGTGGTCTACAGCCTCTCAGCAC-3′ | |||
| R: 5′-GTGTAGTTCAAGTGAGGCAGAG-3′ | |||
| GATA-2 | M77810 | L: | 528 |
| 5′-TCTCAATCACCTCGACTCG-3′ | |||
| R: 5′-GACGGAGAGCATGAAGATG-3′ | |||
| Flk-2/Flt-3 | M64689 | L: | 299 |
| 5′-CTTCTGGTCAAATGCTGTGC-3′ | |||
| R: 5′-GGGAGTTCCGAGAGAGAAC-3′ | |||
| GM-CSF | X03221 | L: | 367 |
| 5′-TGTGGTCTACAGCCTCTCAGCAC-3′ | |||
| R: 5′-GTGTAGTTCAAGTGAGGCAGAG-3′ | |||
| Hox B5 | M26283 | L: | 485 |
| 5′-ATTATGGCAGTGGCAGCTCT-3′ | |||
| R: 5′-TGTGAAGCTTCCTCATCCAG-3′ | |||
| Hox B6 | J30782 | L: | 469 |
| 5′-CTGGAGCTGGAGAAGGAATT-3′ | |||
| R: 5′-TCCGATTCTCAGTTCTCACC-3′ | |||
| Hox B7 | Y00436 | L: | 164 |
| 5′-AAGCGAG&CCGCCAGACCTACACG-3′ | |||
| R: 5′-CCACTTCATGCGCCGGTTCTGAA-3′ | |||
| Hox B8 | X13721 | L: | 489 |
| 5′-AATCCAGGAGTTCTACCACG-3′ | |||
| R: 5′-GCAACTTCTACGGCTACCAC-3′ | |||
| HPRT | J00423 | L: | 229 |
| 5′-CACAGGACTAGAACACCTGC-3′ | |||
| R: 5′-GCTGGTGAAAAGGACCTCT-3′ | |||
| LIF-R | D17444 | L: | 460 |
| 5′-TGACTTCTTCACCTCCTCCT-3′ | |||
| R: 5′-TACAGATGGATGAGAGGACG-3′ | |||
| SCF | M59915 | L: | 320 |
| 5′-GCTTGACTACTCTTCTGGAC-3′ | |||
| R: 5′-GTGACACTGACTCGTGAATC-3′ | |||
| SCF-R | Y00864 | L: | 490 |
| 5′-TGTGTCTACATCCGTGAACTCC-3′ | |||
| R: 5′-GGTATAAGTGCCTCCTTCTGTG-3′ | |||
| CD34 | M81104 | L: | 618 |
| 5′-GTTACCTCTGGGATCCCTTCAGGCTC-3′ | |||
| R: 5′-GMTMCGTAACCAGTGGAGACCTC-3′ |
RT-PCR.mRNA from embryos, fetal liver, spleen, kidney, adult bone marrow, and undifferentiated and differentiating ES cells and PGCs was first isolated using an isolation kit (Stratagene, Heidelberg, Germany) and the mRNA was purified using the QuickPrep Micro mRNA purification kit (Pharmacia, Freiburg, Germany). A complete litter was used to prepare mRNA for each time point. After preparation, the mRNA was immediately aliquoted and frozen at −70°C. Between two and three mRNA samples from each tissue and organ from different litters for each time point were prepared and stored. Experiments were then repeated not only with samples from the same litter, but also from different litters. The mRNA was reverse transcribed into cDNA, which was in turn amplified using the GeneAmp RNA PCR kit (Perkin Elmer, Friedrichshaven, Germany). The PCR was performed using a Gene ATAQ thermocycler controller (Pharmacia) for 35 cycles (except where otherwise stated) using the following parameters: 1 minute at 95°C, 2 minutes at 50°C, and 2 minutes at 72°C. The PCR products were separated by agarose (NuSieve) gel (3%) electrophoresis.
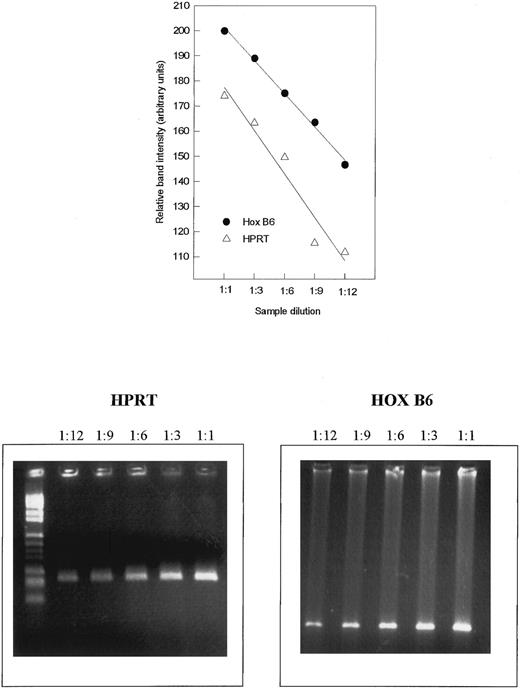
To determine the relative increase and decrease in gene expression, a semiquantitative PCR assay was developed. The mRNA concentration was determined photometrically at 260 nm and 20 ng mRNA was reverse transcribed. The PCR reaction was performed for 30 cycles (1 minute 95°C, 2 minutes at 50°C, and 2 minutes at 72°C). To show that the PCR amplification was linear, the cDNA was diluted 1:1, 1:3, 1:6, 1:9, and 1:12 before amplification was performed. As standard, primers specific for hypoxanthine phosphoribosyltransferase (HPRT) were used. The PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide The intensity of the resulting bands was determined using image analysis software (Sigma Scan Image; Jandel Scientific, Corte Madera, CA). The band intensity is given as arbitrary units.
Oligonucleotides.Oligonucleotides for RT-PCR primers and probes for Southern blotting (where appropriate) were prepared by the Section of Polymer Science (University of Ulm) or Interactiva (Ulm, Germany). Synthesis was performed using a Pharmacia Gene Assembler Plus DNA synthesizer and the oligonucleotides purified over a high-performance liquid chromatography RP18 reverse-phase column (Waters, Milford, ME). The oligonucleotide primer sequences shown in Table 1 were selected using the Primer Designer software package (Sigma) after obtaining the gene sequences from the GenEMBL data bank.
Southern blotting.For Southern blotting of HOX B6 and EPO, the following oligonucleotides were used: HOX B6, 5′-AGAACCGGCGCATGAAGTGGAAAAAGGAGAGCAAACTGCT-3′; and EPO, 5′-ATCAGTGGTCTACGTAGCCTCACTTCACTGCTTCGGGTACTGGGAGCT CAG-3′. These probes were 3′ end-labeled with digoxygenin using the DIG oligonucleotide tailing kit (Boehringer Mannhein, Mannhein, Germany). The ethidium bromide gel was washed for 5 minutes in water, followed by 15 minutes in 0.25 mol/L Hcl and 1 minute in water, and the DNA was denatured for 30 minutes in 0.2 mol/L NaOH and 0.6 mol/L NaCl. The gel was then equilibriated by incubating for 20 minutes in fourfold, 20 minutes in twofold, and three times for 20 minutes in onefold concentration of transfer buffer (19.2 mmol/L sodium dihydrogen phosphate, 36 mmol/L sodium citrate, pH 3.0). The gel was then electroblotted (LKB 2051 Midget Multiblot Electrophoresis Transfer Unit; Pharmacia, Freiburg, Germany) onto a 0.2-μm nitrocellulose filter (BioRad, Richmond, CA) at 4°C using 200 mA for 2 hours. The filter was then prehybridized using a prehybridization buffer containing 0.5% blocking reagent (Boehringer Mannheim) and hybridization using 25 ng/mL of labeled DNA probe. The signal was detected using antidigoxigenin conjugated to alkaline phosphatase according to the according to the DNA Labeling and Detection Kit (Boehringer Mannheim).
Standardization of RT-PCR for semiquantitation. Shown here are samples for the internal standard HPRT and HOX B6. The intensity of the ethidium bromide-stained agarose gels (lower panel) is given as relative units versus sample dilution (upper panel).
Standardization of RT-PCR for semiquantitation. Shown here are samples for the internal standard HPRT and HOX B6. The intensity of the ethidium bromide-stained agarose gels (lower panel) is given as relative units versus sample dilution (upper panel).
RESULTS
In preliminary studies, whole-mount in situ hybridization was used to detect EPO gene expression in the 8.5-day mouse embryo.34 For these experiments, a mixture of four, digoxigenin 3′ end-labeled ologonucleotides (positions: 605-654, 1212-1265, 2356-2406, and 3251-3301) were used. Similar experiments using a mixture of three digoxigenin 3′ end-labeled oligonucleotides (positions: 24-74, 576-629, and 1216-1266) for HOX B6 showed that the spatial expression was exactly the same as that for EPO in 8.5-day mouse embryos (data not shown). In both cases, positive hybridization was observed in the extraembryonic mesoderm of the visceral yolk sac and the primitive streak region. In addition, HOX B6 could additionally be seen in the notochord. No signal was observed in the rest of the embryo proper. Similarly, no signal was obtained for labeled sense probes or sham hybridization. This finding, together with the aforementioned observations by Shen et al,29 led to a more detailed comparison of HOX B6 with HOX B5, B7, and B8 and EPO during development.
HOX B6 gene expression during development.To obtain an indication of the relative amounts of HOX B6 expression occurring during development, the RT-PCR was standardized and semiquantitated using the expression of the household gene HPRT. Figure 1 shows the results when 20 ng of mRNA was first reverse transcribed and PCR was performed for 30 cycles. To show that the PCR was linear under these circumstances, cDNA was diluted 1:1, 1:3, 1:6, and 1:12 before amplification was performed. This was performed for both HPRT as well as HOX B6 (Fig 1, lower panel). The intensity of the ethidium bromide-stained bands produced after electrophoresis was determined by image analysis and the results are graphically depicted as arbitrary units as a function of sample dilution (Fig 1, upper panel). Although not parallel, the linearity indicates that semiquantitation can be performed. This type of analysis was later performed for all primer pairs and samples.
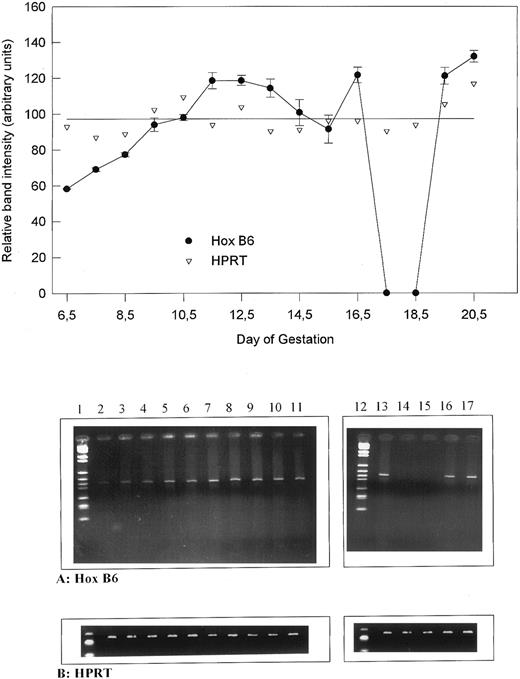
The relative gene expression of HOX B6 compared with HPRT during murine development. (Upper panel) Graphical representation showing the relative band intensity (in arbitrary units) against day of development. The single line shows the mean of all HPRT values. (Lower panel) Ethidium bromide gels for HOX B6 and HPRT from which the graph was obtained. Lane 1, markers; lane 2, 6.5 days; lane 3, 7.5 days; lane 4, 8.5 days; lane 5, 9.5 days; lane 6, 10.5 days; lane 7, 11.5 days; lane 8, 12.5 days; lane 9, 13.5 days; lane 10, 14.5 days; lane 11, 15.5 days; lane 12, markers; lane 13, 16.5 days; lane 14, 17.5 days; lane 15, 18.5 days; lane 16, 19.5 days; and lane 17, 0.5-day neonate.
The relative gene expression of HOX B6 compared with HPRT during murine development. (Upper panel) Graphical representation showing the relative band intensity (in arbitrary units) against day of development. The single line shows the mean of all HPRT values. (Lower panel) Ethidium bromide gels for HOX B6 and HPRT from which the graph was obtained. Lane 1, markers; lane 2, 6.5 days; lane 3, 7.5 days; lane 4, 8.5 days; lane 5, 9.5 days; lane 6, 10.5 days; lane 7, 11.5 days; lane 8, 12.5 days; lane 9, 13.5 days; lane 10, 14.5 days; lane 11, 15.5 days; lane 12, markers; lane 13, 16.5 days; lane 14, 17.5 days; lane 15, 18.5 days; lane 16, 19.5 days; and lane 17, 0.5-day neonate.
When expression of HOX B6 during development was compared with the HPRT control, the results seen in Fig 2 (upper panel) were obtained. To obtain these results, 20 ng mRNA was used to initiate the RT-PCR for both HPRT and HOX B6. The expression of HPRT varied little during development. In contrast, the general expression of HOX B6 showed a definite expression pattern. As expected, HOX B6 gene expression increased from 6.5 to 11.5 days of development. Between 11.5 and 13.5 days, expression remains relatively stable. From 13.5 to 15.5 days, a slight decrease occurs before another increase followed by a decrease to nondetectable levels between 17.5 and 18.5 days. Just before birth, HOX B6 gene expression is increased and remains active after birth. Figure 2 (lower panel) shows the respective HOX B6 and HPRT ethidium bromide gels that produced the graphical results.
Comparison between HOX B6 and HOX B5, B7, and B8 gene expression during development.Table 2 shows a comparison summary of HOX gene expression for B5 (formerly 2.1), B6 (2.2), B7 (2.3), and B8 (2.4). Whereas HOX B8 is only expressed from 7.5 to 11.5 days of gestation, HOX B7 is expressed at all times and in all organs studied. HOX B5 and B6 expression parallels each other with few exceptions, notably in the fetal liver on days 17.5 and 18.5, when HOX B5 is expressed, but not HOX B6.
HOX Gene Expression in the Embryo, Fetus, Neonate, and Adult
| Day . | Organ/Tissue . | HOX B5 . | HOX B6 . | HOX B7 . | HOX B8 . |
|---|---|---|---|---|---|
| 6.5 | Whole embryo | + | + | + | − |
| 7.5 | Whole embryo | + | + | + | (+) |
| 8.5 | Whole embryo | + | + | + | + |
| 9.5 | Whole embryo | + | + | + | + |
| 10.5 | Whole embryo | + | + | + | + |
| 11.5 | Whole embryo | + | + | + | + |
| 12.5 | Fetal liver/embryo | +/+ | +/+ | +/+ | − |
| 13.5 | Fetal liver/embryo | +/+ | +/+ | +/+ | − |
| 14.5 | Fetal liver/embryo | +/+ | +/+ | +/+ | − |
| 15.5 | Fetal liver/embryo | +/+ | +/+ | +/+ | − |
| 16.5 | Fetal liver/embryo | +/+ | +/+ | +/+ | − |
| 17.5 | Fetal liver | + | − | + | − |
| Fetal spleen | + | + | + | − | |
| Fetal kidney | + | + | + | − | |
| Fetal heart | − | + | + | − | |
| Fetal lung | − | + | + | − | |
| 18.5 | Fetal liver | + | − | + | − |
| Fetal spleen | + | + | + | − | |
| Fetal kidney | + | + | + | − | |
| Fetal heart | − | − | + | − | |
| Fetal lung | − | − | + | − | |
| 19.5 | Fetal liver | + | + | + | − |
| Fetal spleen | + | + | + | − | |
| Fetal kidney | + | + | + | − | |
| Fetal heart | − | − | + | − | |
| Fetal lung | − | − | + | − | |
| 0.5-day neonate | Liver | + | + | + | − |
| Spleen | + | + | + | − | |
| Kidney | + | + | + | − | |
| Heart | + | + | + | − | |
| Lung | + | + | + | − | |
| Adult | Liver | + | + | + | − |
| Spleen | + | + | + | − | |
| Kidney | + | + | + | − | |
| Heart | ND | − | + | − | |
| Lung | ND | − | + | − |
| Day . | Organ/Tissue . | HOX B5 . | HOX B6 . | HOX B7 . | HOX B8 . |
|---|---|---|---|---|---|
| 6.5 | Whole embryo | + | + | + | − |
| 7.5 | Whole embryo | + | + | + | (+) |
| 8.5 | Whole embryo | + | + | + | + |
| 9.5 | Whole embryo | + | + | + | + |
| 10.5 | Whole embryo | + | + | + | + |
| 11.5 | Whole embryo | + | + | + | + |
| 12.5 | Fetal liver/embryo | +/+ | +/+ | +/+ | − |
| 13.5 | Fetal liver/embryo | +/+ | +/+ | +/+ | − |
| 14.5 | Fetal liver/embryo | +/+ | +/+ | +/+ | − |
| 15.5 | Fetal liver/embryo | +/+ | +/+ | +/+ | − |
| 16.5 | Fetal liver/embryo | +/+ | +/+ | +/+ | − |
| 17.5 | Fetal liver | + | − | + | − |
| Fetal spleen | + | + | + | − | |
| Fetal kidney | + | + | + | − | |
| Fetal heart | − | + | + | − | |
| Fetal lung | − | + | + | − | |
| 18.5 | Fetal liver | + | − | + | − |
| Fetal spleen | + | + | + | − | |
| Fetal kidney | + | + | + | − | |
| Fetal heart | − | − | + | − | |
| Fetal lung | − | − | + | − | |
| 19.5 | Fetal liver | + | + | + | − |
| Fetal spleen | + | + | + | − | |
| Fetal kidney | + | + | + | − | |
| Fetal heart | − | − | + | − | |
| Fetal lung | − | − | + | − | |
| 0.5-day neonate | Liver | + | + | + | − |
| Spleen | + | + | + | − | |
| Kidney | + | + | + | − | |
| Heart | + | + | + | − | |
| Lung | + | + | + | − | |
| Adult | Liver | + | + | + | − |
| Spleen | + | + | + | − | |
| Kidney | + | + | + | − | |
| Heart | ND | − | + | − | |
| Lung | ND | − | + | − |
Abbreviation: ND, not done.
Comparison of HOX B6 and EPO gene expression during development.Figure 3 shows the ethidium bromide gels and respective Southern blots for HOX B6 and EPO gene expression during development. In this case, similar amounts of mRNA (30 ng) from all developmental stages were used for RT-PCR. The change in tissue/organ expression of the HOX B6 gene can be followed here. As shown above, HOX B6 is detected in the embryo from days 6.5 to 10.5. Thereafter, expression was seen not only in the rest of the fetus, but also in the fetal liver from days 11.5 to 16.5. However, between days 16.5 and 17.5, HOX B6 gene expression is reduced to nondetectable levels in the fetal liver. No HOX B6 gene expression is seen between days 17.5 and 18.5 in this organ (Table 2). In contrast, HOX B6 was expressed in the fetal spleen and fetal kidney (data not shown in this Fig 2 but given in Table 2) on days 17.5 and 18.5. At 19.5 days, HOX B6 gene expression increases in the fetal liver and can be detected after birth in the day-1 neonatal liver (Fig 3A and B).
Ethidium bromide gels (A and C) and Southern blots (B and D) for HOX B6 and EPO compared with HPRT control gene expression (E) in different tissues and organs during murine development. Lane 1, markers; lane 2, 6.5-day embryo; lane 3, 7.5-day embryo; lane 4, 8.5-day embryo; lane 5, 9.5-day embryo; lane 6, 10.5-day embryo; lane 7, 11.5-day embryo; lane 8, 12.5-day fetal liver; lane 9, 13.5-day fetal liver; lane 10, 14.5-day fetal liver; lane 11, 15.5-day fetal liver; lane 12, markers; lane 13, 16.5-day fetal liver; lane 14, 17.5-day fetal liver; lane 15, 17.5-day fetal spleen; lane 16, 18.5-day fetal liver; lane 17, 18.5-day fetal spleen; lane 18, 19.5-day fetal liver; and lane 19, 0.5-day liver.
Ethidium bromide gels (A and C) and Southern blots (B and D) for HOX B6 and EPO compared with HPRT control gene expression (E) in different tissues and organs during murine development. Lane 1, markers; lane 2, 6.5-day embryo; lane 3, 7.5-day embryo; lane 4, 8.5-day embryo; lane 5, 9.5-day embryo; lane 6, 10.5-day embryo; lane 7, 11.5-day embryo; lane 8, 12.5-day fetal liver; lane 9, 13.5-day fetal liver; lane 10, 14.5-day fetal liver; lane 11, 15.5-day fetal liver; lane 12, markers; lane 13, 16.5-day fetal liver; lane 14, 17.5-day fetal liver; lane 15, 17.5-day fetal spleen; lane 16, 18.5-day fetal liver; lane 17, 18.5-day fetal spleen; lane 18, 19.5-day fetal liver; and lane 19, 0.5-day liver.
Figure 3C and D shows the ethidium bromide gels and respective Southern blots for EPO gene expression during development using an initial mRNA of 30 ng throughout. The pattern of gene expression is exactly the same for both EPO and HOX B6. Thus, EPO gene expression occurs in the embryo from 6.5 to 10.5 days. Thereafter, it is transferred exclusively to the fetal liver until 16.5 days. Between days 17.5 and 18.5 in the fetal liver, EPO gene expression could not be detected by RT-PCR. However, as with HOX B6, EPO gene expression is observed in the 17.5- and 18.5-day fetal spleen and is also found in the fetal kidney during this time period (data not shown). In a similar manner to HOX B6, EPO gene expression increased in the 19.5-day fetal liver and was present 1 day after birth in the neonatal liver. This pattern of results was not only repeatable between mRNA aliquots of the same litter sample, but also between litter samples. Figure 3E shows the HPRT controls. These results clearly show a close correlation between HOX B6 and EPO gene expression.
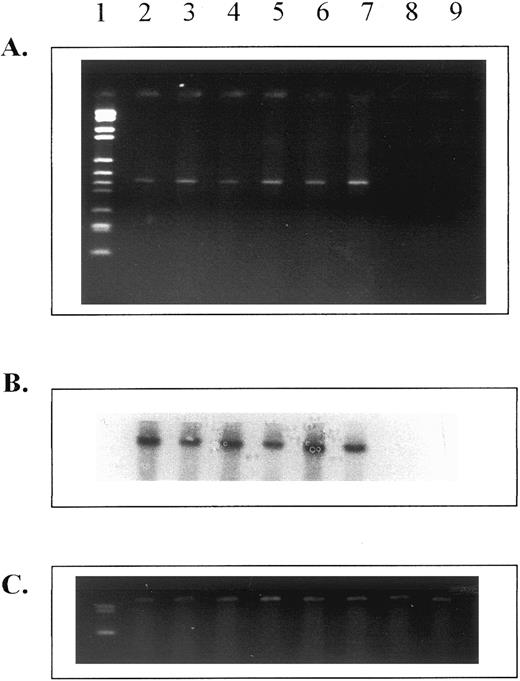
HOX B6 is expressed in erythroid progenitor cells but not in hematopoietic stem cells.The pattern of expression of both HOX B6 and EPO parallels that of erythropoietic ontogeny in many respects. It was therefore of interest to study whether HOX B6 gene expression was present in erythropoietic progenitor cells during development. Figure 4 shows that this is indeed the case. HOX B6 gene expression could be detected in cells from colonies of both CFU-E and BFU-E derived from 8.5-day embryos, 13.5-day fetal liver, and adult bone marrow.
Expression of HOX B6 from in vitro erythropoietic progenitor cell colonies (CFU-E after 48 hours and BFU-E after 10 days incubation) and in colonies derived from HPP-CFC (after 14 days of incubation). (A) Ethidium bromide gel for HOX B6. (B) Southern blot for HOX B6. (C) HPRT control. Lane 1, markers; lane 2, erythroid colonies from 8.5-day embryos; lane 3, erythroid burst colonies from 8.5-day embryos; lane 4, erythroid colonies from 13.5-day fetal liver; lane 5, erythroid burst colonies from 13.5-day fetal liver; lane 6, erythroid colonies from adult bone marrow; lane 7, erythroid burst colonies from adult bone marrow; lane 8, colonies from 13.5-day fetal liver HPP-CFC; and lane 9, colonies from adult bone marrow HPP-CFC.
Expression of HOX B6 from in vitro erythropoietic progenitor cell colonies (CFU-E after 48 hours and BFU-E after 10 days incubation) and in colonies derived from HPP-CFC (after 14 days of incubation). (A) Ethidium bromide gel for HOX B6. (B) Southern blot for HOX B6. (C) HPRT control. Lane 1, markers; lane 2, erythroid colonies from 8.5-day embryos; lane 3, erythroid burst colonies from 8.5-day embryos; lane 4, erythroid colonies from 13.5-day fetal liver; lane 5, erythroid burst colonies from 13.5-day fetal liver; lane 6, erythroid colonies from adult bone marrow; lane 7, erythroid burst colonies from adult bone marrow; lane 8, colonies from 13.5-day fetal liver HPP-CFC; and lane 9, colonies from adult bone marrow HPP-CFC.
HOX B6 gene expression was not found in HPP-CFC colonies derived from fetal liver (lane 8) or 5-FU–treated adult bone marrow (lane 9). Figure 4 shows that, although HPRT gene expression was detected in individual colonies of HPP-CFC, no HOX B6 gene expression was present in these macroscopic colonies. Colonies derived from multipotential stem cells (mixed erythroid colonies) were not used, because these have an erythropoietic component.
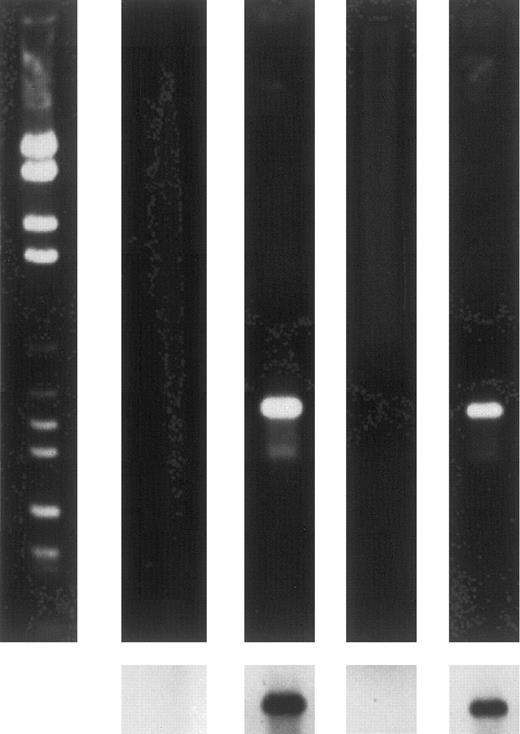
To further ensure that only erythropoietic cells were expressing HOX B6, 13.5-day fetal liver and adult bone marrow cells suspensions were incubated with the erythroid-specific antibody TER-119. Positive and negative cell populations were separated using micro-bead magnetic separation. The resulting TER-119+ population contained greater than 95% erythroid cells, as determined by morphology and benzidine staining. When mRNA from both positive and negative cells populations was subjected to RT-PCR, the results shown in Fig 5 were obtained. These results clearly show that TER-119+ cells from both 13.5-day fetal liver and adult bone marrow express the HOX B6 gene, whereas no expression was detected in the TER-119− cell population. Therefore, it would appear that HOX B6 is expressed only in the erythropoietic lineage. To study this further, the expression of HOX B6 was investigated in two in vitro differentiating systems in which commitment into the erythropoietic lineage can be manipulated.
Ethidium bromide-stained gels (upper panels) and Southern blots (lower panels) showing expression by RT-PCR of HOX B6 in TER-119–separated 13.5-day fetal liver and adult bone marrow cells. From left to right-hand lane: markers; 13.5-day fetal liver TER-119− population; 13.5-day fetal liver TER-119+ population; adult bone marrow TER-119− population; and adult bone marrow TER-119+ population.
Ethidium bromide-stained gels (upper panels) and Southern blots (lower panels) showing expression by RT-PCR of HOX B6 in TER-119–separated 13.5-day fetal liver and adult bone marrow cells. From left to right-hand lane: markers; 13.5-day fetal liver TER-119− population; 13.5-day fetal liver TER-119+ population; adult bone marrow TER-119− population; and adult bone marrow TER-119+ population.
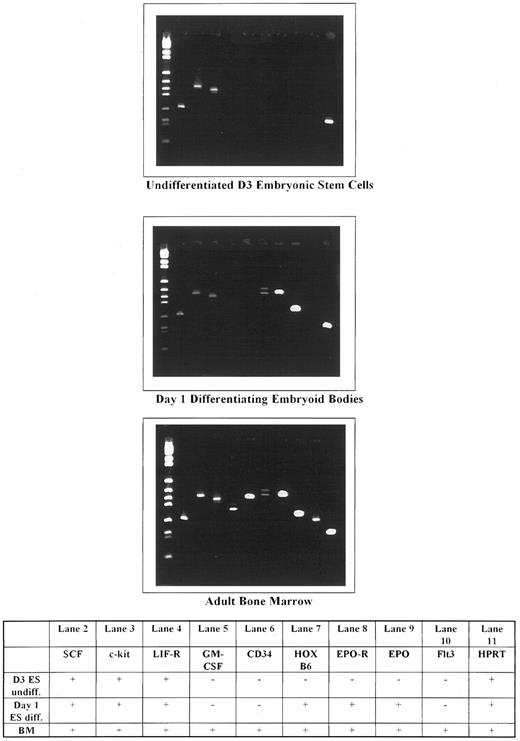
HOX B6 is only expressed in differentiating ES cells.The embryonic stem cell line D3 was used for one of these studies. The cells were collected in their undifferentiated, log phase growing state as well as at various times up to 7 days after LIF was removed from the medium. When LIF is removed from the ES cell culture, the individual cells form an embryoid body in which hematopoiesis can develop.
An array of different genes were examined in undifferentiated and differentiating ES cells. The results are shown in Fig 6. In undifferentiated ES cells, only c-kit, c-kit ligand (SCF ), and the LIF-R were expressed. Neither HOX B6 gene expression nor any mesoderm or erythroid markers were observed. Within 24 hours after removal of LIF, not only could c-kit, SCF, and the LIF-R be identified, but also HOX B6, the EPO-R, and EPO. In addition, the Brachyury (T-gene), GATA-1, and Band 3 were also expressed (results not shown). Interestingly, no CD34 or Flk2/Flt3 could be detected at this time. These results show that HOX B6 is not expressed in a nonhematopoietic stem cell population but is expressed concomitantly with erythroid and mesoderm markers during the spontaneous entry and differentiation into lineage-specific hematopoietic cells.
Gene expression in undifferentiated and day-1 differentiating embryonic, subcloned D3 stem cells, compared with normal adult mouse bone marrow. Molecular weight markers are shown in lane 1.
Gene expression in undifferentiated and day-1 differentiating embryonic, subcloned D3 stem cells, compared with normal adult mouse bone marrow. Molecular weight markers are shown in lane 1.
HOX B6 is only expressed in differentiating primordial germ cells.In addition to ES cells, a primary embryonic, nonhematopoietic stem cell population was also examined for its expression of HOX B6 in both the undifferentiated and differentiated states. We have recently shown that PGC colonies grown in the presence of LIF, SCF, and IL-3 can be induced into the most primitive in vitro hematopoietic stem cell population, the cobblestone area-forming cell (CAFC), multipotential stem cells, and erythropoietic cells.30 Table 3 shows a summary of expression of different genes in undifferentiated and differentiating PGCs.
Gene Expression in Undifferentiated PGC Colonies and Differentiating Colonies Derived From PGCs
| Gene Expression of . | Undifferentiated PGC Colonies . | Differentiating Cells From PGCs . | Adult Bone Marrow . |
|---|---|---|---|
| c-Kit (SCF-R) | ++ | + | + |
| LIF-R | ++ | + | + |
| bFGF-R | ++ | + | + |
| IL-3-R | ++ | + | + |
| EPO-R | − | ++ | ++ |
| GATA-1 | − | ++ | ++ |
| GATA-2 | − | + | + |
| CD 34 | − | ++ | ++ |
| Flk2/Flt3 | − | + | + |
| HOX B6 | − | ++ | + |
| HOX B7 | − | + | + |
| HOX B8 | − | − | − |
| HPRT | + | + | + |
| Gene Expression of . | Undifferentiated PGC Colonies . | Differentiating Cells From PGCs . | Adult Bone Marrow . |
|---|---|---|---|
| c-Kit (SCF-R) | ++ | + | + |
| LIF-R | ++ | + | + |
| bFGF-R | ++ | + | + |
| IL-3-R | ++ | + | + |
| EPO-R | − | ++ | ++ |
| GATA-1 | − | ++ | ++ |
| GATA-2 | − | + | + |
| CD 34 | − | ++ | ++ |
| Flk2/Flt3 | − | + | + |
| HOX B6 | − | ++ | + |
| HOX B7 | − | + | + |
| HOX B8 | − | − | − |
| HPRT | + | + | + |
Undifferentiated PGCs expressed only LIF-R, c-kit, IL-3–R, and basic fibroblast growth factor receptor (bFGF-R). No mesoderm or erythroid markers were detected. In contrast, cells from PGC colonies cultured in methyl cellulose in the presence of EPO and IL-3 not only expressed the above four receptors, but also HOX B6, erythroid-relevant markers and, unlike ES cells, CD34 and Flk2/Flt3. These results show not only that a nonhematopoietic primary ES cell population can be induced into the hematopoietic system and further into erythropoiesis, but also that this cell determination process is accompanied by the expression of hematopoietic stem cell erythropoietic markers. They also show that HOX B6 is not expressed in the undifferentiated state but is associated with lineage-specific differentiation, in this case erythropoiesis.
DISCUSSION
Whole-mount in situ hybridization provided the first indication that a possible link between HOX B6 and EPO gene expression existed, because both genes were observed at the same time in the same areas of the embryo cylinder, namely the extraembryonic mesoderm and the primitive streak region. More detailed studies have now produced results that describe a remarkable correlation both in time and spatial distribution between the expression of HOX B6, EPO, and erythropoiesis during hematopoietic development.
Embryonic hematopoiesis and erythropoiesis in particular can be detected from 6.5 to 10.5 days.30,34,35 Previous studies from this laboratory showed that, in the fetal liver, erythropoiesis, both in vivo and in vitro, increased from 11.5 days to a maximum at 14.5 days, followed thereafter by a decrease.36,37 Fetal splenic erythropoiesis was detected at 17.5 days and reached a maximum 8 days after birth.37 Fetal bone marrow CFU-E could be detected before birth, but numbers rapidly increase 2 days after birth. During erythropoietic ontogeny, EPO has been detected in the macrophages of the fetal liver blood islands.38 The results reported here extend those published previously. They show that EPO is not only expressed as early as 6.5 days, a fact that corroborates previous reports,39 but that it is localized in two areas of the 8.5-day embryo cylinder, namely the visceral yolk sac and the area of the primitive streak. Thus, EPO gene expression has both an extraembryonic and embryonic component. This, in turn, correlates with recent results showing that colonies derived from CFU-E and BFU-E as well as multipotential stem cells can be detected not only in the yolk sac, but also in the embryo proper.30
A graphical summary of the time sequence and pattern of expression of erythropoiesis and EPO and HOX B6 gene expression is shown in Fig 7. At all time points, HOX B6 gene expression precisely follows erythropoietic and EPO ontogeny. Definitive, adult hematopoiesis in both mouse and human is now known to begin in the area of the dorsal aorta.40-42 The detection of transient hematopoietic stem cells in this region occurs 1.5 to 2 days before that in the mouse fetal liver.40,41 However, the presence of erythropoietic progenitor cells and HOX B6 and EPO gene expression in this region was not performed in this study. A critical time point occurs between days 16.5 and 17.5 of gestation. At 17.5 days, with erythropoiesis rapidly decreasing, both EPO and HOX B6 gene expression are undetectable in the fetal liver, but are found in the fetal spleen and kidney. It is at this time point that erythropoiesis is transferred from the fetal liver to the fetal spleen. However, for reasons which are not understood, both EPO and HOX B6 gene expression increase again in the fetal liver at 19.5 days. Erythropoiesis is still present in the fetal liver and continues to be detectable in the early neonatal liver.43 Although the number of hepatocytes in the fetal liver increases rapidly from about 15.5 days onwards, a dilution effect causing EPO and HOX B6 gene expression to become undetectable by RT-PCR is unlikely. In such an event, a decrease in gene expression would be apparent. Although neither Northern blots nor RNase protection assays were performed, the pattern of EPO and HOX B6 gene expression detected by RT-PCR was reproducible. The results imply that, during late gestation, major transitions occur.
Graphical summary of expression of erythropoiesis, and HOX B6 and EPO gene expression from 6.5 to 19.5 days of murine gestation showing how expression is coordinated in all three systems. Note that the decrease in erythropoiesis in the fetal liver is proceeded by a more rapid reduction in HOX B6 and EPO expression. During this time, erythropoiesis is transferred to the fetal spleen and HOX B6 and EPO are transferred to the fetal spleen and fetal kidney.
Graphical summary of expression of erythropoiesis, and HOX B6 and EPO gene expression from 6.5 to 19.5 days of murine gestation showing how expression is coordinated in all three systems. Note that the decrease in erythropoiesis in the fetal liver is proceeded by a more rapid reduction in HOX B6 and EPO expression. During this time, erythropoiesis is transferred to the fetal spleen and HOX B6 and EPO are transferred to the fetal spleen and fetal kidney.
As a transcription factor, it is possible to envisage HOX B6 playing an important role in the determination and/or differentiation of erythropoietic cells in a similar manner to GATA-1 and the SCL transcription factors.44 However, the role played by HOX B6 in EPO gene expression is an open question and it remains to be seen whether this transcription factor plays a pivotal role in the expression and/or modulation of the EPO gene in response to perturbation.
That HOX B6 is specifically involved in the regulation of erythropoiesis was also implied by a series of three different experiments. First, HOX B6 was only detectable in TER-119+ separated erythroid cells from fetal liver and adult bone marrow. Second, in undifferentiated ES cells, no indication of HOX B6 or erythroid-relevant molecules was observed. This is in contrast to a previous report by Heberlein et al,45 who showed that, in several ES and embryonic carcinoma cell lines, GATA-1 and the EPO-R were expressed when maintained in the undifferentiated state by the addition of 40% Buffalo rat liver (BRL)-conditioned medium as a source of LIF. Whether this discrepancy is due to other factors present in the BRL-conditioned medium or is a function of the ES cell lines is unknown. However, within 24 hours after the removal of LIF, HOX B6, the EPO-R, and EPO itself were all expressed. Yet another difference to previous reports was observed in the absence of expression of the mouse equivalent of the CD34 gene and Flk2/Flt3. Both McClanahan et al46 and Krause et al47 found CD34 expressed in ES cells. McClanahan et al46 detected CD34 and Flt2/Flk3 expression in both undifferentiated ES cells and differentiating embryoid bodies, but not in blastocysts. Whether our primers for CD34 and Flk2/Flt3 detect different transcripts in ES cells to those of other investigators is not known, although both CD34 and Flk2/Flt3 could be detected in normal adult bone marrow. Nevertheless, this does not distract from the observation that HOX B6 and erythroid-relevant genes are expressed simultaneously in this model system.
Similar results were also obtained in the third series of experiments using a primary, embryonic, nonhematopoietic stem cell population, ie, the PGCs. We have shown that this population can be expanded in vitro and induced into the hematopoietic system producing both stem and progenitor cell populations.30 From these and more recent studies showing that PGCs can reconstitute lethally irradiated recipients (to be published elsewhere), it was postulated that PGCs could be hematopoietic-initiating cells for both primitive, embryonic, and definitive, adult hematopoiesis.30,48 Because PGCs can be maintained in culture,30 this system also provided a model for studying HOX B6 gene expression. Thus, similar to the ES cells, no HOX B6 gene expression was observed in the undifferentiated PGC-cultured population. In contrast, differentiating PGCs induced into the hematopoietic system expressed an array of different genes. Not only were HOX B6 and B7, EPO-R, and GATA-1 and -2 expressed, but CD34 and Flk2/Flt3 could also be detected. Therefore, using three different model systems, HOX B6 gene expression was shown to be associated specifically with erythropoietic differentiation.
The function of the homeobox gene products is largely unknown. However, their role in the normal development of the animal is unequivacable. In the adult, many HOX genes are ubiquitously expressed, but recently it has become clear that HOX genes appear to play a role in hematopoietic regulation.18 Thus, besides the general observations concerning HOX genes mentioned in the introduction, it is now known that specific mammalian HOX genes are expressed in hematopoietic stem cell populations, whereas others are expressed in progenitor and mature hematopoietic cells.18 Furthermore, different HOX clusters appear to be predominant in specific hematopoietic cell lineages.19-24 However, a recent report has, for the first time, linked the role of HOX genes directly with gene disruption associated with acute leukemias. Thus, when the mixed-lineage leukemia (mll ) gene is knocked out, HOX expression is abolished, leading to embryo lethality, whereas heterozygotes demonstrate several abnormalities, including anemia and B-cell defects.49 Although a knock-out mutant for HOX B6 has been reported showing major skeletal deformations, a study of hematopoiesis in these animals was not undertaken.50
In conclusion, we have shown that HOX B6 expression is intimately associated with erythropoiesis during development. It remains to be seen whether a role for HOX B6 in the ontogeny of EPO production is present and what the function for this transcription factor plays in erythropoiesis. Nevertheless, the fact that HOX B6 is not expressed in stem cells but is expressed in early and late erythropoietic progenitor cells regardless of their source, implicates HOX B6 as a factor that is expressed during erythropoietic differentiation.
ACKNOWLEDGMENT
Many thanks to I. Brackmann for her excellent technical assistance.
Supported by the Deutsche Forschungsgemeinschaft SFB 322, Project A9, and the German Red Cross Blood Bank, Baden Württemburg.
Address reprint requests to Ivan N. Rich, PhD, Professor of Medicine, Director, Stem Cell Research Program, Division of Transplantation Medicine, Center for Cancer Treatment and Research, 7 Richland Medical Park, Columbia, SC 29203.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal