Abstract
Although the liver is the primary site of fibrinogen (FBG) synthesis, epithelial cells from diverse tissues have been shown to express one or more of the FBG Aα, Bβ, and γ chain genes. In contrast, marrow megakaryocytes, which store FBG in the α-granules, are thought not to express the FBG genes. Our earlier studies have shown that epithelial cells in a variety of extrahepatic tissues express the γ chain gene ubiquitously, while the mRNAs for the Aα and Bβ chain genes are essentially undetectable. During systemic inflammation, the liver secretes increased levels of FBG into the circulation, and lung epithelium responds to local inflammation during pulmonary infection by increased transcription of the γ-FBG gene. Therefore, to determine whether extrahepatic epithelial cells express the Aα, Bβ, and γ chain genes in response to proinflammatory mediators, cultured lung epithelial cells were treated with interleukin-6 (IL-6) and dexamethasone (DEX). Northern blot analysis demonstrated that the levels of γ-FBG mRNA in cultured lung (A549) and liver (HepG2) epithelial cells increased 2- to 10-fold in response to treatment. Reverse-transcriptase-polymerase chain reaction amplification demonstrated increased accumulation of steady state levels of FBG Aα, Bβ, and γ chain mRNAs in lung epithelial cells after treatment. The basal level of lung cell γ-FBG gene transcription was not accompanied by appreciable levels of Aα and Bβ chain gene transcription; however, nuclear run-on analysis suggested that the increase in lung cell FBG mRNAs in response to DEX ± IL-6 was due to new transcription. Furthermore, stimulation of lung epithelial cells with IL-6 + DEX resulted in maximal secretion of intact FBG (340 kD) composed of the characteristic Aα, Bβ, and γ chain polypeptides. The data suggest that basal expression of the γ-FBG gene in extrahepatic tissue occurs ubiquitously in the absence of detectable levels of Aα- and Bβ-FBG gene expression, which are then upregulated on induction of an inflammatory response. This would result in local synthesis and secretion of FBG in the injured tissue, supporting the hypothesis that production of FBG by extrahepatic epithelial cells in response to inflammation plays a role in wound repair.
FIBRINOGEN (FBG) functions in primary hemostasis in support of platelet aggregation, in secondary hemostasis in fibrin clot formation at the site of vessel injury, and also functions in the maintenance of homeostasis, as it is one of several hepatic proteins whose plasma levels are upregulated during a systemic inflammatory response.1-4 Historically, the hepatocyte and megakaryocyte (MK) have been considered the sites of Aα, Bβ, and γ chain gene expression and biosynthesis of intact FBG.5-7 However, recent reports suggest that the origin of MK and platelet FBG is by αIIbβ3-mediated endocytosis of plasma FBG and subsequent storage in α-granules.8-12 Endocytosis of plasma FBG into MK α-granules requires the dimeric γA configuration,11 which is consistent with data showing that FBG-mediated platelet aggregation is largely dependent on two intact platelet recognition domains defined by γA chain residues 400-411.13 Yet, rat MKs fixed immediately ex vivo express only γ chain mRNA and only in the most immature MKs,14 suggesting that MK expression of the γ chain gene is developmentally regulated. Furthermore, the extrahepatic expression of FBG mRNAs or polypeptides has been reported. On estrogen stimulation of ovarian granulosa cells, Bβ and γ chain polypeptides are secreted.15 Baumheuter et al,16 demonstrated the expression of Aα, but not Bβ or γ chain mRNA in rat kidney. Comparative analysis of the expression of Aα, Bβ, and γ chain mRNAs in extrahepatic tissues of normal rats revealed that the Aα and Bβ chain mRNAs are not detected in any tissue other than liver.17 In contrast, MKs from patients with high-grade T-cell lymphoma express Aα-FBG mRNA.18 Fibrinogen synthesis occurs in human cervical carcinoma cells, but not primary cervical epithelial cells cultured under basal conditions.19 Moreover, epithelial cells from human intestine respond to interleukin-6 (IL-6) induction by a modest increase in synthesis and secretion of FBG.20
To elucidate the molecular mechanisms involved in tissue-specific regulation of the γ-FBG gene, our previous studies investigated the tissue distribution of γ chain gene expression, the pattern of γ chain pre-mRNA processing,14,17,21,22 and the γ chain polypeptide isoforms produced.23,24 The data indicate that the expression of rat γ-FBG mRNA is tissue-specific (γB) and ubiquitous (γA). The predominant form of the mRNA (γA) is expressed in restricted cell types in many tissues, while the minor, alternatively processed form of the mRNA (γB, last intron retained as an exon) is expressed solely in liver, suggesting that the expression of γA mRNA is constitutive, while the expression of γB mRNA is regulated in liver.22 The expression of the γ chain gene, but not the Aα and Bβ chain genes, in extrahepatic tissues is likely due to activation of the γ-FBG gene by ubiquitous transcription factors.25-28 In contrast, FBG Aα and Bβ chain genes are activated by HNF-1 which, along with other liver transcription factors, contributes to their liver-specific expression.16,29-31 The 5′ flanking regions of all three FBG genes contain IL-6 response elements; IL-6 upregulates expression of hepatic FBG.28,31-34 Furthermore, glucocorticoids act synergistically with IL-6 in upregulation of FBG gene expression.31,32,35 36 These observations indicate that the ubiquitous expression of the γ-FBG gene is not coordinated with expression of the Aα and Bβ chain genes under basal conditions in extrahepatic tissue. Therefore, this study was undertaken to determine whether cultured lung epithelial cells synthesize and secrete intact FBG in response to proinflammatory mediators.
MATERIALS AND METHODS
Cell cultures.Human epithelial cell lines A549, lung adenocarcinoma CCL 185; 786-O, kidney adenocarcinoma CRL 1932; LS123, colon adenocarcinoma CCL 225; 407, embryonic intestine CCL 6; and HepG2, hepatocarcinoma HB 8065, were obtained from the American Type Culture Collection (Rockville, MD) and cultured as recommended. When cells reached confluence, they were treated with 0.1 μmol/L dexamethasone (DEX) (American Reagent Laboratories, Inc, Shirley, NY), 25 U/mL recombinant human IL-6 (Life Technologies) or both for 16 to 18 hours.
RNA isolation, Northern blot analysis, and reverse transcriptase-polymerase chain reaction (RT-PCR).Total RNA and poly (A)+ mRNA isolation, RNA gel electrophoresis and Northern blot analysis were performed as previously described.37 Human FBG γ chain cDNA (pHIγ2), a generous gift from Dr D. W. Chung (University of Washington, Seattle, WA), was labeled to a specific activity of 1.2 to 1.8 × 109 cpm/μg using [α32P]dCTP. For RT-PCR, first strand cDNA synthesis was primed by oligo (dT) using 10 μg of total RNA isolated from control or treated HepG2 or A549 cells. One-tenth of the mRNA:cDNA heteroduplex was used in the PCR performed with the Gene-Amp kit (Perkin-Elmer, Norwalk, CT) and a Cetus-Perkin Elmer PCR machine as described.38 The FBG sense (S) and antisense (AS) primer pairs map to human cDNAs at nucleotide (nt) positions as follows: Aα-S = nt 1426–1457 with Aα-AS = nt 1874–1848; Bβ-S = nt 1021–1049 with Bβ-AS = nt 1320–1291; and γ-S = nt 1022–1050 with γ-AS = nt 1202–1174. A β-actin primer pair derived from rat, which amplifies cDNA from many species,38 was used as a positive control, and primers with no template were used as negative controls.
Nuclear run-on transcription.Analysis of nascent transcripts from A549 and HepG2 cells was performed essentially as described39,40 with modifications. After treatment, cells were lifted by trypsin-EDTA, washed twice in ice-cold phosphate-buffered saline, and lysed in the presence of 5 mmol/L benzamidine and 1 mmol/L PMSF in the lysis buffer.39,40 Run-on transcription was performed using 250 to 330 μCi of [32P]CTP (3,000 Ci/mmol, 10 μCi/μL; DuPont NEN, Boston, MA) for 30 minutes at 30°C with gentle shaking. Elongated transcripts were isolated by DNase and Proteinase K treatments and phenol/chloroform/isoamyl alcohol extraction.39,40 32P-RNA was further purified by two successive alcohol precipitations, dissolved in sterile water, and denatured at 90°C. Probes were diluted to 1.5 × 106 cpm/mL in Northern hybridization solution.37 The inserts from the FBG Aα, Bβ, and γ chain cDNAs (kind gifts from Dr D. W. Chung, University of Washington, Seattle) and human γ-actin (from Dr L. Kedes, Stanford University, Palo Alto, CA) were subcloned into either pGEM3Z or pGEM7Z (Promega, Madison, WI). The unrestricted target DNA was NaOH-heat denatured, neutralized, and immobilized (5 μg per slot) on ZetaProbe membranes (BioRad). The blots were hybridized at 65°C for 72 hours. Relative rates of transcription were assessed by scanning densitometry of the autoradiographs and changes in transcription of nascent RNA were normalized to γ-actin.
Indirect immunofluorescent staining.Cells were grown on glass coverslips then fixed in formalin after treatment. Cells were permeabilized with Triton X-100 before staining23 with rabbit antihuman FBG IgG (Dako, Carpinteria, CA), which was affinity purified as described.41 Purified antihuman FBG IgG or normal rabbit IgG (Dako) was used at a final concentration of 80 μg/mL. Rhodamine-conjugated goat antirabbit IgG (Jackson Scientific, West Grove, PA) was used at a 1/15 dilution. The specificity of anti-FBG staining was confirmed by preincubating the diluted IgG with a fivefold excess of purified human FBG at 4°C overnight, followed by 1 hour of incubation at 22°C then centrifugation at 12,800g for 15 minutes; the supernatant was used for immunostaining. Rhodamine-conjugated wheat germ agglutinin was obtained from Sigma (St Louis, MO).
Metabolic labeling of cultured cells and immunoprecipitation of FBG.Treated and untreated HepG2, A549, 786-O, LS123, and 407 cells were continuously labeled for 16 to 18 hours, concomitantly with DEX and/or IL-6 treatment, with 40 μCi/mL 35S-Met + 35S-Cys (Express Labeling; DuPont NEN). Intracellular and secreted FBG were each immunoprecipitated using antihuman FBG antiserum coupled to Protein A-Sepharose.42 Immunoprecipitated proteins were electrophoresed reduced by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)13 or nonreduced on 2% agarose gels.42 Densitometry was performed on reduced and denatured FBG polypeptides immunoprecipitated from A549 cell culture media after treatment with DEX, IL-6, or both and compared with untreated control cells. The fold-increase in FBG expression was calculated from the total area under the peaks for Aα, Bβ and γ chain polypeptides and normalized to the area for the control.
RESULTS
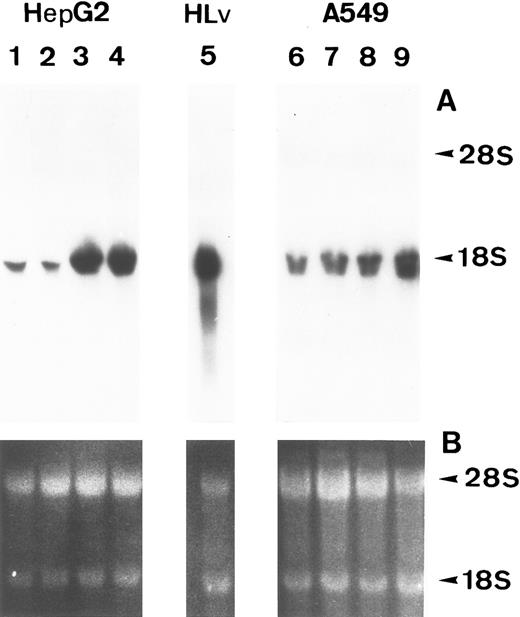
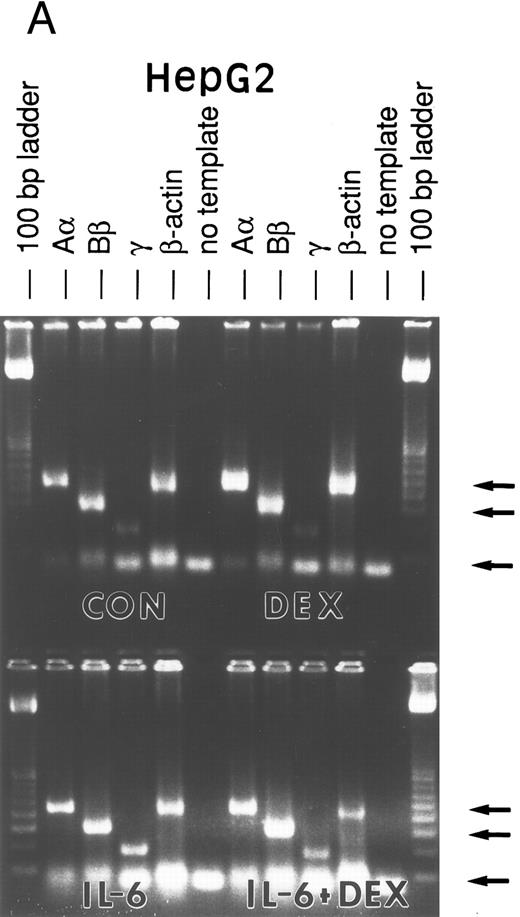
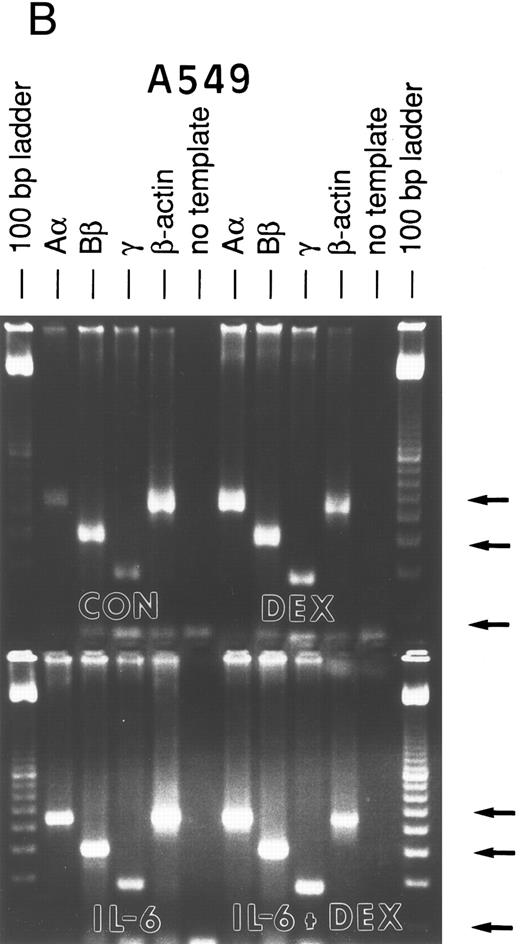
Lung epithelial cells express FBG mRNAs after induction with IL-6 ± DEX.The potential of the human lung epithelial-derived cell line, A549, to express the FBG genes before and after induction with DEX and IL-6 was studied. Human hepatocarcinoma cells, HepG2, were used as the positive control cell line. Northern blot analysis of total mRNA indicated that IL-6 was a potent mediator of γ-FBG gene upregulation in HepG2 cells (Fig 1, lanes 3 and 4). HepG2 cells responded to induction of the γ chain mRNA by IL-6 at levels expected from previous studies. In A549 cells, DEX appeared to induce γ chain mRNA only slightly over control, IL-6 induction was approximately threefold and IL-6 + DEX resulted in induction of γ chain mRNA greater than fourfold over control (Fig 1, lanes 7-9). RT-PCR analysis of A549 and HepG2 total RNA using primers derived from FBG Aα, Bβ, and γ chains resulted in amplification of a cDNA of the expected size for each mRNA: Aα-FBG (449 bp), Bβ-FBG (300 bp), γ-FBG (181 bp), and the expected size for β-actin (443 bp) (Fig 2). Increased amounts of FBG-specific compared to β-actin PCR products were obtained for DEX ± IL-6 treated cells, suggesting induction of FBG gene transcription by proinflammatory mediators. Moreover, results of RT-PCR for the γ chain corroborated the results obtained by Northern blotting (Figs 1 and 2). Lung A549 cells were stimulated by DEX alone, whereas HepG2 cells treated with DEX alone did not show a detectable increase in the amount of γ chain mRNA. The identity of Aα, Bβ, and γ chain mRNAs induced by IL-6 in the A549 cells was confirmed by nucleotide sequencing of the cDNA prepared by RT-PCR (not shown).
Induction of FBG γ chain mRNA expression in HepG2 and A549 cells. HepG2 (lanes 1-4) and A549 (lanes 6-9) cells were treated as follows: lanes 1 and 6, no treatment; lanes 2 and 7, 0.1 μmol/L DEX; lanes 3 and 8, 25 U/mL IL-6; lanes 4 and 9, 0.1 μmol/L DEX + 25 U/mL IL-6. (A) Northern blot of 30 μg total RNA from HepG2 or A549 cells, or 5 μg human liver poly(A)+ mRNA (lane 5) probed with 32P-labeled human FBG γ chain cDNA. (B) Duplicate gel of RNA samples stained with acridine orange to confirm the integrity and amount of each RNA sample loaded. Arrowheads mark the 28S and 18S rRNAs.
Induction of FBG γ chain mRNA expression in HepG2 and A549 cells. HepG2 (lanes 1-4) and A549 (lanes 6-9) cells were treated as follows: lanes 1 and 6, no treatment; lanes 2 and 7, 0.1 μmol/L DEX; lanes 3 and 8, 25 U/mL IL-6; lanes 4 and 9, 0.1 μmol/L DEX + 25 U/mL IL-6. (A) Northern blot of 30 μg total RNA from HepG2 or A549 cells, or 5 μg human liver poly(A)+ mRNA (lane 5) probed with 32P-labeled human FBG γ chain cDNA. (B) Duplicate gel of RNA samples stained with acridine orange to confirm the integrity and amount of each RNA sample loaded. Arrowheads mark the 28S and 18S rRNAs.
RT-PCR amplification of FBG Aα, Bβ, and γ chain mRNAs. HepG2 cells (A) and A549 cells (B) were untreated (CON), or treated with DEX, IL-6, or DEX + IL-6. A control PCR reaction with no template was included. Total mRNA was amplified with FBG Aα, Bβ, and γ chain or β-actin primer pairs. The reaction products were resolved on 1.8% agarose gels in TAE buffer. The 100-, 300-, and 500-bp bands of the 100-bp ladder are denoted from top to bottom (arrows).
RT-PCR amplification of FBG Aα, Bβ, and γ chain mRNAs. HepG2 cells (A) and A549 cells (B) were untreated (CON), or treated with DEX, IL-6, or DEX + IL-6. A control PCR reaction with no template was included. Total mRNA was amplified with FBG Aα, Bβ, and γ chain or β-actin primer pairs. The reaction products were resolved on 1.8% agarose gels in TAE buffer. The 100-, 300-, and 500-bp bands of the 100-bp ladder are denoted from top to bottom (arrows).
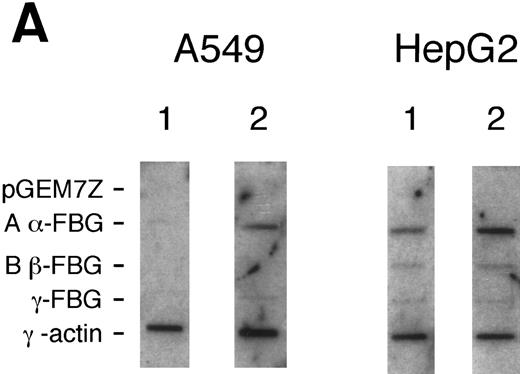
Effects of DEX and IL-6 on transcription of FBG Aα, Bβ, and γ chain genes.To determine whether FBG gene transcripts accumulate in lung epithelial cells because of transcriptional activation, nuclear run-on experiments were performed on cells treated with proinflammatory mediators (Fig 3). In A549 cells, newly synthesized transcripts were detected for FBG Aα, Bβ, and γ chain genes after induction for 16 to 18 hours with DEX + IL-6, whereas levels of new FBG transcripts in controls were barely detectable. In contrast, control HepG2 cells continuously synthesized new transcripts for FBG Aα, Bβ, and γ chain genes. Although the amplitudes of the FBG mRNA signals were very low for both HepG2 and A549 cells in comparison with the γ-actin mRNA signals, changes in the amounts of newly transcribed FBG mRNAs were detectable by densitometric scanning. On induction with DEX + IL-6, both A549 and HepG2 cells were fourfold to fivefold more transcriptionally active (Fig 3). Even though upregulation of the Aα chain appears most dramatic in both the HepG2 and A549 cells, the relative rate of new transcription was similar for each of the FBG transcripts. Results of nuclear run-on experiments with cells treated with DEX or IL-6 (not shown) were intermediate to results obtained with the combined treatment and paralleled results shown by Northern and RT-PCR analyses (Figs 1 and 2). Transcriptional activation resulted in approximately a fourfold to fivefold increase in FBG Aα, Bβ, and γ chain mRNAs. Taken together, the results indicate that cultured lung epithelial cells respond to DEX + IL-6 in a manner similar to liver epithelial cells. The increase in transcription after DEX + IL-6 treatment was similar to the increased levels of steady state mRNAs (Fig 2), indicating that transcription initiation accounts for the inducible expression of FBG mRNA in lung and liver epithelial cells.
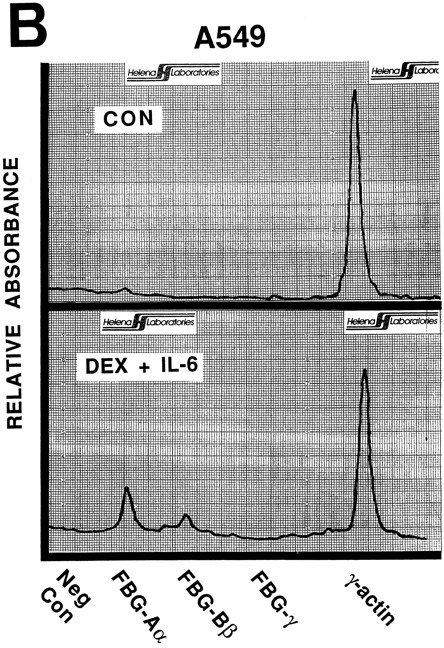
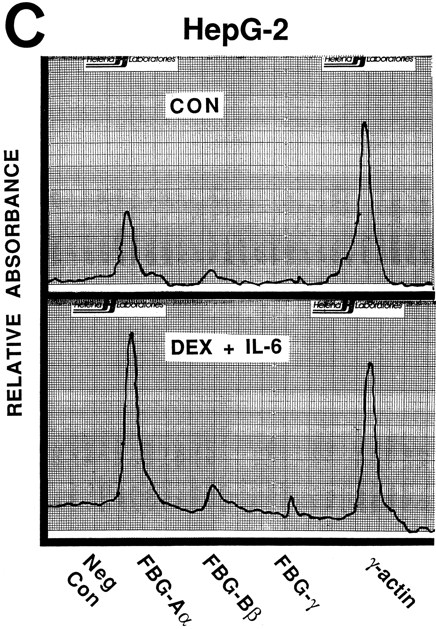
Nuclear run-on transcription of A549 cells compared with HepG2 cells after induction with proinflammatory mediators. Newly synthesized transcripts were labeled with 32P-CTP, the RNA isolated and used to probe denatured target DNA corresponding to FBG Aα, Bβ, and γ chain cDNAs. The autoradiograph of the slot-blots (A) were subjected to densitometric scanning and the tracings of control (CON) and treated (DEX + IL-6) A549 (B) and HepG2 (C) cells are shown. Plasmid pGEM3Z DNA served as negative control (Neg Con), and hybridization to human γ-actin cDNA was used to normalize the data between samples.
Nuclear run-on transcription of A549 cells compared with HepG2 cells after induction with proinflammatory mediators. Newly synthesized transcripts were labeled with 32P-CTP, the RNA isolated and used to probe denatured target DNA corresponding to FBG Aα, Bβ, and γ chain cDNAs. The autoradiograph of the slot-blots (A) were subjected to densitometric scanning and the tracings of control (CON) and treated (DEX + IL-6) A549 (B) and HepG2 (C) cells are shown. Plasmid pGEM3Z DNA served as negative control (Neg Con), and hybridization to human γ-actin cDNA was used to normalize the data between samples.
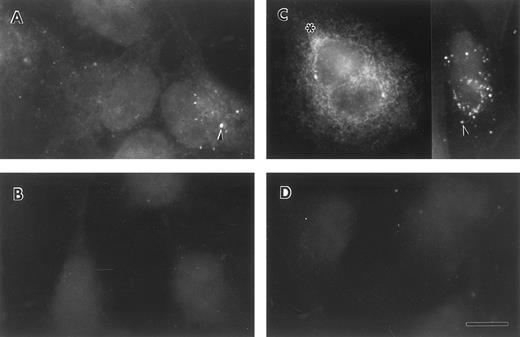
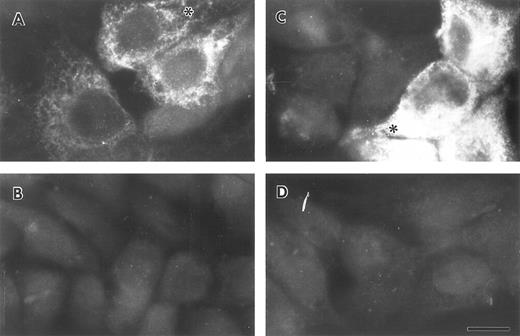
Lung epithelial cells synthesize and secrete FBG.To determine whether lung epithelial cells synthesize immunoreactive FBG, control and treated A549 cells were stained with antihuman FBG IgG and visualized by indirect immunofluorescence. Control A549 cells demonstrated limited positive staining (Fig 4A); however, after treatment with DEX + IL-6, the degree of positive staining increased significantly and was localized to the cytoplasm in a network pattern suggestive of endoplasmic reticulum, and in discrete patches suggestive of Golgi apparatus (Fig 4C).43 Staining of both cell types with rhodamine-conjugated wheat germ agglutinin (not shown), a marker of terminal N-glycosylation occurring in the Golgi,43 was noted in similar discrete patches, consistent with transport and processing of a secreted glycoprotein such as FBG. HepG2 cells produced immunoreactive FBG under basal conditions (Fig 5A) that increased after DEX + IL-6 treatment (Fig 5C). The FBG-specific staining was inhibited completely by preincubation of the anti-FBG IgG with excess purified FBG (Figs 4 and 5, B and D). Staining of all treatment groups with normal rabbit IgG was negative (not shown). These results demonstrate that FBG synthesis is induced in cultured lung epithelial cells after treatment with IL-6 + DEX.
Indirect immunofluorescent staining of control and DEX + IL-6 stimulated A549 cells. Untreated (A and B) and treated (C and D) lung epithelial cells were stained with purified anti-FBG IgG (A and C) or anti-FBG IgG pre-adsorbed with excess purified FBG (B and D). Asterisks denote positive staining in a spidery network pattern and arrowheads denote positive staining in discrete patches.43 The bar = 10 μm.
Indirect immunofluorescent staining of control and DEX + IL-6 stimulated A549 cells. Untreated (A and B) and treated (C and D) lung epithelial cells were stained with purified anti-FBG IgG (A and C) or anti-FBG IgG pre-adsorbed with excess purified FBG (B and D). Asterisks denote positive staining in a spidery network pattern and arrowheads denote positive staining in discrete patches.43 The bar = 10 μm.
Indirect immunofluorescent staining of control and DEX + IL-6 stimulated HepG2 cells. Untreated (A and B) and treated (C and D) liver epithelial cells were stained with purified anti-FBG IgG (A and C) or anti-FBG IgG pre-adsorbed with excess purified FBG (B and D). Asterisks denote positive staining in a spidery network pattern of positive staining.43 The bar = 10 μm.
Indirect immunofluorescent staining of control and DEX + IL-6 stimulated HepG2 cells. Untreated (A and B) and treated (C and D) liver epithelial cells were stained with purified anti-FBG IgG (A and C) or anti-FBG IgG pre-adsorbed with excess purified FBG (B and D). Asterisks denote positive staining in a spidery network pattern of positive staining.43 The bar = 10 μm.
To determine whether lung epithelial cells secrete FBG after stimulation with DEX, IL-6, or both, metabolic labeling of A549 cells and immunoprecipitation of FBG was performed. Very little FBG was secreted constitutively by A549 cells; however, after stimulation with proinflammatory mediators, FBG synthesis and secretion increased dramatically (Fig 6A, media). Similar results were obtained from 12 separate experiments. In all cases, DEX or IL-6 treatment resulted in increased synthesis, secretion, and accumulation of intact FBG in the medium compared with the untreated control, and the combination of DEX + IL-6 resulted in the highest level of intact FBG expression. The secreted FBG was of the expected molecular weight of 340 kD (Fig 6B). As expected for a rapidly secreted protein,44-46 the intracellular levels of metabolically labeled FBG did not accumulate (Fig 6A, cells). Using normal rabbit IgG in place of anti-FBG IgG for immunoprecipitation resulted in only fibronectin binding nonspecifically (not shown). Comparison of densitometric tracings of the amounts of FBG polypeptides in treated or control lanes indicated from 2.5- to 10-fold increases in secretion of intact FBG after treatment (not shown), which is consistent with increased hepatic production of FBG during systemic inflammation. To determine whether other carcinoma or epithelial-derived cell lines also make FBG either constitutively or inducibly, intestinal LS123 and 407, and kidney 786-O cell lines were treated with DEX + IL-6 for 16 to 18 hours and the conditioned media subjected to immunopurification by anti-FBG IgG-Protein A-Sepharose. In comparison with A549 lung epithelial cells, the intestinal adenocarcinoma LS123 made approximately 50-fold less FBG, although DEX + IL-6 treatment appeared to induce FBG expression several-fold in the LS123 cells. In contrast, the kidney adenocarcinoma cell line 786-O did not make FBG in detectable levels, nor did the embryonic intestinal epithelial cell line (407) either with or without DEX + IL-6 treatment (not shown).
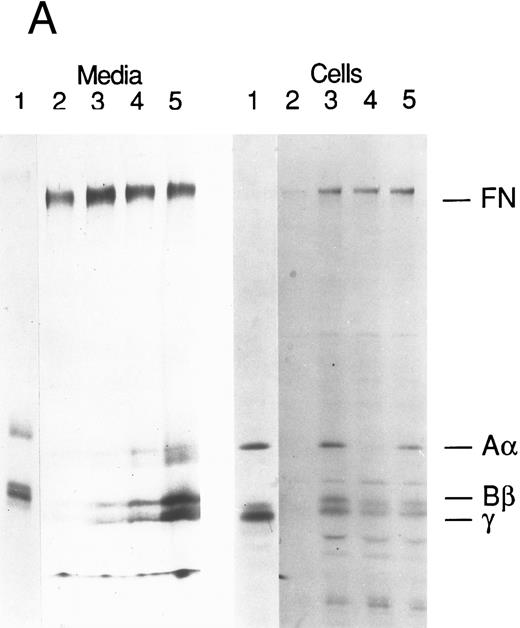
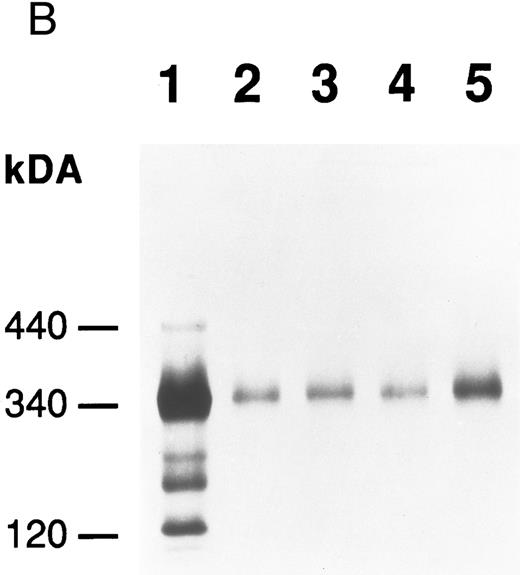
Production of FBG by lung epithelial cells stimulated with pro-inflammatory mediators. Immunoprecipitation of secreted (media) and intracellular (cells) FBG was performed using purified rabbit antihuman FBG IgG coupled to Protein A-Sepharose. (A) A fluorograph of immunoprecipitated proteins electrophoresed on a 5% to 15% polyacrylamide gel after reduction and denaturation. Untreated HepG2 cells served as the positive control cell line for FBG biosynthesis. In lane 1, untreated HepG2 cells; lane 2, untreated A549 cells; lane 3, DEX-treated A549 cells; lane 4, IL-6–treated A549 cells; lane 5, DEX + IL-6–treated A549 cells. The position of migration of the FBG Aα, Bβ, and γ chain polypeptides is denoted. Fibronectin (FN) bound nonspecifically to anti-FBG IgG-Protein A-Sepharose. (B) A 2% agarose gel electrophoresis of nonreduced FBG immunoprecipitated from media of metabolically labeled A549 cells treated as described for (A), lanes 2-5; lane 1, nonreduced FBG immunoprecipitated from untreated, metabolically labeled HepG2 cell conditioned media. Molecular weight markers are denoted in the left margin.
Production of FBG by lung epithelial cells stimulated with pro-inflammatory mediators. Immunoprecipitation of secreted (media) and intracellular (cells) FBG was performed using purified rabbit antihuman FBG IgG coupled to Protein A-Sepharose. (A) A fluorograph of immunoprecipitated proteins electrophoresed on a 5% to 15% polyacrylamide gel after reduction and denaturation. Untreated HepG2 cells served as the positive control cell line for FBG biosynthesis. In lane 1, untreated HepG2 cells; lane 2, untreated A549 cells; lane 3, DEX-treated A549 cells; lane 4, IL-6–treated A549 cells; lane 5, DEX + IL-6–treated A549 cells. The position of migration of the FBG Aα, Bβ, and γ chain polypeptides is denoted. Fibronectin (FN) bound nonspecifically to anti-FBG IgG-Protein A-Sepharose. (B) A 2% agarose gel electrophoresis of nonreduced FBG immunoprecipitated from media of metabolically labeled A549 cells treated as described for (A), lanes 2-5; lane 1, nonreduced FBG immunoprecipitated from untreated, metabolically labeled HepG2 cell conditioned media. Molecular weight markers are denoted in the left margin.
DISCUSSION
The acute phase response to inflammation is conserved across species, with FBG being universally upregulated 2- to 10-fold in rat, mouse, human, rabbit, and ferret. Increased levels of circulating FBG and its subsequent conversion to fibrin serve to restore homeostasis by providing a provisional matrix to promote wound healing and tissue remodeling.47 The data reported in this work demonstrate the inducible expression of FBG Aα, Bβ, and γ chain mRNAs and secretion of intact FBG polypeptides in response to proinflammatory mediators by lung epithelial cells, and provide evidence that extrahepatic epithelial cells contribute to changes in acute phase proteins during an inflammatory response. Furthermore, the results of the nuclear run-on analysis indicate that the upregulation of FBG gene expression in lung epithelial cells is due to transcription initiation, suggesting coordinated expression of FBG in response to inflammation in extrahepatic tissue. Presently, the question of whether or not lung epithelium expresses the FBG genes in vivo in response to inflammation remains, as yet, unanswered, in particular since the in vitro expression of FBG occurs in carcinoma epithelial cell lines [intestinal Caco-220; cervical ME-18019; lung A549 (this study); and, liver HepG244,45]. Analysis of other adenocarcinoma cell lines from kidney or colon epithelium indicated that the transformed phenotype does not always result in detectable levels of FBG synthesis (this study). In addition, our previous in situ and Northern hybridization analyses14,17 22 demonstrated abundant, steady state levels of γ-FBG mRNA in epithelial cells of lung and brain and in marrow MKs. Together, these data indicate that the transformed epithelial cell phenotype is not required to produce FBG in vitro, and that extrahepatic γ-FBG gene expression occurs in vivo in the absence of cellular transformation.
Moreover, additional studies in the laboratory provide evidence to suggest that, indeed, lung epithelium upregulates γ-FBG gene expression in response to proinflammatory mediators produced in lung tissue.48,49 We use an animal model of Pneumocystis carinii pneumonia as an in vivo model of lung inflammation. Recently, we cloned and characterized a lung-specific cDNA encoding γ-FBG identical in sequence to hepatic-derived γ-FBG, demonstrating conclusively that γ-FBG mRNA is expressed in bronchial and bronchiolar epithelial cells in vivo.48 Northern blotting indicated that the steady-state levels of γ chain mRNA are elevated fivefold in the P carinii infected lung compared with uninfected lung.49 Furthermore, the induction of a systemic acute phase response during P carinii pneumonia occurs as demonstrated by upregulation of γ chain gene expression in liver and a corresponding increase in the amount of circulating plasma FBG.49 Thus, in an animal model of lung inflammation, lung epithelium responds by upregulating γ-FBG gene expression. These observations are consistent with prior reports demonstrating the induction of an acute phase response50 and elevated IL-6 production in lung and serum during P carinii pneumonia.51 However, we were unable to prove that lung epithelial cells synthesize FBG polypeptides in vivo due to the inherent difficulty in distinguishing newly synthesized FBG from plasma FBG deposited in the interstitium. Fibrin (ogen) deposition has been demonstrated in the interstitium of normal tissue biopsies,52,53 normal rat lungs (P.J.S.-H., unpublished observation, June 1992), and severely injured lung.54,55 Although the abundant levels of γ, but not Aα and Bβ, chain mRNA in extrahepatic tissues suggest constitutive expression of the γ chain gene, the likelihood of synthesis and secretion of only γ chain polypeptides from lung cells is remote, as hepatic FBG is secreted only after assembly of all six chains into the dimeric, disulfide-bonded whole molecule.44-46 We are currently performing immunoelectron microscopy studies to determine whether FBG can be found in the secretory pathway of inflamed lung epithelium in vivo.
The mechanisms controlling the coordinated upregulation of the FBG genes during a systemic (hepatic) inflammatory response are not fully understood, although recent studies have shown that FBG gene expression in response to IL-6 induction involves several different transcription factors.26-34 Intensive effort has been applied to studying the upstream regions of the FBG Bβ chain gene for sequences that are responsible for regulation of FBG expression. The reasons for this are twofold: (1) translation of the nascent Bβ chain of FBG is the rate limiting step in human FBG biosynthesis and assembly,44-46 and (2) the organization of the FBG genes is unusual for a multisubunit protein (Fig 7). The γ chain gene is transcribed toward the Aα chain gene, while the Bβ chain gene is transcribed from the bottom strand toward the Aα chain gene.56,57 These studies indicate that IL-6 and glucocorticoid response elements are important; however, the direct contribution of Bβ chain 5′ flanking regions to coordinating Aα and γ chain gene expression has not been elucidated. The results of this study, together with the evidence from our earlier work,14,17,21,22 suggest that the low, basal level of γ chain mRNA expression is driven by its ubiquitously activated promoter in epithelial cells of diverse tissues. Under basal conditions, the transcription factor HNF-1 exhibits a limited tissue distribution with high levels of HNF-1 mRNA in liver and kidney, but comparatively little in lung.58 However, after induction by IL-6 and glucocorticoids, not only is the γ chain gene upregulated in lung epithelial cells, but the Aα and Bβ chain genes are transcriptionally activated as well. This result is consistent with the data of Roy et al,46 who showed that the overexpression of any FBG chain specifically elevates the expression of the other two chains. Together, the data suggest that cis-acting regulatory regions of the γ chain may function in conjunction with cell-type specific trans-acting factors that are activated by a proinflammatory response pathway, resulting in the upregulation of all three FBG genes in extrahepatic tissues. Indeed, tissue-specific transcription requires a battery of transcription factors that are transcriptionally regulated themselves.
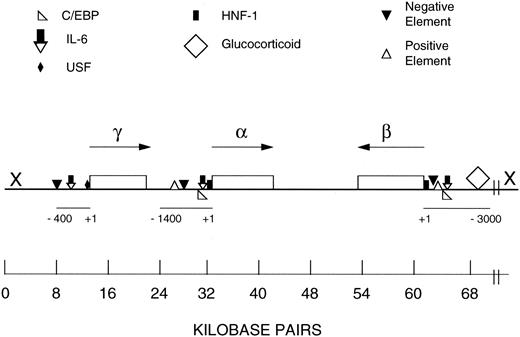
Schematic representation of the human FBG locus and regulatory regions of the Aα, Bβ, and γ chain genes. FBG Aα, Bβ, and γ chain genes are single copy, and each primary transcript is produced from a single transcription initiation event.2 The FBG locus is found on chromosome 4q23-q32. The genes are drawn approximately to scale (kb bar); however, the scale of the 5′ flanking region of each gene is expanded as noted by the scale bar, which is numbered in bp from +1 to −400/−1400/−3000. The direction of transcription of each gene is indicated by the arrow. The symbol X at the 5′ end of the β chain gene represents the hypothetical location of the putative regulatory region for the coordinated expression of the FBG genes based on earlier studies.31,32 56 The results of this study suggest that further analysis of distal sequences of the 5′ flanking regions of the γ chain gene (X) may aid in elucidation of the mechanism for coordinated induction of the FBG genes in response to IL-6 in extrahepatic tissues. The map of the FBG locus and regulatory elements were derived from other references.2,28,31,32
Schematic representation of the human FBG locus and regulatory regions of the Aα, Bβ, and γ chain genes. FBG Aα, Bβ, and γ chain genes are single copy, and each primary transcript is produced from a single transcription initiation event.2 The FBG locus is found on chromosome 4q23-q32. The genes are drawn approximately to scale (kb bar); however, the scale of the 5′ flanking region of each gene is expanded as noted by the scale bar, which is numbered in bp from +1 to −400/−1400/−3000. The direction of transcription of each gene is indicated by the arrow. The symbol X at the 5′ end of the β chain gene represents the hypothetical location of the putative regulatory region for the coordinated expression of the FBG genes based on earlier studies.31,32 56 The results of this study suggest that further analysis of distal sequences of the 5′ flanking regions of the γ chain gene (X) may aid in elucidation of the mechanism for coordinated induction of the FBG genes in response to IL-6 in extrahepatic tissues. The map of the FBG locus and regulatory elements were derived from other references.2,28,31,32
The origin of MK and platelet FBG is now thought to be derived entirely from endocytosis of plasma FBG,8-12 despite the reports documenting the synthesis of FBG in marrow MKs.5-7 It is enigmatic that MKs express the genes for other α-granule constituent adhesive glycoproteins, such as fibronectin and von Willebrand factor,9,14 but not consistently for FBG.8,12,14 However, in light of the present study, it is clear that epithelial cells of extrahepatic origin are able to express the FBG genes, and that intact FBG is secreted from these cells. Taken together, the data demonstrate that, while hepatocytes synthesize and secrete FBG constitutively and on demand, lung epithelial cells synthesize and secrete little intact FBG constitutively; however, on induction with proinflammatory mediators, significant levels of FBG are synthesized and secreted. These studies suggest that the basal expression of the γ chain gene occurs ubiquitously and independently of expression of the Aα and Bβ chain genes in extrahepatic tissue. The synthesis of FBG in extrahepatic tissue may serve to restore homeostasis by aiding in wound repair or extracellular matrix remodeling after injury. This is consistent with our recent demonstration that FBG is secreted basolaterally from A549 lung epithelial cells after induction with DEX + IL-6 (manuscript submitted). However, excessive production of FBG and formation of fibrin, and subsequent fibrinolysis locally in tissue at sites of inflammation, may further enhance production of proinflammatory cytokines and pathophysiologic events.59,60 Finally, the data in this report and others15 18-22 support the hypothesis that epithelial cells in different tissues respond to local injury by upregulating FBG synthesis and secretion, and perhaps other acute phase proteins, via proinflammatory mediators produced in injured or infected tissue.
ACKNOWLEDGMENT
The expert technical assistance of Barbara J. Earnest, Sarah O. Lawrence, and Enzie N. Briskie is gratefully acknowledged. The author thanks Dr Victor J. Marder for his helpful discussions regarding these studies, and Drs Francis Gigliotti and Constantine G. Haidaris for their collaborations on the animal model of Pneumocystis carinii pneumonia, which led to the study of γ-FBG gene regulation in lung tissue. The author also thanks Dr Haidaris and Gayle Guadiz for their editorial contributions to the manuscript.
Supported in part by a grant from the Strong Children's Research Center, University of Rochester, and Grants No. HL50615 and HL30616 from the National Institutes of Health.
Address reprint requests to Patricia J. Simpson Haidaris, PhD, Hematology Unit — Department of Medicine, PO Box 610, University of Rochester School of Medicine and Dentistry, 601 Elmwood Ave, Rochester, NY 14642.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal