Abstract
Granulocyte-macrophage colony-stimulating factor (GM-CSF ) and Steel factor (SLF ) synergistically stimulate Raf-1 kinase activity, protein synthesis, and proliferation in hematopoietic MO7e cells; synergistic action of these factors is blocked by the suppressive chemokines macrophage inflammatory protein-1α (MIP-1α) and interferon-inducible protein 10 (IP-10; Aronica et al, J Biol Chem 270:21998, 1995). We assessed the potential for both stimulatory and inhibitory factors to act through the MAP kinase signaling pathway by studying the effects of growth factors and chemokines on MAP kinase activation. Also, because activation of kinase signaling pathways and stimulation of protein synthesis by peptide growth factors are associated with increased phosphorylation of eukaryotic initiation factor 4E (eIF-4E) and the translational repressor 4E-binding protein 1 (4E-BP1) in some target cells, we investigated whether growth factor treatment could alter eIF-4E or 4E-BP1 phosphorylation state in MO7e cells. We report that treatment of MO7e cells with GM-CSF and SLF stimulated significant, greater-than-additive increases in MAP kinase activity and the phosphorylation of both eIF-4E and 4E-BP1. Increased 4E-BP1 phosphorylation correlated with a decrease in the association of 4E-BP1 with eIF-4E. Growth factor-induced phosphorylation of 4E-BP1 and dissociation of 4E-BP1 from eIF-4E was blocked in cells treated with rapamycin, wortmannin, or PD098059. Treatment of cells with IP-10 or MIP-1α blocked the stimulatory effects of GM-CSF and SLF, resulting in suppression of MAP kinase activity, eIF-4E and 4E-BP1 phosphorylation, and eIF-4E/4E-BP1 dissociation. Our results suggest that GM-CSF and SLF exert part of their combined growth-promoting effects on MO7e cells through activation of MAP kinase and enhancement of eIF-4E and 4E-BP1 phosphorylation and dissociation and that suppression of growth factor-induced protein synthesis by MIP-1α and IP-10 involves translational repression at the level of eIF-4E.
GROWTH OF hematopoietic progenitor cells is coordinated by a number of stimulatory and inhibitory cytokines.1,2 Several cytokines, including granulocyte-macrophage colony-stimulating factor (GM-CSF ) and Steel factor (SLF ) promote the growth of hematopoietic progenitor cells in a synergistic manner when administered in combination.3,4 Activation of cytokine receptors leads to the activation of receptor-associated proteins and the subsequent activation of several intracellular signaling pathways.3,5 Although the activation and use of the Ras/Raf-1/MAP kinase signaling pathway has been related directly to the control of proliferation in some cell systems,3,5-7 mechanisms responsible for regulation of synergistically induced cell proliferation are not completely known. The chemokine family of cytokines, which include macrophage inflammatory protein-1α (MIP-1α), interferon-inducible protein 10 (IP-10), platelet factor 4 (PF4), and interleukin-8 (IL-8), suppress the synergistic action of combination of cytokines on hematopoietic progenitor cell growth.2,8 The myelosuppressive effects shown for members of the chemokine family have been well characterized. However, the cellular mechanisms through which growth inhibition is mediated by chemokines have only just begun to be elucidated.9-11
The rate-limiting step in eukaryotic translation is the binding of mRNA to the 40s subunit of the ribosomal complex.12 Most eukaryotic mRNA species possess a 7-methylguanosine-containing group, or cap structure, at their 5′ terminus that is bound by the cap-binding protein complex eukaryotic initiation factor 4F (eIF-4F ) during the initiation of protein synthesis.13,14 eIF-4F is composed of three subunits: eIF-4A, eIF-4G (p220), and eIF-4E.14 Association of eIF-4E with the cap structure of mRNA is required for the initiation of protein synthesis and serves as one key point of regulation within the protein synthesis machinery of the cell.13,14 In growth-arrested cells, in which protein synthesis levels are markedly reduced, eIF-4E exists in a predominantly underphosphorylated state.15,16 As the rate and degree of eIF-4E phosphorylation increases within target cells, both the activity of eIF-4E and the level of protein synthesis within these cells increases.15-18 Stimulation of cell growth by several peptide growth factors, including insulin and epidermal growth factor (EGF ), which are known to promote protein synthesis in target cells, evokes a rapid increase in eIF-4E phosphorylation.14-18 Stimulation of the insulin receptor through ligand binding has been correlated with the phosphorylation of eIF-4E binding protein 1 (4E-BP1) and the dissociation of 4E-BP1 from eIF-4E.19 The dissociation of 4E-BP1 from eIF-4E is thought to allow eIF-4E to join with eIF-4A and eIF-4G in the formation of an active eIF-4F initiation complex.14 Phosphorylation of 4E-BP1 and its dissociation from eIF-4E correlates with the activation of p70 S6K protein kinase (p70S6K) and MAP kinase in target cells treated with insulin and peptide growth factors.14,18-20 Recent reports have shown that the regulation of 4E-BP1 phosphorylation and the ability of 4E-BP1 to serve as an inhibitor of eIF-4E function in nonhematopoietic systems involves the action of a rapamycin- and wortmannin-sensitive signaling pathway in which both p70S6K and 4E-BP1 are phosphorylated downstream of PI3 kinase by a currently unknown protein kinase.14,21 22 Evidence providing a direct link between MAP kinase activity and the regulation of eIF-4E activation by 4E-BP1 has yet to be demonstrated.
We have recently shown that treatment of the human growth factor-dependent hematopoietic cell line MO7e with GM-CSF in combination with SLF promotes synergistic increases in Raf-1 kinase activity and protein synthesis levels, both of which are blocked upon pretreatment of MO7e cells with MIP-1α or IP-10.9 Because Raf-1 kinase plays a central role in the MAP kinase signaling cascade,23 we investigated whether MAP kinase activity in MO7e cells could be influenced by growth factors in the presence or absence of chemokines. In addition, we set out to determine whether the synergistic stimulation of protein synthesis evoked by GM-CSF and SLF and the suppression of this synergism by various chemokines9 was associated with the phosphorylation of eIF-4E and 4E-BP1 proteins in MO7e cells.
MATERIALS AND METHODS
Materials.RPMI cell culture medium was purchased from Biowhittaker (Walkersville, MD). Fetal bovine serum (FBS) was purchased from Hyclone Laboratories (Logan, UT). 7-methyl GTP-Sepharose 4B was purchased from Pharmacia Biotech Inc (Piscataway, NJ). Protein G-Sepharose 4B was purchased from Zymed Laboratories, Inc (South San Francisco, CA). [32P] orthophosphate and [γ-32P]adenosine triphosphate (ATP) were purchased from DuPont-New England Nuclear (Boston, MA). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) reagents and prestained protein molecular weight markers were purchased from Bio-Rad (Hercules, CA). Agarose-conjugated myelin basic protein (MBP) was provided by Kinetek Biotech (Vancouver, British Columbia, Canada). The Mek1 (MAP kinase kinase) inhibitor PD09059 was obtained from New England Biolabs (Beverly, MA). All other reagents, including 7-methylguanosine 5′-triphosphate (m7GTP), rapamycin, and wortmannin, were purchased from Sigma Chemical Co (St Louis, MO).
Cytokines, antibodies, and kinase substrate.Purified recombinant human (rhu) GM-CSF and rhuSLF were kindly provided by Immunex Corp (Seattle, WA). Recombiant murine (rmu) MIP-1α was obtained from R&D Systems (Minneapolis, MN). We have shown that rhu and rmu preparations of MIP-1α were equally suppressive on human hematopoietic progenitor cells.8 rhuIP-10 was purified as described24 and kindly provided by Dr Andreas Sarris (MD Anderson Cancer Center, Houston, TX). rhuPF4 was a kind gift provided by Repligen (Cambridge, MA). rhuIL-8 was purchased from Peprotech Inc (Rocky Hills, NJ). All chemokines were resuspended in phosphate-buffered saline (PBS). Murine monoclonal antibodies (MoAbs) directed against eIF-4E and rabbit polyclonal antisera directed against murine eIF-4E and human 4E-BP1 were prepared as described.25 26 MoAbs directed against MAP kinase (ERK2) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). MBP synthetic peptide for quantitative MAP kinase activity assays was purchased from Upstate Biological (Lake Placid, NY).
Cells.The human factor-dependent hematopoietic cell line, MO7e, was obtained from Genetics Institute (Boston, MA). Biologic characterisitcs and culture conditions for the MO7e cell line have been described.4,9,11 MO7e cells were maintained in RPMI 1640 culture medium supplemented with 20% FBS and 100 U/mL rhuGM-CSF. Before growth factor or chemokine treatment, MO7e cells were washed with RPMI 1640 and factor-starved (growth arrested) in serum-free RPMI 1640 supplemented with 0.5% bovine serum albumin for 16 to 18 hours at 37°C.4 9
MAP kinase activity assays.Factor-starved MO7e cells resuspended at 2 × 106 cells/mL were exposed to cytokines at various times throughout a 4-hour treatment period. For studies involving chemokines, cells were treated with each chemokine for 1 hour before growth factor treatment. We have shown that this pretreatment period is required for inhibitory chemokines to exert their maximal suppressive effects on cell proliferation.9 11 For qualitative analysis of MAP kinase activity, cell lysates containing equivalent amounts of total protein were combined with agarose-conjugated MBP and 100 μCi/mL [32P] ATP in reaction buffer (100 mmol/L Tris, pH 7.0, 0.4 mmol/L EGTA, 0.4 mmol/L NaVO3 , 40 mmol/L Mg acetate) for 15 minutes. Proteins were separated by SDS-PAGE and transfered to polyvinylidene difluoride (PVDF ) membranes, and [32P]-labeling of MBP was determined by autoradiography. MAP kinase protein content was determined by probing PVDF membranes with antibodies that recognize multiple forms of MAP kinase (anti-PAN MAP kinase antibodies) and visualized using enhanced chemiluminescense (ECL) reagents for nonradioisotypic determination of protein content. For quantitative analysis of MAP kinase activity, the p42 (ERK2) isoform of MAP kinase was immunoprecipitated from cell lysates containing equal amounts of total protein using anti-MAPK (ERK2) antibodies (Upstate Biotechnology Inc, Lake Placid, NY) and combined with an MBP substrate peptide (Upstate Biotechnology Inc) and 100 μCi/mL [τ-32P] ATP in reaction buffer (100 mmol/L Tris, pH 7.0, 0.4 mmol/L EGTA, 0.4 mmol/L NaVO3 , 40 mmol/L Mg acetate, 1 mmol/L unlabeled ATP) for 15 minutes at room temperature. Immunoblot analysis showed that equivalent amounts of ERK2 protein were immunoprecipitated from cell lysates using this procedure (data not shown). Reactions were terminated upon the addition of 40% trichloroacetic acid (TCA), and aliquots were spotted onto P81 phosphocellulose filter discs (Whatman Inc, Hillsboro, OR). Filters were washed twice with 0.75% phosphoric acid and once with acetone. Air-dried filters were placed in scintillation cocktail and counted on a Beckman scintillation counter (Beckman Instruments Inc, Irvine, CA). Mock assay controls containing no MAPK immunoprecipitate were used to determine nonspecific binding of [32P] onto filter disks.
Immunoprecipitation and immunodetection of phosphorylated eIF-4E and 4E-BP1 proteins.Factor-starved MO7e cells (3 × 106 cells/mL) resuspended in serum-free, phosphate-free RPMI supplemented with 100 μCi/mL [32P]orthophosphate were treated with growth factors, chemokines, or combinations of factors for various times over a 12-hour treatment duration. Cells were harvested, washed in PBS, and resuspended in 1% NP-40 lysis buffer9 containing 0.1 mmol/L phenylmethylsulfonyl fluoride, 10 μg/mL aprotinin, 10 μg/mL leupeptin, and 0.4 mmol/L sodium orthovanadate. Cell lysates containing equivalent amounts of TCA-precipitable counts per minute were incubated with 3 μg of eIF-4E polyclonal or MoAbs directed against eIF-4E on ice for 1 hour, followed by protein G-Sepharose 4B beads (Transduction Laboratories, Lexington, KY). MoAbs or polyclonal serum directed against eIF-4E were equally effective at isolating eIF-4E proteins (data not shown). 4E-BP1 proteins were isolated from precleared cell lysates using anti–4E-BP1 polyclonal antibodies (4°C for 1 hour) followed by protein G-Sepharose 4B beads. Beads were collected, washed twice with lysis buffer and twice with PBS, and then resuspended in SDS-containing sample buffer by heating to 100°C for 5 to 10 minutes. Isolated proteins were separated by 12% SDS-PAGE under reducing conditions and transferred to PVDF membranes. Phosphorylation of protein bands was determined by exposing PVDF membranes to film and analyzing the intensity of [32P]-labeled protein bands through autoradiography and densitometry. Scanning analysis of autoradiograms was conducted using a ScanMaker optical scanner (Microtek, Hsinchu, Taiwan, Republic of China) and Sigmagel (Jandel Scientific, San Rafael, CA) and Photoshop imaging analysis software (Adobe Systems Inc, Mountain View, CA). The same PVDF membranes were immunoblotted with eIF-4E or 4E-BP1 antibodies and ECL reagents (Amersham, Arlington Heights, IL) for nonradioisotypic determination of protein content.
Affinity chromatographic isolation of eIF-4E.Factor-starved MO7e cells were washed, resuspended at 3 × 106 cells/mL in serum-free, phosphate-free RPMI 1640 medium supplemented with 100 μCi/mL carrier-free [32P] orthophosphate, and treated with growth factors or chemokines for specified time points over a 12-hour treatment duration. Cells were treated with each chemokine (MIP-1α, IP-10, IL-8, and PF4) for 1 hour before growth factor treatment. eIF-4E proteins were isolated from cell lysates using m7GTP affinity chromatography, as described.16 Briefly, cell lysates containing equal amounts of TCA-precipitable [32P]-radiolabeled proteins (counts per minute) were passed through 0.2-mL columns containing m7GTP-Sepharose 4B beads in B20 buffer (20 mmol/L HEPES, 20 mmol/L KOAc, 0.1 mmol/L EDTA). Columns were washed with B20 buffer containing 1 mmol/L ATP and 0.2 mmol/L GTP, followed by B20 buffer without nucleotides. eIF-4E proteins were eluted from the chromatography columns with B20 buffer containing 100 μmol/L m7GTP. This method has been shown to be effective in isolating eIF-4E proteins, because the m7GTP resin of the column effectively mimicks the cap structure of mRNA.15-17 Both unphosphorylated and phosphorylated forms of eIF-4E are bound by the column resin, with the phosphorylated form of the protein having a higher affinity for the m7GTP moiety. Isolated proteins were separated by 12% SDS-PAGE, transferred to PVDF membrane, and exposed to film for autoradiography. The relative degree of 32P incorporation, as a measure of eIF-4E phosphorylation state at a particular time of treatment, was determined by subjecting autoradiograms to densitometric analysis. The same PVDF membranes were immunoblotted with eIF-4E antibodies, and 4E-BP1 antibodies in some cases, and ECL reagents (Amersham) for nonradioisotypic determination of eIF-4E protein content or 4E-BP1 content.
Proliferation of MO7e colony-forming cells (CFCs).Factor-starved MO7e cells were pretreated at 37°C for 1 hour with control medium, rapamycin (20 ng/mL), wortmannin (10−7 mol/L), or the Mek1 inhibitor PD098059 (50 μmol/L). Cells were washed twice with culture medium and plated at 1 × 103 cells/mL in 0.3% agar culture medium containing 10% FBS, 100 U/mL rhuGM-CSF, and 50 ng/mL rhuSLF, as previously described.9 Colonies were scored after 7 days of incubation at 5% CO2 and lowered (5%) O2 .
Statistical analysis.The statistical significance between treatment groups were determined by Student's t-test.
RESULTS
GM-CSF and SLF stimulate synergistic increases in MAP kinase activity.Factor-starved MO7e cells were treated with GM-CSF (100 U/mL), SLF (50 ng/mL), or the combination of GM-CSF plus SLF for 10 minutes or were treated with MIP-1α (50 ng/mL), IP-10 (50 ng/mL), or PD098059 (50 μmol/L) for 1 hour before treatment with GM-CSF and SLF for 10 minutes. Cell lysates were combined with MBP in the presence of [32P]ATP and MAP kinase activity assessed by SDS-PAGE and autoradiography, as described in the Materials and Methods (Fig 1). Combined treatment of MO7e cells with GM-CSF and SLF stimulated MAP kinase activity, as indicated by [32P]-labeling of MBP (Fig 1, upper panel, lane 7), to a much greater extent than that evoked by treatment with either cytokine alone (Fig 1, upper panel, lanes 3 and 4). Pretreatment of MO7e cells with MIP-1α, IP-10, or PD098059 suppressed the increase in MAP kinase activity stimulated by GM-CSF in combination with SLF (Fig 1, upper panel, lanes 8 through 10). MAP kinase protein content was not altered by any of the treatment conditions (Fig 1, lower panel).
GM-CSF and SLF combined treatment stimulates MAP kinase activity in factor-starved MO7e cells. Factor-starved MO7e cells were treated with GM-CSF (100 U/mL), SLF (50 ng/mL), or the combination of GM-CSF plus SLF for 10 minutes or were treated with MIP-1α (50 ng/mL), IP-10 (50 ng/mL), or PD098059 (50 μmol/L) for 1 hour before treatment with GM-CSF plus SLF for 10 minutes. Cell lysates containing eqivalent amounts of total protein were combined with MBP in the presence of [32P]ATP and MAP kinase activity assessed by SDS-PAGE and autoradiography, as described in the Materials and Methods. Combined treatment of MO7e cells with GM-CSF and SLF stimulated an increase in MAP kinase activity (upper panel, lane 7) to a greater extent than that evoked by treatment with either cytokine alone (upper panel, lanes 3 and 4). Pretreatment with MIP-1α, IP-10, or PD098059 suppressed MAP kinase activity stimulated by GM-CSF in combination with SLF (upper panel, lanes 8, 9, and 10). Two predominant isoforms of MAP kinase, noted to the right of the lower panel as ERK1 and ERK2, were detected in MO7e lysates. ERK1 and ERK2 protein content were not altered by any of the treatments indicated (lower panel). Similar results were obtained in two additional experiments.
GM-CSF and SLF combined treatment stimulates MAP kinase activity in factor-starved MO7e cells. Factor-starved MO7e cells were treated with GM-CSF (100 U/mL), SLF (50 ng/mL), or the combination of GM-CSF plus SLF for 10 minutes or were treated with MIP-1α (50 ng/mL), IP-10 (50 ng/mL), or PD098059 (50 μmol/L) for 1 hour before treatment with GM-CSF plus SLF for 10 minutes. Cell lysates containing eqivalent amounts of total protein were combined with MBP in the presence of [32P]ATP and MAP kinase activity assessed by SDS-PAGE and autoradiography, as described in the Materials and Methods. Combined treatment of MO7e cells with GM-CSF and SLF stimulated an increase in MAP kinase activity (upper panel, lane 7) to a greater extent than that evoked by treatment with either cytokine alone (upper panel, lanes 3 and 4). Pretreatment with MIP-1α, IP-10, or PD098059 suppressed MAP kinase activity stimulated by GM-CSF in combination with SLF (upper panel, lanes 8, 9, and 10). Two predominant isoforms of MAP kinase, noted to the right of the lower panel as ERK1 and ERK2, were detected in MO7e lysates. ERK1 and ERK2 protein content were not altered by any of the treatments indicated (lower panel). Similar results were obtained in two additional experiments.
For quantitative measurement of kinase activity, factor-starved MO7e cells were treated with GM-CSF (100 U/mL), SLF (50 ng/mL), or GM-CSF in combination with SLF for various times over a 4-hour treatment duration. These cytokine concentrations were chosen as they yield maximal increases in cell proliferation when used alone and synergistic increases when used together4,9,11 (data not shown). MAP kinase (ERK2) proteins were immunoprecipitated from cell lysates using MAP kinase-specific antibodies and combined with an MBP substrate peptide in the presence of [32P]ATP. The p42 ERK2 isoform of MAP kinase was chosen because previous studies indicated that ERK2 was the predominant form of MAP kinase phosphorylated in response to GM-CSF and SLF in MO7e cells.7 Incorporation of [32P]-phosphate into the MBP substrate peptide, as a measure of ERK2 MAP kinase activity, was determined by scintillation counting as described in the Materials and Methods (Fig 2A and B). GM-CSF or SLF treatment evoked significant twofold to fourfold increases in MAP kinase activity above control levels within 15 minutes (Fig 2A). Treatment of MO7e cells with GM-CSF in combination with SLF stimulated rapid, sevenfold to ninefold increases in MAP kinase activity within 15 minutes, levels that were significantly greater than MAP kinase activity levels evoked by single factors or control cells, and were greater-than-additive when compared with the effects of GM-CSF and SLF administered alone (Fig 2A). MAP kinase activity levels stimulated by GM-CSF plus SLF declined slowly within the first hour after treatment, but remained significantly higher than control levels at 4 hours after treatment (Fig 2B).
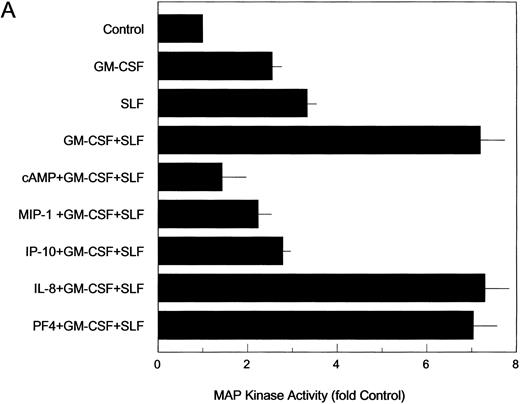
IP-10 and MIP-1α inhibit the stimulatory effects of GM-CSF and SLF on ERK2 MAP kinase activity in MO7e cells. Factor-starved MO7e cells were treated with GM-CSF, SLF, or the combination of GM-CSF and SLF (GM + SLF ) for 15 minutes (A) or for various times throughout a 4-hour treatment period (B and C) or were treated with 8-bromo-cAMP (cAMP + GM-CSF + SLF ) (A) or with MIP-1α (MIP-1α + G + S), IP-10 (IP-10 + G + S), PF4 (PF4 + G + S), or IL-8 (IL-8 + G + S) for 1 hour before combined treatment with GM-CSF and SLF for 15 minutes (A) or for various times throughout a 4-hour treatment period (B and C). MAP kinase proteins were immunoprecipitated from cell lysates and the relative kinase activity, as indicated by phosphorylation of the MBP peptide substrate, was determined for each treatment group as described in the Materials and Methods. Control cells received vehicle alone (control, A through C). Values are expressed as the fold increase above control levels. Each bar (A) represents the mean ± the standard deviation of four determinations obtained in separate experiments. Each point (B and C) represents the mean of four determinations obtained in separate experiments. MAP kinase levels for the separate GM-CSF and SLF treatment groups were significantly higher than controls at 15 minutes, 30 minutes, and 1 hour (P < .05). MAP kinase levels for the GM-CSF + SLF group were significantly higher than those for control, GM-CSF, SLF, cAMP + GM-CSF + SLF, MIP-1α + GM-CSF + SLF, and IP-10 + GM-CSF + SLF treatment groups at all time points studied (P < .05) and were significantly higher to a greater-than-additive extent above the combined mean MAP kinase activity for the individual GM-CSF and SLF groups when administered separately (P < .05).
IP-10 and MIP-1α inhibit the stimulatory effects of GM-CSF and SLF on ERK2 MAP kinase activity in MO7e cells. Factor-starved MO7e cells were treated with GM-CSF, SLF, or the combination of GM-CSF and SLF (GM + SLF ) for 15 minutes (A) or for various times throughout a 4-hour treatment period (B and C) or were treated with 8-bromo-cAMP (cAMP + GM-CSF + SLF ) (A) or with MIP-1α (MIP-1α + G + S), IP-10 (IP-10 + G + S), PF4 (PF4 + G + S), or IL-8 (IL-8 + G + S) for 1 hour before combined treatment with GM-CSF and SLF for 15 minutes (A) or for various times throughout a 4-hour treatment period (B and C). MAP kinase proteins were immunoprecipitated from cell lysates and the relative kinase activity, as indicated by phosphorylation of the MBP peptide substrate, was determined for each treatment group as described in the Materials and Methods. Control cells received vehicle alone (control, A through C). Values are expressed as the fold increase above control levels. Each bar (A) represents the mean ± the standard deviation of four determinations obtained in separate experiments. Each point (B and C) represents the mean of four determinations obtained in separate experiments. MAP kinase levels for the separate GM-CSF and SLF treatment groups were significantly higher than controls at 15 minutes, 30 minutes, and 1 hour (P < .05). MAP kinase levels for the GM-CSF + SLF group were significantly higher than those for control, GM-CSF, SLF, cAMP + GM-CSF + SLF, MIP-1α + GM-CSF + SLF, and IP-10 + GM-CSF + SLF treatment groups at all time points studied (P < .05) and were significantly higher to a greater-than-additive extent above the combined mean MAP kinase activity for the individual GM-CSF and SLF groups when administered separately (P < .05).
Growth factor-stimulated increases in MAP kinase activity are suppressed by IP-10, MIP-1α, and cAMP.To determine whether chemokine treatment could block the stimulatory effect of growth factors on MAP kinase activity, MO7e cells were exposed to IP-10 (50 ng/mL) or MIP-1α (50 ng/mL) for 1 hour at 37°C before combined treatment with GM-CSF and SLF. Pretreatment with either chemokine was sufficient to block the increase in ERK2 MAP kinase activity stimulated by GM-CSF and SLF down to a level of kinase activity consistent with the action of either cytokine alone (Fig 2A and C). The suppressive activity of MIP-1α and IP-10 remained consistent throughout the 4-hour treatment duration (Fig 2C). Consistent with previous studies in which cAMP was shown to block growth factor stimulation of Raf-1 kinase in MO7e cells,9 pretreatment of cells for 1 hour with 8-bromo-cAMP (50 μmol/L), a potent analog of cAMP, blocked the stimulatory action of GM-CSF and SLF on MAP kinase activity (Fig 2A). PF4 and IL-8 suppress proliferation of MO7e cells, but the mechanisms of action studied thus far suggest that these effects are mediated differently from that of MIP-1α and IP-10.9 Prior exposure of MO7e cells for 1 hour with 50 ng/mL PF4 or IL-8 failed to suppress the stimulatory effects of GM-CSF plus SLF on MAP kinase activity within the first hour of growth factor treatment (Fig 2A and C). MAP kinase activity levels for cells treated with PF4 or IL-8 before GM-CSF and SLF were significantly higher than control levels at 4 hours after treatment and statistically the same as levels obtained from cells cotreated with GM-CSF and SLF in the absence of chemokines (Fig 2C). Treatment of MO7e cells with MIP-1α, IP-10, IL-8, or PF4 in the absence of GM-CSF and/or SLF did not alter MAP kinase levels from the basal levels of untreated control MO7e cells (data not shown).
Increased eIF-4E phosphorylation stimulated by growth factors is antagonized by chemokine pretreatment.We have shown that combined treatment with GM-CSF and SLF results in synergistic stimulation of protein synthesis levels and proliferation in MO7e cells.9 Because increased phosphorylation, and hence activation, of the eukaryotic initiation factor-4E (eIF-4E) has been shown to play a critical role in the regulation of protein synthesis,14,16,19 we set out to determine whether treatment with the growth factors GM-CSF and SLF, alone or in combination with various chemokines, could alter the phosphorylation state of eIF-4E. Factor-starved MO7e cells maintained in phosphate-free medium were incubated with [32P]orthophosphate in the presence of various growth factors and/or chemokines. 32P-labeled eIF-4E was immunoprecipitated from whole cell lysates using anti–eIF-4E antibodies or was isolated by m7GTP affinity column chromatography and analyzed by SDS-PAGE and autoradiography, as described in the Materials and Methods. eIF-4E appears as a single band at an approximate molecular weight of 26 to 28-kD, as indicated by the arrow (Fig 3A through E). For some experiments in which eIF-4E proteins were eluted from m7GTP columns, additional phosphorylated proteins were detected as coisolating with eIF-4E, including a protein band that migrates at approximately 80 to 90 kD (Fig 3B). This has been reported by others using the m7GTP isolation method27 and may correspond to eIF-4B, a separate component of the translation initiation complex. Treatment of MO7e cells with either GM-CSF (100 U/mL) or SLF (50 ng/mL) stimulated significant, fourfold to sevenfold increases in eIF-4E phosphorylation within 2 hours when compared with control cells (Fig 3A and Table 1). Coadministration of GM-CSF and SLF evoked rapid, statistically significant and greater-than-additive increases in eIF-4E phosphorylation when compared with the levels of phosphorylation stimulated by either cytokine alone (Fig 3A and Table 1). Increases in eIF-4E phosphorylation evoked by GM-CSF and SLF coadministration were observed as rapidly as 30 minutes and remained high 12 hours after treatment (Fig 3A and B).
Effect of chemokine and cytokine treatment on eIF-4E phosphorylation in MO7e cells. Factor-starved MO7e cells maintained in phosphate-free RPMI supplemented with [32P] orthophosphate were treated with GM-CSF, SLF, or a combination of GM-CSF and SLF or with MIP-1α (B and D), IL-8 (B), IP-10 (C and D), or PF4 (D) for 1 hour before treatment with GM-CSF + SLF for various times over a 12-hour period. Control cells (lane 1, A through E) received vehicle only. eIF-4E proteins were isolated from cell lysates by immunoprecipitation with anti–eIF-4E antibodies (A, C, and D) or by m7GTP column chromatography (B), as described in the Materials and Methods. Proteins were separated by 12% SDS-PAGE and transfered to PVDF membranes, and the intensity of 32P-labeling was visualized by autoradiography. At left is indicated the position of the molecular weight markers. eIF-4E appears as a single band at approximately 27 to 29 kD, as indicated by the position of the arrow. (E) eIF-4E protein content was determined by immunoblotting PVDF membranes with anti–eIF-4E antibodies and horseradish peroxidase-linked protein G. eIF-4E proteins were visualized upon exposure of ECL-treated membranes to film. Cells were pretreated with chemokines for 1 hour before 2 hours of treatment with GM-CSF and SLF.
Effect of chemokine and cytokine treatment on eIF-4E phosphorylation in MO7e cells. Factor-starved MO7e cells maintained in phosphate-free RPMI supplemented with [32P] orthophosphate were treated with GM-CSF, SLF, or a combination of GM-CSF and SLF or with MIP-1α (B and D), IL-8 (B), IP-10 (C and D), or PF4 (D) for 1 hour before treatment with GM-CSF + SLF for various times over a 12-hour period. Control cells (lane 1, A through E) received vehicle only. eIF-4E proteins were isolated from cell lysates by immunoprecipitation with anti–eIF-4E antibodies (A, C, and D) or by m7GTP column chromatography (B), as described in the Materials and Methods. Proteins were separated by 12% SDS-PAGE and transfered to PVDF membranes, and the intensity of 32P-labeling was visualized by autoradiography. At left is indicated the position of the molecular weight markers. eIF-4E appears as a single band at approximately 27 to 29 kD, as indicated by the position of the arrow. (E) eIF-4E protein content was determined by immunoblotting PVDF membranes with anti–eIF-4E antibodies and horseradish peroxidase-linked protein G. eIF-4E proteins were visualized upon exposure of ECL-treated membranes to film. Cells were pretreated with chemokines for 1 hour before 2 hours of treatment with GM-CSF and SLF.
Densitometric Analysis of eIF-4E Phosphorylation Data
| Treatments . | Mean Area* . | |||
|---|---|---|---|---|
| . | 30 min . | 1 h . | 2 h . | 4 h . |
| Control | 2.04 ± 1.1 | 2.93 ± 2.3 | 2.46 ± 1.2 | 3.02 ± 0.7 |
| GM-CSF | 5.80 ± 1.8† | 9.81 ± 2.3† | 4.36 ± 0.4† | 7.19 ± 3.1† |
| SLF | 6.10 ± 2.6† | 8.14 ± 3.4† | 5.26 ± 0.9† | 6.14 ± 2.7† |
| GM-CSF + SLF | 19.05 ± 2.1†‡ | 24.75 ± 5.1†‡ | 15.53 ± 2.1†‡ | 21.48 ± 4.6†‡ |
| MIP-1α + G + S | 7.87 ± 2.2ρ | 4.56 ± 0.7ρ | 3.92 ± 1.4ρ | 5.37 ± 1.2ρ |
| IP-10 + G + S | 8.39 ± 2.6ρ | 7.49 ± 1.3ρ | 4.48 ± 0.7ρ | 6.39 ± 2.3ρ |
| CT + G + S | 2.50 ± 0.8ρ | 5.38 ± 0.6ρ | 5.78 ± 1.2ρ | 8.39 ± 3.2ρ |
| PF4 + G + S | 18.17 ± 4.1 | 19.54 ± 2.1 | 15.82 ± 1.1 | 19.45 ± 0.9 |
| IL-8 + G + S | 21.08 ± 3.7 | 21.58 ± 4.5 | 13.71 ± 3.0 | 18.86 ± 2.6 |
| Treatments . | Mean Area* . | |||
|---|---|---|---|---|
| . | 30 min . | 1 h . | 2 h . | 4 h . |
| Control | 2.04 ± 1.1 | 2.93 ± 2.3 | 2.46 ± 1.2 | 3.02 ± 0.7 |
| GM-CSF | 5.80 ± 1.8† | 9.81 ± 2.3† | 4.36 ± 0.4† | 7.19 ± 3.1† |
| SLF | 6.10 ± 2.6† | 8.14 ± 3.4† | 5.26 ± 0.9† | 6.14 ± 2.7† |
| GM-CSF + SLF | 19.05 ± 2.1†‡ | 24.75 ± 5.1†‡ | 15.53 ± 2.1†‡ | 21.48 ± 4.6†‡ |
| MIP-1α + G + S | 7.87 ± 2.2ρ | 4.56 ± 0.7ρ | 3.92 ± 1.4ρ | 5.37 ± 1.2ρ |
| IP-10 + G + S | 8.39 ± 2.6ρ | 7.49 ± 1.3ρ | 4.48 ± 0.7ρ | 6.39 ± 2.3ρ |
| CT + G + S | 2.50 ± 0.8ρ | 5.38 ± 0.6ρ | 5.78 ± 1.2ρ | 8.39 ± 3.2ρ |
| PF4 + G + S | 18.17 ± 4.1 | 19.54 ± 2.1 | 15.82 ± 1.1 | 19.45 ± 0.9 |
| IL-8 + G + S | 21.08 ± 3.7 | 21.58 ± 4.5 | 13.71 ± 3.0 | 18.86 ± 2.6 |
Factor-starved MO7e cells maintained in phosphate-free medium and supplemented with [32P]orthophosphate were treated with growth factors for the times indicated or were pretreated with MIP-1α, IP-10, PF4, IL-8, or cholera toxin (CT) for 1 hour before combined treatment with GM-CSF and SLF for the times indicated. eIF-4E proteins immunoprecipitated from cell lysates were separated by 12% SDS-PAGE and blotted to PVDF membranes, as described in the Materials and Methods. Intensity of 32P-labeled proteins were visualized by autoradiography and differences between groups were determined by densitometry. eIF-4E protein content was determined by immunoblotting PVDF membranes with anti–eIF-4E antibodies and horseradish peroxidase-linked protein G. ECL-treated membranes were exposed to film, and autoradiograms were analyzed by densitometry.
Densitometric measurements for each group are expressed as the mean area under the peak, corresponding to band intensity on the autoradiogram, expressed as a percentage of the total area under the curve analyzed by the densitometer within each experiment. Values were normalized to protein content before statistical analysis. Each value represents the mean ± the standard deviation for five to six determinations within each treatment group obtained in separate experiments.
Significant difference from control mean (P < .05).
Significant difference from combined means of individual GM-CSF and SLF treatment groups within same treatment duration (P < .05).
ρ Significant difference from GM-CSF + SLF mean (P < .05).
Pretreatment of MO7e cells with 50 ng/mL MIP-1α or IP-10 blocked the stimulatory effects of GM-CSF and SLF, resulting in phosphorylation levels of eIF-4E consistent with stimulation evoked by single cytokines or down to levels comparable to those of untreated control cells (Fig 3B through D and Table 1). Pretreatment of cells with 50 ng/mL PF4 or IL-8 failed to suppress the stimulatory effects of combined GM-CSF and SLF treatment on eIF-4E phosphorylation (Fig 3B and D and Table 1). Treatment of MO7e cells with MIP-1α, IP-10, PF4, or IL-8 in the absence of GM-CSF and SLF did not significantly alter eIF-4E phosphorylation levels from basal, control cell levels (Fig 3C; data not shown). Pretreatment of MO7e cells with cholera toxin, which raises intracellular levels of cAMP, blocked the stimulatory effects of GM-CSF in combination with SLF on eIF-4E phosphorylation (Table 1; autoradiogram not shown). Similar results were obtained when cells were pretreated with 50 μmol/L forskolin or 10−5 mol/L 8-bromo-cAMP (data not shown). Results of densitometric analysis of phosphorylated bands obtained in six separate experiments are presented in the summary statistics of Table 1. Effects of cytokine or chemokine treatment on eIF-4E phosphorylation were the same irrespective of the procedure used to isolate eIF-4E proteins from cell lysates. The level of eIF-4E protein in MO7e cells was not altered by any cytokine or chemokine treatment by 2 hours of treatment (Fig 3E).
IP-10 and MIP-1α inhibit phosphorylation of 4E-BP1 stimulated by GM-CSF and SLF.A current model of insulin action proposes that stimulatory signals triggered by insulin binding to its receptor result in increased phosphorylation of 4E-BP1 and the dissociation of this translational repressor from eIF-4E.21,22 We set out to determine whether growth factors or chemokines could alter the phosphorylation state of 4E-BP1 in MO7e cells. Factor-starved MO7e cells maintained in phosphate-free medium were treated with various growth factors or chemokines in the presence of [32P]orthophosphate. 4E-BP1 was immunoprecipitated from cell lysates using anti–4E-BP1 antibodies and analyzed by SDS-PAGE and autoradiography. In MO7e cells, 4E-BP1 appears in two major forms, migrating as a pair of bands at approximately 16 to 17 kD, as indicated by the arrow (Fig 4A). The smaller molecular weight form (lower 4E-BP1 band) is less abundant (Fig 4A). The presence of a 4E-BP1 doublet is consistent with previous reports on 4E-BP1,18 whereas recent studies have identified three forms of 4E-BP1 in several nonhematopoietic tissues, including rat adipocytes.22 Treatment of MO7e cells with SLF stimulated significant increases in the phosphorylation of the larger form of 4E-BP1, whereas GM-CSF treatment evoked significant increases in the phosphorylation of both 4E-BP1 bands (Fig 4B and C). Costimulation with GM-CSF and SLF significantly increased phosphorylation of both 4E-BP1 bands (Fig 4B and C). Pretreatment of cells with 50 ng/mL MIP-1α or IP-10 inhibited the stimulatory effects of GM-CSF and SLF, resulting in decreased phosphorylation of both 4E-BP1 bands (Fig 4B and C). In contrast, pretreatment of cells with 50 ng/mL IL-8 or PF4 resulted in a significant block to phosphorylation of the smaller 4E-BP1 protein band, but failed to block, and in some cases augmented, the increase in phosphorylation of the upper 4E-BP1 band stimulated by GM-CSF and SLF (Fig 4B and C). Treatment of MO7e cells with GM-CSF and SLF, alone or in combination, or with any of the chemokines did not alter 4E-BP1 protein content (Fig 4A).
Effect of chemokine and cytokine treatment on the phosphorylation of 4E-BP1 in MO7e cells. (A) Factor-starved MO7e cells were treated with SLF (lane 2), GM-CSF (lane 3), or SLF + GM-CSF (lane 4) for 1 hour or with MIP-1α (lane 5), PF4 (lane 6), IP-10 (lane 7), or IL-8 (lane 8) for 1 hour before cotreatment with GM-CSF and SLF for 1 hour. 4E-BP1 proteins were immunoprecipitated from cell lysates using anti–4E-BP1 antibodies, separated by 12% SDS-PAGE, and transferred to PVDF membranes. 4E-BP1 protein content was determined as described in the Materials and Methods. 4E-BP1 proteins were visualized upon exposure of ECL-treated membranes to film. 4E-BP1 appears as two protein bands migrating with an approximate molecular weight of 16 to 17 kD, as indicated by the position of the arrow. At left is the position of the molecular weight markers. (B) Factor-starved MO7e cells were cultured with [32P]orthophosphate in phosphate-free RPMI. Treatments are the same as those described in (A). Control cells (lane 1) received vehicle alone. 4E-BP1 proteins were immunoprecipitated from cell lysates using anti–4E-BP1 antibodies, as described in the Materials and Methods. Proteins were separated by 12% SDS-PAGE and transfered to PVDF membranes, and the intensity of 32P-labeling was visualized by autoradiography. At left is the position of the molecular weight markers. Protein content was determined by probing PVDF membranes with anti–4E-BP1 antibodies and visualization with ECL reagents, as described in the Materials and Methods (data not shown). (C) Scanning analysis of autoradiograms for determination of phosphorylation level for both forms of 4E-BP1. Phosphorylation levels of the larger form of 4E-BP1 were significantly higher than control levels for all treatment groups tested (P < .05). Phosphorylation levels corresponding to the smaller form of 4E-BP1 (*) were significantly higher than controls for the GM-CSF and SLF + GM-CSF treatment groups (P < .05). For both forms of 4E-BP1, phosphorylation levels evoked by combined GM-CSF and SLF treatment (**) were significantly higher than levels corresponding to the GM-CSF, SLF, MIP-1α + SLF + GM-CSF, and IP-10 + SLF + GM-CSF treatment groups (P < .05). Each bar represents the mean ± the standard deviation for three determinations obtained in separate experiments. Phosphorylation levels for each band were normalized to 4E-BP1 protein content before statistical analysis. Treatment groups are the same as described in (A). (□) 4E-BP1, larger form; (▪) 4E-BP1, smaller form.
Effect of chemokine and cytokine treatment on the phosphorylation of 4E-BP1 in MO7e cells. (A) Factor-starved MO7e cells were treated with SLF (lane 2), GM-CSF (lane 3), or SLF + GM-CSF (lane 4) for 1 hour or with MIP-1α (lane 5), PF4 (lane 6), IP-10 (lane 7), or IL-8 (lane 8) for 1 hour before cotreatment with GM-CSF and SLF for 1 hour. 4E-BP1 proteins were immunoprecipitated from cell lysates using anti–4E-BP1 antibodies, separated by 12% SDS-PAGE, and transferred to PVDF membranes. 4E-BP1 protein content was determined as described in the Materials and Methods. 4E-BP1 proteins were visualized upon exposure of ECL-treated membranes to film. 4E-BP1 appears as two protein bands migrating with an approximate molecular weight of 16 to 17 kD, as indicated by the position of the arrow. At left is the position of the molecular weight markers. (B) Factor-starved MO7e cells were cultured with [32P]orthophosphate in phosphate-free RPMI. Treatments are the same as those described in (A). Control cells (lane 1) received vehicle alone. 4E-BP1 proteins were immunoprecipitated from cell lysates using anti–4E-BP1 antibodies, as described in the Materials and Methods. Proteins were separated by 12% SDS-PAGE and transfered to PVDF membranes, and the intensity of 32P-labeling was visualized by autoradiography. At left is the position of the molecular weight markers. Protein content was determined by probing PVDF membranes with anti–4E-BP1 antibodies and visualization with ECL reagents, as described in the Materials and Methods (data not shown). (C) Scanning analysis of autoradiograms for determination of phosphorylation level for both forms of 4E-BP1. Phosphorylation levels of the larger form of 4E-BP1 were significantly higher than control levels for all treatment groups tested (P < .05). Phosphorylation levels corresponding to the smaller form of 4E-BP1 (*) were significantly higher than controls for the GM-CSF and SLF + GM-CSF treatment groups (P < .05). For both forms of 4E-BP1, phosphorylation levels evoked by combined GM-CSF and SLF treatment (**) were significantly higher than levels corresponding to the GM-CSF, SLF, MIP-1α + SLF + GM-CSF, and IP-10 + SLF + GM-CSF treatment groups (P < .05). Each bar represents the mean ± the standard deviation for three determinations obtained in separate experiments. Phosphorylation levels for each band were normalized to 4E-BP1 protein content before statistical analysis. Treatment groups are the same as described in (A). (□) 4E-BP1, larger form; (▪) 4E-BP1, smaller form.
MIP-1α and IP-10 inhibit growth factor-stimulated dissociation of 4E-BP1 from eIF-4E proteins.We set out to determine whether the stimulatory action of GM-CSF and SLF, in the presence or absence of suppressive chemokines, could affect the association of 4E-BP1 with eIF-4E. Factor-starved MO7e cells were treated with GM-CSF (100 U/mL), SLF (50 ng/mL), or the combination of GM-CSF and SLF for 1 hour or were treated with 50 ng/mL of MIP-1α, IP-10, PF4, or IL-8 for 1 hour before 1 hour of treatment with GM-CSF in combination with SLF. eIF-4E was immunoprecipitated from whole cell lysates using polyclonal and monoclonal anti–eIF-4E antibodies and protein G-sepharose and analyzed by SDS-PAGE. The presence of 4E-BP1 in the eIF-4E immunoprecipitates was determined using anti–4E-BP1 antibodies and ECL reagents, as described in the Materials and Methods. For some experiments, the smaller isoform of 4E-BP1 was undetectable (Fig 5A). Although reactive to the 4E-BP1 antibodies, the faint band migrating below 4E-BP1 in each lane is thought to represent a smaller eIF binding protein, 4E-BP2. The amount of 4E-BP1 protein associated with eIF-4E proteins isolated from cells treated with GM-CSF in combination with SLF (lane 4) was significantly less than that observed for control cells or from cells treated with either GM-CSF or SLF alone (Fig 5A and B). Sample lanes derived from cells pretreated with MIP-1α or IP-10 (Fig 5A, lanes 5 and 6) contained amounts of 4E-BP1 protein significantly greater than levels from control, vehicle-treated cells (Fig 5B). In contrast, lanes corresponding to cells pretreated with PF4 or IL-8 (Fig 5A, lanes 7 and 8) contained 4E-BP1 protein levels significantly lower than controls and statistically the same as those determined for cells coincubated with GM-CSF and SLF (Fig 5B).
Effects of chemokine and cytokine treatment on the association of 4E-BP1 with eIF-4E proteins. Factor-starved MO7e cells were treated with control vehicle (lane 1), GM-CSF (lane 2), SLF (lane 3), or GM-CSF + SLF (lane 4) for 1 hour or with MIP-1α (lane 5), IP-10 (lane 6), IL-8 (lane 7), or PF4 (lane 8) for 1 hour before 1 hour of treatment with GM-CSF + SLF. (A) eIF-4E proteins were immunoprecipitated from cell lysates and separated by 15% SDS-PAGE, as described in the Materials and Methods. 4E-BP1 protein content associated with eIF-4E in each treatment group was determined by immunoblotting PVDF membranes with anti–4E-BP1 antibodies and visualized upon exposing ECL-treated membranes to film. The position of the molecular weight marker is indicated to the left. The position of 4E-BP1 is indicated to the right of (B). Scanning analysis of autoradiograms was performed to determine relative 4E-BP1 protein content, as described in the Materials and Methods. (*) 4E-BP1 protein content was significantly higher than control levels for the MIP-1α and IP-10 pretreatment groups (P < .05). (**) Protein content for 4E-BP1 corresponding to the GM-CSF + SLF, IL-8 + GM-CSF + SLF, and PF4 + GM-CSF + SLF treatment groups was significantly less than control levels (P < .05). Each bar represents the mean ± the standard deviation for three determinations obtained in separate experiments. Treatment groups are the same as those for (A).
Effects of chemokine and cytokine treatment on the association of 4E-BP1 with eIF-4E proteins. Factor-starved MO7e cells were treated with control vehicle (lane 1), GM-CSF (lane 2), SLF (lane 3), or GM-CSF + SLF (lane 4) for 1 hour or with MIP-1α (lane 5), IP-10 (lane 6), IL-8 (lane 7), or PF4 (lane 8) for 1 hour before 1 hour of treatment with GM-CSF + SLF. (A) eIF-4E proteins were immunoprecipitated from cell lysates and separated by 15% SDS-PAGE, as described in the Materials and Methods. 4E-BP1 protein content associated with eIF-4E in each treatment group was determined by immunoblotting PVDF membranes with anti–4E-BP1 antibodies and visualized upon exposing ECL-treated membranes to film. The position of the molecular weight marker is indicated to the left. The position of 4E-BP1 is indicated to the right of (B). Scanning analysis of autoradiograms was performed to determine relative 4E-BP1 protein content, as described in the Materials and Methods. (*) 4E-BP1 protein content was significantly higher than control levels for the MIP-1α and IP-10 pretreatment groups (P < .05). (**) Protein content for 4E-BP1 corresponding to the GM-CSF + SLF, IL-8 + GM-CSF + SLF, and PF4 + GM-CSF + SLF treatment groups was significantly less than control levels (P < .05). Each bar represents the mean ± the standard deviation for three determinations obtained in separate experiments. Treatment groups are the same as those for (A).
cAMP pretreatment blocks growth factor-stimulated increases in 4E-BP1 phosphorylation.A recent report has shown that cAMP can suppress the phosphorylation of 4E-BP1 in rat adipocytes.21 Because we had shown that cAMP could mimick the action of chemokines by blocking the stimulatory effects of GM-CSF and SLF,9 we determined whether cAMP pretreatment could affect the ability of cytokines or chemokines to alter 4E-BP1 phosphorylation levels in MO7e cells. Factor-starved MO7e cells maintained in phosphate-free medium were treated with various growth factors, chemokines, or 8-bromo-cAMP in the presence of [32P]orthophosphate. 4E-BP1 was immunoprecipitated from whole cell lysates using anti–4E-BP1 polyclonal antibodies. Isolated proteins were separated by 12% SDS-PAGE, transferred to PVDF membranes, and exposed to film. As previously observed, only the larger form of 4E-BP1 appears phosphorylated in control, untreated cells (Fig 6, lane 1). Costimulation with GM-CSF plus SLF increased phosphorylation of both 4E-BP1 bands (Fig 6, lanes 2, 4, and 6). Pretreatment of MO7e cells with 50 μmol/L 8-bromo-cAMP blocked growth factor-stimulated increases in 4E-BP1 phosphorylation on both bands (Fig 6, lane 3). Suppression of 4E-BP1 phosphorylation evoked by 8-bromo-cAMP pretreatment was similar to that mediated by MIP-1α and IP-10 pretreatment of MO7e cells (Fig 6, lanes 5 and 7, respectively).
Effect of cAMP pretreatment on 4E-BP1 phosphorylation levels. Factor-starved MO7e cells were cultured with [32P]orthophosphate in phosphate-free RPMI, as described in the Materials and Methods. Cells were treated with SLF in combination with GM-CSF (lanes 2, 4, and 6) for 1 hour or with 8-bromo-cAMP (lane 3), MIP-1α (lane 5), or IP-10 (lane 7) for 1 hour before cotreatment with GM-CSF and SLF for 1 hour. Control cells (lane 1) received vehicle alone. Immunoprecipitated 4E-BP1 proteins were separated by 12% SDS-PAGE and transfered to PVDF membranes, and the intensity of 32P-labeling was visualized by autoradiography. At left is the position of the molecular weight markers. 4E-BP1 appears as a doublet of two protein bands migrating with an approximate molecular weight of 16 to 17 kD, as indicated to the right of the figure. Similar results were obtained in each of two additional experiments.
Effect of cAMP pretreatment on 4E-BP1 phosphorylation levels. Factor-starved MO7e cells were cultured with [32P]orthophosphate in phosphate-free RPMI, as described in the Materials and Methods. Cells were treated with SLF in combination with GM-CSF (lanes 2, 4, and 6) for 1 hour or with 8-bromo-cAMP (lane 3), MIP-1α (lane 5), or IP-10 (lane 7) for 1 hour before cotreatment with GM-CSF and SLF for 1 hour. Control cells (lane 1) received vehicle alone. Immunoprecipitated 4E-BP1 proteins were separated by 12% SDS-PAGE and transfered to PVDF membranes, and the intensity of 32P-labeling was visualized by autoradiography. At left is the position of the molecular weight markers. 4E-BP1 appears as a doublet of two protein bands migrating with an approximate molecular weight of 16 to 17 kD, as indicated to the right of the figure. Similar results were obtained in each of two additional experiments.
Rapamycin, wortmannin, and PD098059 inhibit growth factor-stimulated MO7e colony formation.To assess the potential role of various kinase pathways in the control of MO7e cell proliferation, we pretreated factor-starved MO7e cells with rapamycin (20 ng/mL), wortmannin (10−7 mol/L), or PD098059 (50 μmol/L) for 1 hour before plating in 0.3% agar in the presence of GM-CSF and SLF, as described in the Materials and Methods. Inhibitor concentrations were selected based on data obtained in MO7e growth curves for viability in response to drug concentration (data not shown). After 7 days in culture, colony number and size for each treatment group was assessed. Treatment of MO7e cells with GM-CSF in combination with SLF promoted growth of MO7e colonies. Colony numbers arising from MO7e cells pretreated with rapamycin, wortmannin, or PD098059 were all significantly less than those pretreated with control medium (P < .001; Fig 7). Interestingly, rapamycin pretreatment yielded significantly less colonies in comparison to cells pretreated with PD098059 (P < .05). Rapamycin pretreatment was also more potent in suppressing stimulation of MO7e colony formation evoked by GM-CSF or SLF when administered separately (data not shown). In addition, colonies arising from MO7e cells treated with rapamycin or wortmannin were visually smaller than those pretreated with PD098059 or control cells treated with GM-CSF + SLF in the absence of inhibitors (data not shown).
Effect of rapamycin, wortmannin, and PD98059 on growth factor-stimulated MO7e colony formation. Factor-starved MO7e cells were pulse treated with rapamycin (20 ng/mL), wortmannin (10−7 mol/L), or PD098059 (50 μmol/L) for 1 hour, washed twice with culture medium, and plated in 0.3% agar in the presence of GM-CSF and SLF, as described in the Materials and Methods. After 7 days in culture, the colony number for each treatment group was assessed. Treatment of MO7e cells with GM-CSF in combination with SLF promoted growth of MO7e colonies, ranging from 60 to 170 colonies/plate. Colony numbers arising from MO7e cells pretreated with rapamycin, wortmannin, or PD098059 (*) were all significantly less than those treated with GM-CSF + SLF in the absence of inhibitors (P < .001). Rapamycin pretreatment yielded significantly less colonies in comparison to cells pretreated with PD098059 (**P < .05). Each experiment was conducted in triplicate. Each bar depicted represents the mean ± SEM colony number obtained from three individual experiments.
Effect of rapamycin, wortmannin, and PD98059 on growth factor-stimulated MO7e colony formation. Factor-starved MO7e cells were pulse treated with rapamycin (20 ng/mL), wortmannin (10−7 mol/L), or PD098059 (50 μmol/L) for 1 hour, washed twice with culture medium, and plated in 0.3% agar in the presence of GM-CSF and SLF, as described in the Materials and Methods. After 7 days in culture, the colony number for each treatment group was assessed. Treatment of MO7e cells with GM-CSF in combination with SLF promoted growth of MO7e colonies, ranging from 60 to 170 colonies/plate. Colony numbers arising from MO7e cells pretreated with rapamycin, wortmannin, or PD098059 (*) were all significantly less than those treated with GM-CSF + SLF in the absence of inhibitors (P < .001). Rapamycin pretreatment yielded significantly less colonies in comparison to cells pretreated with PD098059 (**P < .05). Each experiment was conducted in triplicate. Each bar depicted represents the mean ± SEM colony number obtained from three individual experiments.
Rapamycin, wortmannin, and PD098059 inhibit 4E-BP1 phosphorylation and growth factor-stimulated dissociation of 4E-BP1 from eIF-4E.Given that MAP kinase activity in MO7e cells is significantly stimulated by combined GM-CSF and SLF treatment and that a block of the MAP kinase pathway by PD098059 suppresses the growth of MO7e colony-forming cells in response to GM-CSF and SLF, we set out to determine whether kinase inhibitors could block phosphorylation of eIF-4E and 4E-BP1 evoked by GM-CSF and SLF. MO7e cells were treated with GM-CSF and SLF for 15 minutes or were pretreated with rapamycin, wortmannin, or PD098059 for 1 hour before 15 minutes of treatment with GM-CSF + SLF in the presence of [32P]orthophosphate. eIF-4E and 4E-BP1 proteins were isolated from cell lysates using anti–eIF-4E and anti–4E-BP1 antibodies, respectively; separated by SDS-PAGE; and transferred to PVDF membranes. The intensity of [32P] radiolabeling was determined by autoradiography. Treatment of MO7e cells with GM-CSF in combination with SLF evoked increases in eIF-4E phosphorylation (Fig 8A) and phosphorylation of both forms of 4E-BP1 (Fig 8B). Pretreatment of cells with rapamycin, wortmannin, and PD098059 blocked the stimulatory effects of GM-CSF and SLF on 4E-BP1 phosphorylation (Fig 8B), whereas only wortmannin and PD98059 appeared to suppress growth factor stimulation of eIF-4E phosphorylation (Fig 8A).
Effect of rapamycin, wortmannin, and PD098059 on eIF-4E/4E-BP1 phosphorylation and association in MO7e cells. (A and B) Factor-starved MO7e cells were treated with GM-CSF (100 U/mL) and SLF (50 ng/mL) for 15 minutes or were pulse treated with rapamycin, wortmannin, or PD098059 for 1 hour and washed twice with culture medium before 15 minutes of treatment with GM-CSF and SLF in the presence of [32P]orthophosphate. eIF-4E (A) and 4E-BP1 (B) proteins were isolated from cell lysates using anti–eIF-4E and anti–4E-BP1 antibodies, respectively, separated by SDS-PAGE, and transferred to PVDF membranes, and the intensity of [32P] radiolabeling was determined by autoradiography. Combined treatment of MO7e cells with GM-CSF and SLF increased eIF-4E phosphorylation (A) and phosphorylation of both forms of 4E-BP1 (B). Pretreatment of cells with rapamycin, wortmannin, and PD098059 blocked the stimulatory effects of GM-CSF and SLF on 4E-BP1 phosphorylation, whereas only wortmannin and PD098059 suppressed growth factor stimulation of eIF-4E phosphorylation (A). eIF-4E and 4E-BP1 content were not altered by any of the treatments indicated (data not shown). Similar results were obtained in two additional experiments. (C) Factor-starved MO7e cells were pretreated with 50 ng/mL MIP-1α, IP-10, IL-8, or PF4 or 20 ng/mL rapamycin, 10−7 mol/L wortmannin, or 50 μmol/L PD098059 for 1 hour before combined GM-CSF and SLF treatment for 15 minutes. eIF-4E proteins isolated from cell lysates by m7GTP affinity column chromatography were separated by SDS-PAGE, transferred to PVDF membranes, and probed for eIF-4E and 4E-BP1/BP2 content using anti–eIF-4E and anti–4E-BP1/BP2 antibodies, as described in the Materials and Methods. 4E-BP1 coisolated with eIF-4E in control, quiescent cells (lane 1). Treatment of MO7e cells with GM-CSF and SLF results in the dissociation of 4E-BP1 from eIF-4E, as indicated by the lack of 4E-BP1 content in lane 2 (lower panel). Pretreatment of cells with MIP-1α, IP-10, rapamycin, wortmannin, and, to a lesser extent, PD098059 suppressed the dissociation of eIF-4E from 4E-BP1 in the presence of GM-CSF and SLF (lower panel, lanes 3, 5, 7, 8, and 9). In contrast, 4E-BP1 dissociated from eIF-4E in MO7e cells pretreated with IL-8 or PF4 (lanes 4 and 6). Similar results were obtained in two separate experiments.
Effect of rapamycin, wortmannin, and PD098059 on eIF-4E/4E-BP1 phosphorylation and association in MO7e cells. (A and B) Factor-starved MO7e cells were treated with GM-CSF (100 U/mL) and SLF (50 ng/mL) for 15 minutes or were pulse treated with rapamycin, wortmannin, or PD098059 for 1 hour and washed twice with culture medium before 15 minutes of treatment with GM-CSF and SLF in the presence of [32P]orthophosphate. eIF-4E (A) and 4E-BP1 (B) proteins were isolated from cell lysates using anti–eIF-4E and anti–4E-BP1 antibodies, respectively, separated by SDS-PAGE, and transferred to PVDF membranes, and the intensity of [32P] radiolabeling was determined by autoradiography. Combined treatment of MO7e cells with GM-CSF and SLF increased eIF-4E phosphorylation (A) and phosphorylation of both forms of 4E-BP1 (B). Pretreatment of cells with rapamycin, wortmannin, and PD098059 blocked the stimulatory effects of GM-CSF and SLF on 4E-BP1 phosphorylation, whereas only wortmannin and PD098059 suppressed growth factor stimulation of eIF-4E phosphorylation (A). eIF-4E and 4E-BP1 content were not altered by any of the treatments indicated (data not shown). Similar results were obtained in two additional experiments. (C) Factor-starved MO7e cells were pretreated with 50 ng/mL MIP-1α, IP-10, IL-8, or PF4 or 20 ng/mL rapamycin, 10−7 mol/L wortmannin, or 50 μmol/L PD098059 for 1 hour before combined GM-CSF and SLF treatment for 15 minutes. eIF-4E proteins isolated from cell lysates by m7GTP affinity column chromatography were separated by SDS-PAGE, transferred to PVDF membranes, and probed for eIF-4E and 4E-BP1/BP2 content using anti–eIF-4E and anti–4E-BP1/BP2 antibodies, as described in the Materials and Methods. 4E-BP1 coisolated with eIF-4E in control, quiescent cells (lane 1). Treatment of MO7e cells with GM-CSF and SLF results in the dissociation of 4E-BP1 from eIF-4E, as indicated by the lack of 4E-BP1 content in lane 2 (lower panel). Pretreatment of cells with MIP-1α, IP-10, rapamycin, wortmannin, and, to a lesser extent, PD098059 suppressed the dissociation of eIF-4E from 4E-BP1 in the presence of GM-CSF and SLF (lower panel, lanes 3, 5, 7, 8, and 9). In contrast, 4E-BP1 dissociated from eIF-4E in MO7e cells pretreated with IL-8 or PF4 (lanes 4 and 6). Similar results were obtained in two separate experiments.
To assess the ability of chemokines and kinase inhibitors to block growth factor-stimulated dissociation of 4E-BP1 from eIF-4E, MO7e cells were pretreated with 50 ng/mL MIP-1α, IP-10, IL-8, or PF4 or 20 ng/mL rapamycin, 10−7 mol/L wortmannin or 50 μmol/L PD098059 for 1 hour before combined GM-CSF and SLF treatment for 15 minutes. eIF-4E proteins isolated from cell lysates by m7GTP affinity column chromatography were separated by SDS-PAGE, transferred to PVDF membranes, and probed for eIF-4E and 4E-BP1/BP2 content using anti–eIF-4E and anti–4E-BP1/BP2 antibodies and ECL detection reagents, as described in the Materials and Methods. As shown in Fig 8C, 4E-BP1 coisolated with eIF-4E in control, quiescent cells. Treatment of MO7e cells with GM-CSF and SLF resulted in the dissociation of 4E-BP1 from eIF-4E, as indicated by the lack of 4E-BP1 content in lane 2 (Fig 8C, lower panel). Pretreatment of cells with MIP-1α, IP-10, rapamycin, wortmannin, and PD098059 suppressed the dissociation of eIF-4E from 4E-BP1 in the presence of GM-CSF and SLF (Fig 8C, lanes 3, 5, 7, 8, and 9). In contrast, less 4E-BP1 colocalized with eIF-4E obtained from MO7e cells pretreated with IL-8 or PF4 (Fig 8C, lanes 4 and 6).
DISCUSSION
The MAP kinase cascade plays a central role in linking membrane-associated events evoked by stimulatory factors to the regulation of cell proliferation and differentiation.7,28-30 We present here our findings showing that the activity level of MAP kinase in hematopoietic MO7e cells is increased in a significant, greater-than-additive manner in response to combined treatment with the hematopoietic growth factors GM-CSF and SLF. Although our results are consistent with those reported in previous studies for stimulation of MAP kinase by single cytokines in hematopoietic cells,7,28,30 this is the first time that direct, greater-than-additive activation of MAP kinase by combined cytokine treatment has been reported. Evidence providing a direct link between MAP kinase activation and stimulation of proliferation in response to the cytokine thrombopoietin has very recently been shown in MO7e cells.31 Our observations that the Mek1 inhibitor PD098059 blocks MAP kinase activity and significantly suppresses growth factor-stimulated colony formation in MO7e cells, taken together with results of our recent study of Raf-1 kinase activity in MO7e cells,9 lend further support to the hypothesis that the synergistic interaction of GM-CSF and SLF in stimulating cell proliferation is mediated, at least in part, through the activation of the Ras/Raf-1/MAP kinase signalling cascade. This is consistent with our observation that the chemokines MIP-1α and IP-10, which suppress synergistic cell proliferation and activation of Raf-1 kinase by GM-CSF and SLF, also block the activation of MAP kinase stimulated by GM-CSF and SLF.
Stimulation of signal transduction pathways by mitogenic growth factors is associated with increased protein synthesis in many cell systems.13,14 Insulin treatment stimulates increased phosphorylation of the translational repressor 4E-BP1 and its dissociation from eIF-4E in target cells.14,19,21,22 Insulin also induces phosphorylation of eIF-4E itself, increasing eIF-4E affinity towards eIF-4G and the cap structure of mRNA.22 Consistent with this mechanism of action proposed for insulin, EGF, platelet-derived growth factor (PGDF), and insulin-like growth factor-I (IGF-I),19,21,22 our results indicate that treatment of MO7e cells with GM-CSF in combination with SLF stimulates increased phosphorylation of eIF-4E and both forms of 4E-BP1. Our observation that GM-CSF and SLF administered separately can stimulate eIF-4E phosphorylation is consistent with a recent report showing enhancement of eIF-4E phosphorylation in erythroblasts in response to the cytokine erythropoietin.32 In contrast to the action of insulin, PDGF, and erythropoietin in other cell systems, enhancement of 4E-BP1 phosphorylation and dissociation of the 4E-BP1/eIF-4E complex in MO7e cells appears maximal when individual cytokines are combined. Similar to the observations made for MAP kinase activation in MO7e cells, enhancement of eIF-4E phosphorylation evoked by GM-CSF and SLF cotreatment is prolonged in duration when compared with induction evoked by single cytokines. Whether the duration or the degree of activation of MAP kinase or eIF-4E phosphorylation relates directly to synergistic stimulation of MO7e cell proliferation by combinations of cytokines has yet to be determined. However, the degree of Raf-1 activation has been reported as correlating with a graded proliferative response in at least one myeloid cell system.33
Previous reports have indicated that the larger form of 4E-BP1 binds primarily to eIF-4E and that the phosphorylation state of this form of 4E-BP1 is most closely related to 4E-BP1/eIF-4E complex formation and translational repression.14,19,22 Consistent with reports that indicate that phosphorylated 4E-BP1 does not form a complex with eIF-4E, we observed that increases in 4E-BP1 phosphorylation correlated with a decreased association between 4E-BP1 and eIF-4E in MO7e cells treated with GM-CSF and SLF. This may help to explain why PF4 and IL-8, which block phosphorylation of the smaller form of 4E-BP1 but not of the larger form, are less effective than MIP-1α and IP-10 at blocking the dissociation of 4E-BP1 from eIF-4E and suppressing increases in protein synthesis in growth factor-stimulated cells.9 The cellular content of both forms of 4E-BP1 was not affected by cytokine or chemokine treatment, indicating that the differences we observed in phosphorylation intensity were not due to changes in protein content or to simple conversion of one form of 4E-BP1 to the other. Although changes in phosphorylation intensity were readily visible on 4E-BP1 autoradiograms, we were unable to detect changes in 4E-BP1 mobility in most phosphorylation experiments. It may be possible that the methodology we used was not sensitive enough to detect slight differences in molecular weight or conformational change in 4E-BP1 proteins, although alterations in SDS-PAGE gel length, acrylamide percentage, buffer conditions, running voltage, and running time were tested (data not shown). Although purely speculative, it may be possible that the regulation pattern for 4E-BP1 present in MO7e cells may involve phosphorylation or dephosphorylation events within the 4E-BP1 molecule, including a change in overall phosphate turnover rate, which do not sufficiently alter the conformation of the protein and/or its migration in our SDS-PAGE system.
We report here that 8-bromo-cAMP can mimic the action of MIP-1α and IP-10 by blocking growth factor stimulation of MAP kinase in MO7e cells. This is consistent with results of our previous studies in which MIP-1α and IP-10 were shown to increase cellular levels of cAMP and blocked the combined action of GM-CSF and SLF on Raf-1 kinase activity apparently through activation of cAMP-dependent protein kinase A.9,11 The observations that cAMP can mimic chemokine action in the suppression of Raf-1 kinase9 and MAP kinase activity (herein), eIF-4E phosphorylation (herein), protein synthesis,9 and proliferation of MO7e cells9 suggests that MIP-1α and IP-10 may suppress cell growth through combined modulation of several metabolic pathways within target cells, including modulation of cAMP. A role for cAMP as an inhibitor of protein synthesis at the level of the eIF-4E complex has recently been shown in rat aortic smooth muscle cells, in which forskolin, which increases intracellular cAMP levels, decreases phosphorylation of rat 4E-BP1 and increases the association of 4E-BP1 with eIF-4E.21 This is consistent with our observation that pretreatment of MO7e cells with 8-bromo-cAMP blocks the stimulatory actions of GM-CSF and SLF by inhibiting phosphorylation of eIF-4E and both forms of 4E-BP1.
Whereas growth factor-induced MAP kinase activity has been correlated with the dissociation of the 4E-BP1/eIF-4E complex in many cell systems,14,22 recent evidence has shown that MAP kinase itself fails to dissociate the 4E-BP1/eIF-4E complex in vivo and suggest that the action of MAP kinase within target cell systems is independent of the inhibitory effect of 4E-BP1 on eIF-4E.22 Phosphorylation of 4E-BP1 and its dissociation from eIF-4E is thought to be accomplished by a currently unidentified rapamycin- and wortmannin-sensitive protein kinase that lies downstream from PI3 kinase and recognizes both 4E-BP1 and p70S6K as substrates.14,25,34-36 In contrast to studies that separate the action of MAP kinase from regulation of 4E-BP1 activity, the results we obtained for MO7e CFC and eIF-4E/4E-BP1 phosphorylation and dissociation studies conducted with rapamycin, wortmannin, and PD098059 strongly suggests a link between MAP kinase activation and the regulation of 4E-BP1 activity, at least in the case of MO7e cells. Our observation that rapamycin fails to alter growth factor stimulation of eIF-4E phosphorylation while suppressing 4E-BP1 phosphorylation and blocking the dissociation of 4E-BP1 from eIF-4E is consistent with the results of a recent study conducted in Chinese hamster ovary (CHO) cells37 and suggests that phosphorylation of eIF-4E and 4E-BP1 are mediated by separate and distinct pathways. The observation that 4E-BP1 in MO7e cells pretreated with rapamycin did not dissociate from eIF-4E indicates that phosphorylation of eIF-4E is not a sufficient signal to promote dissociation of the eIF-4E/4E-BP1 complex, as reported in other cell systems.14 Determination of whether the results we obtained with the M07e cell line reflects a form of regulation of 4E-BP1 and eIF-4E distinctive to hematopoietic-derived cells will require further study. However, it is interesting to note that, in both our system and studies conducted in CHO.T cells, in which MAP kinase activity appears linked to phosphorylation of eIF-4E and 4E-BP1, 4E-BP1 appears predominantly as two forms rather than as three, suggesting alternate pathways for the regulation of 4E-BP1 activity.37
Our findings suggest that GM-CSF and SLF exert part of their combined stimulatory effects on hematopoietic cell growth through rapid and sustained activation of MAP kinase, stimulation of eIF-4E and 4E-BP1 phosphorylation, and the dissociation of the 4E-BP1/eIF-4E complex. Experimental results indicate that the stimulatory actions of GM-CSF and SLF on MO7e cell growth involve the activation of rapamycin- and wortmannin-sensitive pathways and that the action of these growth factors are blocked by the suppresive cytokines MIP-1α and IP-10. Our observations suggest that PF4 and IL-8, which fail to inhibit growth factor stimulation of MAP kinase activity and the phosphorylation and dissociation of eIF-4E and 4E-BP1, exert their suppressive effects within MO7e cells through mechanisms distinct from those used by IP-10 and MIP-1α.
ACKNOWLEDGMENT
We thank Dr Andreas Sarris for the kind gift of rhuIP-10.
Supported by Public Health Service Grants No. R01HL56416, R37CA36464, R01HL49202, and R01HL54037 to H.E.B. and by a grant from MRC to N.S. S.M.A. is supported by National Institutes of Health Training Grant No. T32DK07519 to H.E.B. and A.C.G. is supported by an NSERC 67 studentship.
Address reprint requests to Susan M. Aronica, PhD, Walther Oncology Center, Indiana University School of Medicine, 975 W Walnut St, Room 501, Indianapolis, IN 46202-5121.

![Fig. 1. GM-CSF and SLF combined treatment stimulates MAP kinase activity in factor-starved MO7e cells. Factor-starved MO7e cells were treated with GM-CSF (100 U/mL), SLF (50 ng/mL), or the combination of GM-CSF plus SLF for 10 minutes or were treated with MIP-1α (50 ng/mL), IP-10 (50 ng/mL), or PD098059 (50 μmol/L) for 1 hour before treatment with GM-CSF plus SLF for 10 minutes. Cell lysates containing eqivalent amounts of total protein were combined with MBP in the presence of [32P]ATP and MAP kinase activity assessed by SDS-PAGE and autoradiography, as described in the Materials and Methods. Combined treatment of MO7e cells with GM-CSF and SLF stimulated an increase in MAP kinase activity (upper panel, lane 7) to a greater extent than that evoked by treatment with either cytokine alone (upper panel, lanes 3 and 4). Pretreatment with MIP-1α, IP-10, or PD098059 suppressed MAP kinase activity stimulated by GM-CSF in combination with SLF (upper panel, lanes 8, 9, and 10). Two predominant isoforms of MAP kinase, noted to the right of the lower panel as ERK1 and ERK2, were detected in MO7e lysates. ERK1 and ERK2 protein content were not altered by any of the treatments indicated (lower panel). Similar results were obtained in two additional experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/10/10.1182_blood.v89.10.3582/3/m_bl_0049f1.jpeg?Expires=1767809062&Signature=BehPgzQognumJXc-JhLM5QK0fZWO8N6kSmwktOFr9qQ5iw7qczZQiAhpYhn~XfSgY5JcPMmHY-SuQ2mflDaGDd7SBtAcMF927PZfaQ2i~lFKPfDmOScnvQWkvKZi~afFC~GjrmzIVX9yeOEQmGfmmkhTxgh7IESlY9HD7p8KXG98tiKNvBdyweHlVqmYaS7nJAuzX2J-leEhUVnNRhhCFRfyozl5MFjnceCQxog8T2bjKb2Y~CYVsgu-oUyeTj3-BNxvwbpqSzze~DXzhpSoyNCk97Z9cM8LJMUW4R4ymbYrJic1CmYOqBtRJ4tU9WW9EUZuCLSJotAB2zOM6KKB6Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 3. Effect of chemokine and cytokine treatment on eIF-4E phosphorylation in MO7e cells. Factor-starved MO7e cells maintained in phosphate-free RPMI supplemented with [32P] orthophosphate were treated with GM-CSF, SLF, or a combination of GM-CSF and SLF or with MIP-1α (B and D), IL-8 (B), IP-10 (C and D), or PF4 (D) for 1 hour before treatment with GM-CSF + SLF for various times over a 12-hour period. Control cells (lane 1, A through E) received vehicle only. eIF-4E proteins were isolated from cell lysates by immunoprecipitation with anti–eIF-4E antibodies (A, C, and D) or by m7GTP column chromatography (B), as described in the Materials and Methods. Proteins were separated by 12% SDS-PAGE and transfered to PVDF membranes, and the intensity of 32P-labeling was visualized by autoradiography. At left is indicated the position of the molecular weight markers. eIF-4E appears as a single band at approximately 27 to 29 kD, as indicated by the position of the arrow. (E) eIF-4E protein content was determined by immunoblotting PVDF membranes with anti–eIF-4E antibodies and horseradish peroxidase-linked protein G. eIF-4E proteins were visualized upon exposure of ECL-treated membranes to film. Cells were pretreated with chemokines for 1 hour before 2 hours of treatment with GM-CSF and SLF.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/10/10.1182_blood.v89.10.3582/3/m_bl_0049f3a.jpeg?Expires=1767809062&Signature=4mUjbJC3YZkYCJ9HDowVkfG9ifWZUCxoqjR1VKTksO-wffLVHZOiHyXdYE9ZNVTD6rgvmTNudE0QmFCB4HBvbqLB7uwYhqrGiDqnYgUNDhs~bivgs1jLyQJjOQnSMPgLLLhoN7jI9scCBphffGSQ22EDhJhHbxQUHYrtenHQiuFpUJHU9ioapYroxQTTjYloZ49pMPtPgUEi-8HLIHLLQfdqNzRRxAyvzh9I80Tcl1MsnK5Iny8jJ7uQZNt3tRIbgV3aUKyMJxCMjk1Nma8UIWYJlH1oThZvm0tFQF94RzMT0DPanGbATkdO-YstQ35JuTLI8WbGX5GIS7ge6ZI1Bg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Effect of chemokine and cytokine treatment on eIF-4E phosphorylation in MO7e cells. Factor-starved MO7e cells maintained in phosphate-free RPMI supplemented with [32P] orthophosphate were treated with GM-CSF, SLF, or a combination of GM-CSF and SLF or with MIP-1α (B and D), IL-8 (B), IP-10 (C and D), or PF4 (D) for 1 hour before treatment with GM-CSF + SLF for various times over a 12-hour period. Control cells (lane 1, A through E) received vehicle only. eIF-4E proteins were isolated from cell lysates by immunoprecipitation with anti–eIF-4E antibodies (A, C, and D) or by m7GTP column chromatography (B), as described in the Materials and Methods. Proteins were separated by 12% SDS-PAGE and transfered to PVDF membranes, and the intensity of 32P-labeling was visualized by autoradiography. At left is indicated the position of the molecular weight markers. eIF-4E appears as a single band at approximately 27 to 29 kD, as indicated by the position of the arrow. (E) eIF-4E protein content was determined by immunoblotting PVDF membranes with anti–eIF-4E antibodies and horseradish peroxidase-linked protein G. eIF-4E proteins were visualized upon exposure of ECL-treated membranes to film. Cells were pretreated with chemokines for 1 hour before 2 hours of treatment with GM-CSF and SLF.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/10/10.1182_blood.v89.10.3582/3/m_bl_0049f3b.jpeg?Expires=1767809062&Signature=i6cRKeB8zrF4ks6pjqHGamEkEPQ8BYqBXBYHGYoS7UfZbVvBYgOL8gsKcm1NMJi0fG7YycA~p7eGkfrEvW7eLFXrq9m32o1Q2wCqZtGGxrO2yokiebClyFKZ7o-iITdGf6~IuU3So1yIlXtRVZA95jqXtvCFWRTO~YTM~hDR8ZN2fMBCNVeywVJkvwnD9SNA5UpyO8VpDpP41jAXspNiM27momPGIzFLpqZ5fPYIcMNbEKzJga7cQQyCb~RtYeujFdvO5tCaR75RogDRsGghswawmMvKMUqG5L6OpAj8GSTZh75OM1tjlf04dS2ma-bs0jQQPeTdQppdCnkAyD92Sw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Effect of chemokine and cytokine treatment on eIF-4E phosphorylation in MO7e cells. Factor-starved MO7e cells maintained in phosphate-free RPMI supplemented with [32P] orthophosphate were treated with GM-CSF, SLF, or a combination of GM-CSF and SLF or with MIP-1α (B and D), IL-8 (B), IP-10 (C and D), or PF4 (D) for 1 hour before treatment with GM-CSF + SLF for various times over a 12-hour period. Control cells (lane 1, A through E) received vehicle only. eIF-4E proteins were isolated from cell lysates by immunoprecipitation with anti–eIF-4E antibodies (A, C, and D) or by m7GTP column chromatography (B), as described in the Materials and Methods. Proteins were separated by 12% SDS-PAGE and transfered to PVDF membranes, and the intensity of 32P-labeling was visualized by autoradiography. At left is indicated the position of the molecular weight markers. eIF-4E appears as a single band at approximately 27 to 29 kD, as indicated by the position of the arrow. (E) eIF-4E protein content was determined by immunoblotting PVDF membranes with anti–eIF-4E antibodies and horseradish peroxidase-linked protein G. eIF-4E proteins were visualized upon exposure of ECL-treated membranes to film. Cells were pretreated with chemokines for 1 hour before 2 hours of treatment with GM-CSF and SLF.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/10/10.1182_blood.v89.10.3582/3/m_bl_0049f3c.jpeg?Expires=1767809062&Signature=2sKFP54-rVU8lMqJzxp2dJ8NY03PmJLOqL6vUpo3~SLnBDhV4iXqkNFcJBGu~hnQ2ENJn86Y62EJhB6hMm3O7L5y1exI5jzFUktnLD1NRamwC8kXHKF5G4fPtyc3k1bsAEMBse9w55coNRiMIRzHosz-FbyxKjHUSXvtcQtr9~6FDwwSlRsyuGIqnK6DhdJ2C0FgWvcbDCBIZlOzganssQJ7AuEnhmgd0Dp4~WduAki7ZA2~GsSgzRgxxPSFzdScMS8NFas03~SG9Jf7mcxjWkDuK53w97VWpGeIdLshoZYdl~yZsMAOfohJPI8~6r5RTZdPHWT29CqPKlpLyr-lZg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Effect of chemokine and cytokine treatment on eIF-4E phosphorylation in MO7e cells. Factor-starved MO7e cells maintained in phosphate-free RPMI supplemented with [32P] orthophosphate were treated with GM-CSF, SLF, or a combination of GM-CSF and SLF or with MIP-1α (B and D), IL-8 (B), IP-10 (C and D), or PF4 (D) for 1 hour before treatment with GM-CSF + SLF for various times over a 12-hour period. Control cells (lane 1, A through E) received vehicle only. eIF-4E proteins were isolated from cell lysates by immunoprecipitation with anti–eIF-4E antibodies (A, C, and D) or by m7GTP column chromatography (B), as described in the Materials and Methods. Proteins were separated by 12% SDS-PAGE and transfered to PVDF membranes, and the intensity of 32P-labeling was visualized by autoradiography. At left is indicated the position of the molecular weight markers. eIF-4E appears as a single band at approximately 27 to 29 kD, as indicated by the position of the arrow. (E) eIF-4E protein content was determined by immunoblotting PVDF membranes with anti–eIF-4E antibodies and horseradish peroxidase-linked protein G. eIF-4E proteins were visualized upon exposure of ECL-treated membranes to film. Cells were pretreated with chemokines for 1 hour before 2 hours of treatment with GM-CSF and SLF.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/10/10.1182_blood.v89.10.3582/3/m_bl_0049f3d.jpeg?Expires=1767809062&Signature=eSh~DmqcnM77cKTL1Z0lbLmFiW3qDHTVMTXHpIQjf9eFYQyMTobYP9yJJ9zsZfl3AoNL-Llaw0~nWSHfZR67YpVZibbgLLskVR9DCEDSD6Cr-qNHqtdXJCU0b2qLlP7fYlwcE1hT3VI5jSHoz01fqutsyRyWHtXCLIwApxxrRkwK1yHxYBDB27WUyFrlJE6-3d2O-D1FckdHn0GZrQc3t4WqIy7a4aTobBaQY9fFonyL35LQqgFUVgGbk0juVT2kfkw8teWaJSEiso4E8ybx96nm-f43Vhsa8HUGFdDftheZbhDY0eqno9n-zeD3uAX5m~AN0yivzshfU0jNsI5G6g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Effect of chemokine and cytokine treatment on eIF-4E phosphorylation in MO7e cells. Factor-starved MO7e cells maintained in phosphate-free RPMI supplemented with [32P] orthophosphate were treated with GM-CSF, SLF, or a combination of GM-CSF and SLF or with MIP-1α (B and D), IL-8 (B), IP-10 (C and D), or PF4 (D) for 1 hour before treatment with GM-CSF + SLF for various times over a 12-hour period. Control cells (lane 1, A through E) received vehicle only. eIF-4E proteins were isolated from cell lysates by immunoprecipitation with anti–eIF-4E antibodies (A, C, and D) or by m7GTP column chromatography (B), as described in the Materials and Methods. Proteins were separated by 12% SDS-PAGE and transfered to PVDF membranes, and the intensity of 32P-labeling was visualized by autoradiography. At left is indicated the position of the molecular weight markers. eIF-4E appears as a single band at approximately 27 to 29 kD, as indicated by the position of the arrow. (E) eIF-4E protein content was determined by immunoblotting PVDF membranes with anti–eIF-4E antibodies and horseradish peroxidase-linked protein G. eIF-4E proteins were visualized upon exposure of ECL-treated membranes to film. Cells were pretreated with chemokines for 1 hour before 2 hours of treatment with GM-CSF and SLF.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/10/10.1182_blood.v89.10.3582/3/m_bl_0049f3e.jpeg?Expires=1767809062&Signature=RFcZXgr7N1Nf7XZpzSusWq462Tf-mBjF-beOCIjDk8I1RjKCyt53oPKQi04bh5RN0Yd9lEtQZ7GfQRdBJCREgpQNBHAVMTqx0KHQibHrcj2Vu5EpHIGb4qe3zaF1CHyNeiIatD~OK~7IInaNA5Ytgik1~Cf2NXEdVyCaLK8ngNjyXImcg8KjaTulMq5eprX8NHiAMRIASUzUFysxVVKc-UxqtPovzRv70e14KuBB-fmkjl1e2~1F5Yg05OpXTvPIQfYqR-K0NMdfdITl7~DF-~OADWI3MmQpTa6b0C-gZnMJXk6tw1eysw8PbyUmFpa1Hk3iQyeOqxZiVuPfXE53bA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Effect of chemokine and cytokine treatment on the phosphorylation of 4E-BP1 in MO7e cells. (A) Factor-starved MO7e cells were treated with SLF (lane 2), GM-CSF (lane 3), or SLF + GM-CSF (lane 4) for 1 hour or with MIP-1α (lane 5), PF4 (lane 6), IP-10 (lane 7), or IL-8 (lane 8) for 1 hour before cotreatment with GM-CSF and SLF for 1 hour. 4E-BP1 proteins were immunoprecipitated from cell lysates using anti–4E-BP1 antibodies, separated by 12% SDS-PAGE, and transferred to PVDF membranes. 4E-BP1 protein content was determined as described in the Materials and Methods. 4E-BP1 proteins were visualized upon exposure of ECL-treated membranes to film. 4E-BP1 appears as two protein bands migrating with an approximate molecular weight of 16 to 17 kD, as indicated by the position of the arrow. At left is the position of the molecular weight markers. (B) Factor-starved MO7e cells were cultured with [32P]orthophosphate in phosphate-free RPMI. Treatments are the same as those described in (A). Control cells (lane 1) received vehicle alone. 4E-BP1 proteins were immunoprecipitated from cell lysates using anti–4E-BP1 antibodies, as described in the Materials and Methods. Proteins were separated by 12% SDS-PAGE and transfered to PVDF membranes, and the intensity of 32P-labeling was visualized by autoradiography. At left is the position of the molecular weight markers. Protein content was determined by probing PVDF membranes with anti–4E-BP1 antibodies and visualization with ECL reagents, as described in the Materials and Methods (data not shown). (C) Scanning analysis of autoradiograms for determination of phosphorylation level for both forms of 4E-BP1. Phosphorylation levels of the larger form of 4E-BP1 were significantly higher than control levels for all treatment groups tested (P < .05). Phosphorylation levels corresponding to the smaller form of 4E-BP1 (*) were significantly higher than controls for the GM-CSF and SLF + GM-CSF treatment groups (P < .05). For both forms of 4E-BP1, phosphorylation levels evoked by combined GM-CSF and SLF treatment (**) were significantly higher than levels corresponding to the GM-CSF, SLF, MIP-1α + SLF + GM-CSF, and IP-10 + SLF + GM-CSF treatment groups (P < .05). Each bar represents the mean ± the standard deviation for three determinations obtained in separate experiments. Phosphorylation levels for each band were normalized to 4E-BP1 protein content before statistical analysis. Treatment groups are the same as described in (A). (□) 4E-BP1, larger form; (▪) 4E-BP1, smaller form.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/10/10.1182_blood.v89.10.3582/3/m_bl_0049f4a.jpeg?Expires=1767809062&Signature=TqZ2H-hCdTKzR6CvuF1-OK5d3o90jzM2bTs2IqebhL6T9E5TD0RV4liFNrydxkMhqvXIdG5ye2WpifBeUqKXspfzBYtCZmwvmpaD2f4lSmx1AjfHqalr1Jl342vFZ5tBz3X92Hk5B8mJE-tLLr-NtuwK4IjolEWKPUDd86LaKCs6xQncRF1JFrAEEOhmw0pGpLl~~PmhsDGHJrPQxSuiInirk70LZPlBGXhFzsMb9fbvOAbBQBaswdOBkqqIsCVWPYQT4dZQanLj1MHG0uh~v1o-xsZ-sKU7SSo2Yubb7Rtc19Aa2ITfvrJ~~nSJSC4IKCZl40LqdOay9kwM36aZbg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Effect of chemokine and cytokine treatment on the phosphorylation of 4E-BP1 in MO7e cells. (A) Factor-starved MO7e cells were treated with SLF (lane 2), GM-CSF (lane 3), or SLF + GM-CSF (lane 4) for 1 hour or with MIP-1α (lane 5), PF4 (lane 6), IP-10 (lane 7), or IL-8 (lane 8) for 1 hour before cotreatment with GM-CSF and SLF for 1 hour. 4E-BP1 proteins were immunoprecipitated from cell lysates using anti–4E-BP1 antibodies, separated by 12% SDS-PAGE, and transferred to PVDF membranes. 4E-BP1 protein content was determined as described in the Materials and Methods. 4E-BP1 proteins were visualized upon exposure of ECL-treated membranes to film. 4E-BP1 appears as two protein bands migrating with an approximate molecular weight of 16 to 17 kD, as indicated by the position of the arrow. At left is the position of the molecular weight markers. (B) Factor-starved MO7e cells were cultured with [32P]orthophosphate in phosphate-free RPMI. Treatments are the same as those described in (A). Control cells (lane 1) received vehicle alone. 4E-BP1 proteins were immunoprecipitated from cell lysates using anti–4E-BP1 antibodies, as described in the Materials and Methods. Proteins were separated by 12% SDS-PAGE and transfered to PVDF membranes, and the intensity of 32P-labeling was visualized by autoradiography. At left is the position of the molecular weight markers. Protein content was determined by probing PVDF membranes with anti–4E-BP1 antibodies and visualization with ECL reagents, as described in the Materials and Methods (data not shown). (C) Scanning analysis of autoradiograms for determination of phosphorylation level for both forms of 4E-BP1. Phosphorylation levels of the larger form of 4E-BP1 were significantly higher than control levels for all treatment groups tested (P < .05). Phosphorylation levels corresponding to the smaller form of 4E-BP1 (*) were significantly higher than controls for the GM-CSF and SLF + GM-CSF treatment groups (P < .05). For both forms of 4E-BP1, phosphorylation levels evoked by combined GM-CSF and SLF treatment (**) were significantly higher than levels corresponding to the GM-CSF, SLF, MIP-1α + SLF + GM-CSF, and IP-10 + SLF + GM-CSF treatment groups (P < .05). Each bar represents the mean ± the standard deviation for three determinations obtained in separate experiments. Phosphorylation levels for each band were normalized to 4E-BP1 protein content before statistical analysis. Treatment groups are the same as described in (A). (□) 4E-BP1, larger form; (▪) 4E-BP1, smaller form.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/10/10.1182_blood.v89.10.3582/3/m_bl_0049f4b.jpeg?Expires=1767809062&Signature=kzGi4ibj1fnn8GDXWeANZAh-R1OVwVJkineumumA5MTaOsmz4Bz4N9XuvtYx~Bg4v0hvLYfLHMmuytH-P20W0cBAUyZCtbF39e4eu45FQbm3N6ImsoeGhfBdNyxrBEFSUjZ1S8QT5Nr6Oeu0Q5yiPfpRrz6wP1WI8l13KaU6Iof3kulAT-8Ubq-r5yYyvhdZlGWl-DnN8nIPPTpr375xpjBHRIvb0o2d0UafCZ0whIKWgHOox6E6WPA8F6LqTCo~05LBt~BX3OmLF7-1oGaRrLjJNWIJcU3ijYdgj0WUwaIrNBICxfW0QB7LESmnIT40uKrfRuI4VaQNi8l-IuqF0g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Effect of chemokine and cytokine treatment on the phosphorylation of 4E-BP1 in MO7e cells. (A) Factor-starved MO7e cells were treated with SLF (lane 2), GM-CSF (lane 3), or SLF + GM-CSF (lane 4) for 1 hour or with MIP-1α (lane 5), PF4 (lane 6), IP-10 (lane 7), or IL-8 (lane 8) for 1 hour before cotreatment with GM-CSF and SLF for 1 hour. 4E-BP1 proteins were immunoprecipitated from cell lysates using anti–4E-BP1 antibodies, separated by 12% SDS-PAGE, and transferred to PVDF membranes. 4E-BP1 protein content was determined as described in the Materials and Methods. 4E-BP1 proteins were visualized upon exposure of ECL-treated membranes to film. 4E-BP1 appears as two protein bands migrating with an approximate molecular weight of 16 to 17 kD, as indicated by the position of the arrow. At left is the position of the molecular weight markers. (B) Factor-starved MO7e cells were cultured with [32P]orthophosphate in phosphate-free RPMI. Treatments are the same as those described in (A). Control cells (lane 1) received vehicle alone. 4E-BP1 proteins were immunoprecipitated from cell lysates using anti–4E-BP1 antibodies, as described in the Materials and Methods. Proteins were separated by 12% SDS-PAGE and transfered to PVDF membranes, and the intensity of 32P-labeling was visualized by autoradiography. At left is the position of the molecular weight markers. Protein content was determined by probing PVDF membranes with anti–4E-BP1 antibodies and visualization with ECL reagents, as described in the Materials and Methods (data not shown). (C) Scanning analysis of autoradiograms for determination of phosphorylation level for both forms of 4E-BP1. Phosphorylation levels of the larger form of 4E-BP1 were significantly higher than control levels for all treatment groups tested (P < .05). Phosphorylation levels corresponding to the smaller form of 4E-BP1 (*) were significantly higher than controls for the GM-CSF and SLF + GM-CSF treatment groups (P < .05). For both forms of 4E-BP1, phosphorylation levels evoked by combined GM-CSF and SLF treatment (**) were significantly higher than levels corresponding to the GM-CSF, SLF, MIP-1α + SLF + GM-CSF, and IP-10 + SLF + GM-CSF treatment groups (P < .05). Each bar represents the mean ± the standard deviation for three determinations obtained in separate experiments. Phosphorylation levels for each band were normalized to 4E-BP1 protein content before statistical analysis. Treatment groups are the same as described in (A). (□) 4E-BP1, larger form; (▪) 4E-BP1, smaller form.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/10/10.1182_blood.v89.10.3582/3/m_bl_0049f4c.jpeg?Expires=1767809062&Signature=l2xcgpwFxOGdwLj9T4J6YoDIEPKRrlF34UBI51KvjUvDZBRtkw14aYab4wCNfbAuvV7uW6mjC8DQeq9VNcs5pUrhXGVSrn4yYp9GoYrnBsjPPkIY7q~ZkKxqT8rsCrkg3vm44n8OKMIPBm4xYn9IKzhxmunIJPd~cFMUOcXpPZW~KtNlsENcJh7X94~Mb0X0estAa~spOfDjbUayPxEQ4YGeXdX7vdX6-KmA0uOfm8~IsoFwZKtbVPVUecYsgOPuFzf7FB6C9o~USw~rTsaJ16ihsURHZB14YpQUFU7SPfGM01-W5eWlR1ZN706U4Mx2Lk-3-NwOlkiBAmV8WKnWMQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
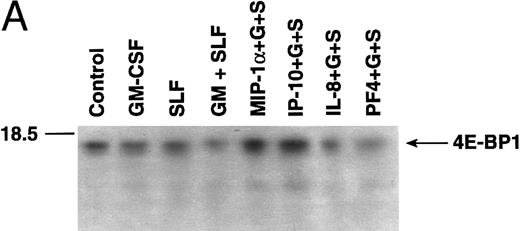

![Fig. 6. Effect of cAMP pretreatment on 4E-BP1 phosphorylation levels. Factor-starved MO7e cells were cultured with [32P]orthophosphate in phosphate-free RPMI, as described in the Materials and Methods. Cells were treated with SLF in combination with GM-CSF (lanes 2, 4, and 6) for 1 hour or with 8-bromo-cAMP (lane 3), MIP-1α (lane 5), or IP-10 (lane 7) for 1 hour before cotreatment with GM-CSF and SLF for 1 hour. Control cells (lane 1) received vehicle alone. Immunoprecipitated 4E-BP1 proteins were separated by 12% SDS-PAGE and transfered to PVDF membranes, and the intensity of 32P-labeling was visualized by autoradiography. At left is the position of the molecular weight markers. 4E-BP1 appears as a doublet of two protein bands migrating with an approximate molecular weight of 16 to 17 kD, as indicated to the right of the figure. Similar results were obtained in each of two additional experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/10/10.1182_blood.v89.10.3582/3/m_bl_0049f6.jpeg?Expires=1767809062&Signature=p5oOukqr8D2bXVG3Qto4Z4dJW1Lj~OxeKX5SnDBcDQsvsIunaSBSefcJdZPq8VrN44onUCyQAc9dp65VJQ2Br-ZQ6JAFXnEjruyBk-Jd7kKrr87aK1lmoN3BQC4jyWguieYamLkONSZykivk7EnDO9pdq59-z7Ioz1ZGzToKgor7MOtwhzRsjOUiRWYIXj4BCyTiQ51NzG58E8Qk9SzCEvp6EZYridwNc6jG4UiD7jRpmTSqQ8E29Wi8D57ni63NE3KKn4x9LfsPOjD5k~78ReadjT9SEEIhXgoXbOGaVzc-0jXufJ4WWY3TtLEgKQ4wfhIOn7YFMFqdSpWlKaziUg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 8. Effect of rapamycin, wortmannin, and PD098059 on eIF-4E/4E-BP1 phosphorylation and association in MO7e cells. (A and B) Factor-starved MO7e cells were treated with GM-CSF (100 U/mL) and SLF (50 ng/mL) for 15 minutes or were pulse treated with rapamycin, wortmannin, or PD098059 for 1 hour and washed twice with culture medium before 15 minutes of treatment with GM-CSF and SLF in the presence of [32P]orthophosphate. eIF-4E (A) and 4E-BP1 (B) proteins were isolated from cell lysates using anti–eIF-4E and anti–4E-BP1 antibodies, respectively, separated by SDS-PAGE, and transferred to PVDF membranes, and the intensity of [32P] radiolabeling was determined by autoradiography. Combined treatment of MO7e cells with GM-CSF and SLF increased eIF-4E phosphorylation (A) and phosphorylation of both forms of 4E-BP1 (B). Pretreatment of cells with rapamycin, wortmannin, and PD098059 blocked the stimulatory effects of GM-CSF and SLF on 4E-BP1 phosphorylation, whereas only wortmannin and PD098059 suppressed growth factor stimulation of eIF-4E phosphorylation (A). eIF-4E and 4E-BP1 content were not altered by any of the treatments indicated (data not shown). Similar results were obtained in two additional experiments. (C) Factor-starved MO7e cells were pretreated with 50 ng/mL MIP-1α, IP-10, IL-8, or PF4 or 20 ng/mL rapamycin, 10−7 mol/L wortmannin, or 50 μmol/L PD098059 for 1 hour before combined GM-CSF and SLF treatment for 15 minutes. eIF-4E proteins isolated from cell lysates by m7GTP affinity column chromatography were separated by SDS-PAGE, transferred to PVDF membranes, and probed for eIF-4E and 4E-BP1/BP2 content using anti–eIF-4E and anti–4E-BP1/BP2 antibodies, as described in the Materials and Methods. 4E-BP1 coisolated with eIF-4E in control, quiescent cells (lane 1). Treatment of MO7e cells with GM-CSF and SLF results in the dissociation of 4E-BP1 from eIF-4E, as indicated by the lack of 4E-BP1 content in lane 2 (lower panel). Pretreatment of cells with MIP-1α, IP-10, rapamycin, wortmannin, and, to a lesser extent, PD098059 suppressed the dissociation of eIF-4E from 4E-BP1 in the presence of GM-CSF and SLF (lower panel, lanes 3, 5, 7, 8, and 9). In contrast, 4E-BP1 dissociated from eIF-4E in MO7e cells pretreated with IL-8 or PF4 (lanes 4 and 6). Similar results were obtained in two separate experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/10/10.1182_blood.v89.10.3582/3/m_bl_0049f8a.jpeg?Expires=1767809062&Signature=JwLapgHRvt8d1168eciF4FscRZ6mNKQuBa6wncIeVIBJNFrYoyOfeG1mIOaHYFPWyb2nqnBMp7LQ6~jdU3En7203bRnzfb~zRKnCrHaQ9oaXmVe4oz8pyU2Ky3N0A1di6y2V0t4FaYaeSvIVJINjVbq7YzZFhQ5Z~oexq6w26PBcFSNqkgGGzsa9iY0D3SKZYcPh7iggR6k186cF4TDxsmbetYh31o9feJI--wQppi~QAeUcJ7IOCK5qsu2MqdaXr3sGMEi-VdTbtrJBdS0FfynhKRP0URivnzfXAGMtouxLofVYynuGNj~IoHTz1YEkvXLWjm8ysFgB52hs8TQ67Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 8. Effect of rapamycin, wortmannin, and PD098059 on eIF-4E/4E-BP1 phosphorylation and association in MO7e cells. (A and B) Factor-starved MO7e cells were treated with GM-CSF (100 U/mL) and SLF (50 ng/mL) for 15 minutes or were pulse treated with rapamycin, wortmannin, or PD098059 for 1 hour and washed twice with culture medium before 15 minutes of treatment with GM-CSF and SLF in the presence of [32P]orthophosphate. eIF-4E (A) and 4E-BP1 (B) proteins were isolated from cell lysates using anti–eIF-4E and anti–4E-BP1 antibodies, respectively, separated by SDS-PAGE, and transferred to PVDF membranes, and the intensity of [32P] radiolabeling was determined by autoradiography. Combined treatment of MO7e cells with GM-CSF and SLF increased eIF-4E phosphorylation (A) and phosphorylation of both forms of 4E-BP1 (B). Pretreatment of cells with rapamycin, wortmannin, and PD098059 blocked the stimulatory effects of GM-CSF and SLF on 4E-BP1 phosphorylation, whereas only wortmannin and PD098059 suppressed growth factor stimulation of eIF-4E phosphorylation (A). eIF-4E and 4E-BP1 content were not altered by any of the treatments indicated (data not shown). Similar results were obtained in two additional experiments. (C) Factor-starved MO7e cells were pretreated with 50 ng/mL MIP-1α, IP-10, IL-8, or PF4 or 20 ng/mL rapamycin, 10−7 mol/L wortmannin, or 50 μmol/L PD098059 for 1 hour before combined GM-CSF and SLF treatment for 15 minutes. eIF-4E proteins isolated from cell lysates by m7GTP affinity column chromatography were separated by SDS-PAGE, transferred to PVDF membranes, and probed for eIF-4E and 4E-BP1/BP2 content using anti–eIF-4E and anti–4E-BP1/BP2 antibodies, as described in the Materials and Methods. 4E-BP1 coisolated with eIF-4E in control, quiescent cells (lane 1). Treatment of MO7e cells with GM-CSF and SLF results in the dissociation of 4E-BP1 from eIF-4E, as indicated by the lack of 4E-BP1 content in lane 2 (lower panel). Pretreatment of cells with MIP-1α, IP-10, rapamycin, wortmannin, and, to a lesser extent, PD098059 suppressed the dissociation of eIF-4E from 4E-BP1 in the presence of GM-CSF and SLF (lower panel, lanes 3, 5, 7, 8, and 9). In contrast, 4E-BP1 dissociated from eIF-4E in MO7e cells pretreated with IL-8 or PF4 (lanes 4 and 6). Similar results were obtained in two separate experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/10/10.1182_blood.v89.10.3582/3/m_bl_0049f8b.jpeg?Expires=1767809062&Signature=SVSEYulUGL-x7L1XRMij7LH1ZiD8dKnRmJp0ewEDK3sLUPicIpD6SaMLEFDqLqPrOp5AszznXiQ-3PkOy0cqG3o06nWAV2HNTVPtzxRnWxIHoHAYM6htjL4lRTyQ8yT2CH2Qth5IPY2XzKwCXxwwqgNYjqy6cNXtY2pxy1hC8JPWmmHE8j69ivouQPb~tHNAv5nv81RXhNdOWS09qH7n0pqCEyYKDvSW2lMIcU6n9tm6llGh0HZZBqMWLU2nCb09zA6~UwiU3CxEqkxTc6Z8S4iu6TrklWGYyckM9URvrcKmxovo2FMklajnvVqT5gRpohWp~cbr5Tkhl33gWtcfKg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 8. Effect of rapamycin, wortmannin, and PD098059 on eIF-4E/4E-BP1 phosphorylation and association in MO7e cells. (A and B) Factor-starved MO7e cells were treated with GM-CSF (100 U/mL) and SLF (50 ng/mL) for 15 minutes or were pulse treated with rapamycin, wortmannin, or PD098059 for 1 hour and washed twice with culture medium before 15 minutes of treatment with GM-CSF and SLF in the presence of [32P]orthophosphate. eIF-4E (A) and 4E-BP1 (B) proteins were isolated from cell lysates using anti–eIF-4E and anti–4E-BP1 antibodies, respectively, separated by SDS-PAGE, and transferred to PVDF membranes, and the intensity of [32P] radiolabeling was determined by autoradiography. Combined treatment of MO7e cells with GM-CSF and SLF increased eIF-4E phosphorylation (A) and phosphorylation of both forms of 4E-BP1 (B). Pretreatment of cells with rapamycin, wortmannin, and PD098059 blocked the stimulatory effects of GM-CSF and SLF on 4E-BP1 phosphorylation, whereas only wortmannin and PD098059 suppressed growth factor stimulation of eIF-4E phosphorylation (A). eIF-4E and 4E-BP1 content were not altered by any of the treatments indicated (data not shown). Similar results were obtained in two additional experiments. (C) Factor-starved MO7e cells were pretreated with 50 ng/mL MIP-1α, IP-10, IL-8, or PF4 or 20 ng/mL rapamycin, 10−7 mol/L wortmannin, or 50 μmol/L PD098059 for 1 hour before combined GM-CSF and SLF treatment for 15 minutes. eIF-4E proteins isolated from cell lysates by m7GTP affinity column chromatography were separated by SDS-PAGE, transferred to PVDF membranes, and probed for eIF-4E and 4E-BP1/BP2 content using anti–eIF-4E and anti–4E-BP1/BP2 antibodies, as described in the Materials and Methods. 4E-BP1 coisolated with eIF-4E in control, quiescent cells (lane 1). Treatment of MO7e cells with GM-CSF and SLF results in the dissociation of 4E-BP1 from eIF-4E, as indicated by the lack of 4E-BP1 content in lane 2 (lower panel). Pretreatment of cells with MIP-1α, IP-10, rapamycin, wortmannin, and, to a lesser extent, PD098059 suppressed the dissociation of eIF-4E from 4E-BP1 in the presence of GM-CSF and SLF (lower panel, lanes 3, 5, 7, 8, and 9). In contrast, 4E-BP1 dissociated from eIF-4E in MO7e cells pretreated with IL-8 or PF4 (lanes 4 and 6). Similar results were obtained in two separate experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/10/10.1182_blood.v89.10.3582/3/m_bl_0049f8c.jpeg?Expires=1767809062&Signature=u~IbaGDlu1qf~qwcOz5tahYZ-ebXZjbU2NT5AsqRVoBdoVykHneXnpkwklDu6CJohv5kSjKQzNV-CLiz11YCmBTj3TJ-hM2h7ZqXgF40AodUJbyPr3FHwo0BvdzJlkIO10U0aMGUyzJQM2ZtOWRxkgCIGRgIUTHe71~EUbYch3ejRaBq4ECg4Xb2a56~o0g-d1QuJxQHRsmz9WyY5NS4oBvvvJhRzTNldun~5i76gNQd5pUeZAkfRTZXGh12mSb88L0kGXCissc69Jtjq-bPV1i3Db2rPeIEwEVnSS6EAcGEL0lp~Lr~lgeo0azX-ZfYNzcNaLFYwd3TCevXvuz2PA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal