Abstract
To gain some insight into the role of c-fes in macrophage differentiation, we have analyzed the ability of HL60 leukemic promyelocytic cells and FDC-P1/MAC-11 murine myeloid precursor cells to differentiate in response to phorbol esters after inhibition of c-fes function. Fes inactivation has been obtained by using oligodeoxynucleotides (ODN) complementary to the 5′ region of c-fes mRNA and to 5′ splice junctions of c-fes primary transcript. After 5 days (d) in culture, in several separate experiments performed with different ODN preparations, a complete inhibition of c-fes expression was observed in HL60 and in FDC-P1/MAC-11 cells. No perturbation of cell growth was evident in our experimental conditions in both cell lines after c-fes inhibition. Furthermore, in HL60 cells lacking c-fes product, an almost complete downregulation of the α4β1 fibronectin receptor occurred. However, in both cell lines, the induction of macrophage differentiation by phorbol esters resulted in an almost complete maturation arrest as evaluated by morphological, cytochemical, immunological criteria, and by the cytofluorimetric cell cycle analysis. A loss of the adhesion capacity of both myeloid cell lines, when compared to terminally differentated macrophages, was also observed. These results suggest that HL60 and FDC-P1/MAC-11 cells, when treated with phorbol 12-myristate 13-acetate, require c-fes protein expression to activate the genetic program underlying macrophage differentiation.
THE c-fes proto-oncogene encodes a tyrosine kinase protein, which is specifically expressed in immature and differentiated hematopoietic cells of the myeloid lineage.1-3 Expression of c-fes protein is evident in mature granulocytes, monocytes, differentiated HL60 cells, leukemic cell lines such as KG1, THP1, and U937, which can be induced to differentiate along the granulocyte and monocyte pathways, but is absent in K562 cell line which is resistant to myeloid differentiation.4 Fes expression is expecially high in acute and chronic myeloid leukemias.1,5 These studies indicate that the c-fes proto-oncogene is associated with myeloid differentiation. Other experiments suggest that c-fes may be considered a regulatory gene in granulocyte differentiation. In fact it has been shown that transfection of human c-fes gene in K562 leukemic cells restore their capability to undergo myeloid differentiation.6 Furthermore, inhibition of c-fes expression in HL60 cells by AS oligodeoxynucleotides (ODN) leads to a complete block of granulocytic differentiation due to the activation of programmed cell death.7,8 No perturbation of the proliferative capacity of HL60 cells lacking c-fes protein was observed.9 Inactivation of the c-fes product do not interfere with monocytic differentiation induced in HL60 cells by vitamin D3 (Vit D3).10 Instead phorbol 12-myristate 13-acetate (PMA)-treated HL60 cells lacking c-fes protein (c-fes− HL60 cells), never show the plastic-adherent behavior characteristic of terminally differentiated macrophages.7,10 To gain some insight into the function of the c-fes proto-oncogene in macrophage differentiation we have inhibited the expression of this gene in HL60 and in FDC-P1/MAC-11 cells induced to differentiate with PMA, using the methodology of antisense (AS) ODN.11,12 We have monitored the differentiation capacity of these cells, and particularly in HL60 cells the level of expression of several integrins13-15 and of several extracellular matrix proteins.16 Different AS-ODN were used to inactivate c-fes expression both at the mRNA or at the primary transcript levels. Our studies show that the inactivation of c-fes protooncogene in PMA-treated HL60 cells leads to: (1) an almost complete block of macrophage differentiation as evaluated by morphological, cytochemical, immunological criteria, and by flow cytometric cell cycle analysis; (2) an almost complete loss of the adhesion capacity; (3) a complete downregulation of the α4β1 fibronectin receptor (CD49d), but not of other integrins such as CD49e, CD49f, CD51, CD11a and CD11b or extracellular matrix proteins such as fibronectin, laminin and vitronectin. Furthermore, inactivation of c-fes protooncogene in the more physiological context of FDC-P1/MAC-11 myeloid precursor cells confirms the biological behavior observed in HL60 cells, since a complete block of PMA induced macrophage differentiation is observed as evaluated by morphological, immunological, functional, and flow cytometric cell cycle analysis.
MATERIALS AND METHODS
Cell cultures and macrophage differentiation induction.Human HL60 and mouse FDC-P1/MAC-1117 (a kind gift from Dr Teruko Tamura, Inst. fur Virologie, Gessen, Germany) cells were cultured in 24-well tissue culture plates. The cells were seeded at the initial concentration of 5 × 105 cells/mL (HL60) or 5 × 104 cells/mL (FDC-P1/MAC-11 ) in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum (FCS) and 2 mmol/L L-glutamine; furthermore, FDC-P1/MAC-11 cells require 5% of WEHI 3B conditioned medium, interleukin-3 (IL-3) rich (WEHI 3B cells were obtained by DSM, German collection of microorganisms and cell cultures, Braunschweig, Germany). Macrophage differentiation was induced by addition of 16 nmol/L PMA (Sigma Chemical Co, St Louis, MO). In these culture conditions, terminal differentiation was obtained after 48 hours (h).10 Morphology was assayed by cytocentrifugation followed by May Grunwald Giemsa staining. Cytochemical reactions for naphthol-AS-D-chloroacetate esterase, α-naphthyl butyrrate esterase, and myeloperoxydase were performed by standard methods18,19 counting at least 300 cells for each sample. Differentiation was also assayed in HL60 cells by flow cytometric analysis monitoring CD11b, CD11c, and CD14 expression20 and in FDC-P1/MAC-11 cells monitoring the Mac-3 and CD11b expression.17 Purified peripheral blood granulocytes or mononuclear cells were used as positive control for cytochemical and immunological analysis.21 Control cell lines K562 and NIH 3T3, not expressing the c-fes proto-oncogene, were maintained, respectively, in RPMI 1640 and DMEM media supplemented with 10% FCS and 2 mmol/L L-glutamine.
Synthesis and purification of ODN.Phosphodiesters ODN were synthesized on an automated solid-phase synthesizer (DNA/RNA synthesizer, Mod. 394, Applied Biosystems Inc, Foster City, CA) by standard β-cyanoethyl-phosphoramidite chemistry, extracted several times with ammonium-hydroxyde and ethanol precipitated as described.22 We used Phosphodiesters ODN to avoid the aspecific effect of charged ODN such as the Phosphorothioates, whose behavior is that of polyanions.23 All ODN were lyophilized and dissolved directly in cell culture medium. The sequences of ODN and their relationship to the organization of human and mouse c-fes coding regions24-26 are shown (see Tables 1 and 2). All the sequences were compared with the National Institute of Health gene bank by DNAsis software (Hitachi, Brisbaine, CA) to avoid homologies greater than 70% with other gene sequences. Furthermore the 5′ region of the c-fes proto-oncogene, where the AS ODN were selected, can be considered unique.27,28 In fact the comparison with the 5′ regions of genes encoding other tyrosine kinases,29 and particularly that belonging to the src family,30 revealed a low degree of homology. We have synthesized several ODN to inactivate c-fes expression at two different levels: (1) at mRNA level, strategy employed in both cell lines, by using a mixture of 3 ODN complementary to 5′ region of c-fes mRNA, near the ATG codon, labeled respectively hFES-AS1, 2, 3 (HL60 cells), and mFES-AS1, 2, 3 (FDC-P1/MAC-11 cells); (2) at the primary transcript level, strategy employed only in HL60 cells, by using a mixture of 2 ODN complementary to 2 different 5′ splice junctions (exon2-intron2, exon3-intron3), labeled hFES-AS-sj1 and 2. For all the AS ODN we synthesized the corresponding control ODN labeled sense (FES-S) and inverted polarity (FES-IP), see Tables 1 and 2.
Nucleotide Sequence of the Oligomers Used in HL60 Cells
| Oligomer . | Sequence (5′-3′ ) . | Mer . | Target . | Region . |
|---|---|---|---|---|
| hFES-S1 | GGCTTCTCTTCCGAGCTG | 18 | Exon 2 | 79-96* |
| hFES-IP1 | CCGAAGAGAAGGCTCGAC | 18 | Exon 2 | 96-79* |
| hFES-AS1 | CAGCTCGGAAGAGAAGCC | 18 | Exon 2 | 96-79* |
| hFES-S2 | GCGGAACAGCACTATG | 16 | Exon 1/Exon 2 | 63-78* |
| hFES-IP2 | CGCCTTGTCGTGATAC | 16 | Exon 1/Exon 2 | 78-63* |
| hFES-AS2 | CATAGTGCTGTTCCGC | 16 | Exon 1/Exon 2 | 78-63* |
| hFES-S3 | TGCAGCCCCCAGGGC | 15 | Exon 2 | 97-111* |
| hFES-IP3 | ACGTCGGGGGTCCCG | 15 | Exon 2 | 111-97* |
| hFES-AS3 | GCCCTGGGGGCTGCA | 15 | Exon 2 | 111-97* |
| hFES-S-sj1 | CAGCCCCATCAGTCAGGTGGGTCTCTATGGGAC | 33 | Splice junction exon 2/intron 2 | 1211-1243† |
| hFES-IP-sj1 | GTCGGGGTAGTCAGTCCACCCAGAGATACCCTG | 33 | Splice junction exon 2/intron 2 | 1243-1211† |
| hFES-AS-sj1 | GTCCCATAGAGACCCACCTGACTGATGGGGCTG | 33 | Splice junction exon 2/intron 2 | 1243-1211† |
| hFES-S-sj2 | GCAGGAGCTCACCAAGGTGAGCGGGCAGCACT | 32 | Splice junction exon 3/intron 3 | 1537-1568† |
| hFES-IP-sj2 | CGTCCTCGAGTGGTTCCACTCGCCCGTCGTGA | 32 | Splice junction exon 3/intron 3 | 1568-1537† |
| hFES-AS-sj2 | AGTGCTGCCCGCTCACCTTGGTGAGCTCCTGC | 32 | Splice junction exon 3/intron 3 | 1568-1537† |
| Oligomer . | Sequence (5′-3′ ) . | Mer . | Target . | Region . |
|---|---|---|---|---|
| hFES-S1 | GGCTTCTCTTCCGAGCTG | 18 | Exon 2 | 79-96* |
| hFES-IP1 | CCGAAGAGAAGGCTCGAC | 18 | Exon 2 | 96-79* |
| hFES-AS1 | CAGCTCGGAAGAGAAGCC | 18 | Exon 2 | 96-79* |
| hFES-S2 | GCGGAACAGCACTATG | 16 | Exon 1/Exon 2 | 63-78* |
| hFES-IP2 | CGCCTTGTCGTGATAC | 16 | Exon 1/Exon 2 | 78-63* |
| hFES-AS2 | CATAGTGCTGTTCCGC | 16 | Exon 1/Exon 2 | 78-63* |
| hFES-S3 | TGCAGCCCCCAGGGC | 15 | Exon 2 | 97-111* |
| hFES-IP3 | ACGTCGGGGGTCCCG | 15 | Exon 2 | 111-97* |
| hFES-AS3 | GCCCTGGGGGCTGCA | 15 | Exon 2 | 111-97* |
| hFES-S-sj1 | CAGCCCCATCAGTCAGGTGGGTCTCTATGGGAC | 33 | Splice junction exon 2/intron 2 | 1211-1243† |
| hFES-IP-sj1 | GTCGGGGTAGTCAGTCCACCCAGAGATACCCTG | 33 | Splice junction exon 2/intron 2 | 1243-1211† |
| hFES-AS-sj1 | GTCCCATAGAGACCCACCTGACTGATGGGGCTG | 33 | Splice junction exon 2/intron 2 | 1243-1211† |
| hFES-S-sj2 | GCAGGAGCTCACCAAGGTGAGCGGGCAGCACT | 32 | Splice junction exon 3/intron 3 | 1537-1568† |
| hFES-IP-sj2 | CGTCCTCGAGTGGTTCCACTCGCCCGTCGTGA | 32 | Splice junction exon 3/intron 3 | 1568-1537† |
| hFES-AS-sj2 | AGTGCTGCCCGCTCACCTTGGTGAGCTCCTGC | 32 | Splice junction exon 3/intron 3 | 1568-1537† |
Nucleotide Sequence of the Oligomers Used in FDC-P1/MAC-11 Cells
| Oligomer . | Sequence (5′-3′ ) . | Mer . | Region* . |
|---|---|---|---|
| mFES-IP1 | CCGAAGAGAAGTCTCGAC | 18 | 63-80 |
| mFES-AS1 | CAGCTCTGAAGAGAAGCC | 18 | 80-63 |
| mFES-IP2 | GGTCCTGTGGTAATAC | 16 | 47-62 |
| mFES-AS2 | CATAATGGTGTCCTGG | 16 | 62-47 |
| mFES-IP3 | ACGTCGGGGGTCCCG | 15 | 81-95 |
| mFES-AS3 | GCCCTGGGGGCTGCA | 15 | 95-81 |
| Oligomer . | Sequence (5′-3′ ) . | Mer . | Region* . |
|---|---|---|---|
| mFES-IP1 | CCGAAGAGAAGTCTCGAC | 18 | 63-80 |
| mFES-AS1 | CAGCTCTGAAGAGAAGCC | 18 | 80-63 |
| mFES-IP2 | GGTCCTGTGGTAATAC | 16 | 47-62 |
| mFES-AS2 | CATAATGGTGTCCTGG | 16 | 62-47 |
| mFES-IP3 | ACGTCGGGGGTCCCG | 15 | 81-95 |
| mFES-AS3 | GCCCTGGGGGCTGCA | 15 | 95-81 |
GenBank, Accession No. X12616.
Determination of ODN stability.All the experiments were performed with a single lot of FCS which has been inactivated at 65°C for 20 minutes, and selected for minimal nuclease activity. Low specific radioactivity (32P)-end labeled31 oligomer diluted with unlabeled oligomer at a final concentration of 10 μmol/L was incubated in the cell culture for 24, 48, 72, and 96 hours at 37°C. ODN integrity was evaluated by polyacrilamide gel electrophoresis followed by autoradiography. We selected the FCS where the ODN was stable for at least 24 hours.
Delivery system of the c-fes ODN into the cells.Myeloid cells express high levels of transferrin receptor. We used in our experiments a delivery system based on receptor-mediated endocytosis to introduce the c-fes ODN complexed with transferrin-polylysine conjugate into HL60 and FDC-P1/MAC-11 cells as described.32 With this delivery system the ODN uptake efficiency was about ten times higher as compared with noncomplexed ODN. The uptake efficiency was evaluated as described.32 The DNA/RNA duplex resistant to S1 nuclease digestion was detected 4 and 24 hours after culture of HL60 cells in the presence of labeled c-fes AS ODN-transferrin-polylysine complex as described.33
ODN treatment of the cells.FES-S, IP, or AS ODN were added to the cultures at an initial concentration of 10 μmol/L, and 5 μmol/L ODN were added every 24 hours. Other control cultures were left untreated. After 120 hours of ODN treatment the culture medium was replaced with fresh medium, containing 10 μmol/L c-fes ODN and 16 nmol/L PMA.
Detection of c-fes mRNA by reverse transcription-polymerase chain reaction (RT-PCR) analysis.Total cellular RNA was extracted from 5 × 10 5 cells exposed to FES ODN for 120 hours and from untreated cells, in the presence of 10 μg of Escherichia coli tRNA, using a modification of the guanidinium isothiocyanate procedure.34 To avoid genomic DNA contamination all samples were digested with RQ1-DNAse for 30 minutes at 37°C, by adding 10 U of RQ1 (Boehringer, Mannheim, Germany), 40 U RNAsin (Boehringer), 1 × RQ1 buffer (40 mmol/L Tris pH 7.9, 10 mmol/L NaCl, 0.6 mmol/L MgCl 2 , 0.01 mmol/L CaCl2 ), to a final volume of 200 mL. One microgram of total RNA extracted from each sample was reverse transcribed using 400 U of Mu-MLV reverse transcriptase (GIBCO-BRL, Gaithersburg, MD) and 1 μg of Oligo-dT15 primer (Boehringer) for 1 hour at 42°C in 1 × RT buffer (50 mmol/L Tris pH 8.3, 60 mmol/L KCl, 1.5 mmol/L MgCl2 , 10 mmol/L Dithiothreitol, 1 mmol/L dATP, dCTP, dGTP, dTTP, 20 U RNAsin, and 0.1 mg/mL bovine serum albumin), in a total volume of 30 μL. The cDNA was then heated at 95°C for 3 minutes and stored at 4°C. One microliter of cDNA was then amplified by adding 2.5 U of Taq polymerase (Boehringer), 0.5 μg of specific direct and reverse primers (see legend of Figs 1 and 2) in a total volume of 50 μL, in 10 mmol/L Tris pH 8.3, 50 mmol/L KCl, 1.5 mmol/L MgCl2 and 200 μmol/L dNTPs.35
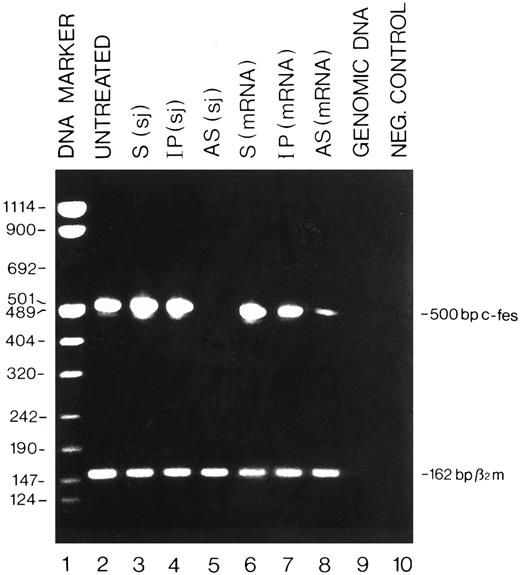
Detection by RT-PCR of the c-fes mRNA in HL60 cells treated with c-fes ODN. Total cellular RNA was extracted after 5 days of c-fes AS ODN treatment as described in Materials and Methods. The 500-bp amplified c-fes fragment was obtained using the oligonucleotide primers hFES-DP (5′-ACTATGGGCTTCTCTTCCGAGCTG-3′ ) and hFES-RP (5′-TCACGGTCCTTGTCTTTGCTGGCCT-3′ ). The 162-bp human β2-microglobulin fragment, used as a quantitative control, was obtained using the oligonucleotide primers hβ2m-DP (5′-CTCGCGCTACTCTCTCTTTCT-3′ ) and hβ2m-RP (5′-TCCATTCTTCAGTAAGTCAACT-3′ ); Lane 1, DNA mol wt marker VIII (Boehringer); lane 2, untreated HL60 cells; lane 3, HL60 cells treated with the mixture of hFES-S-sj1 and 2; lane 4, HL60 cells treated with the mixture of hFES-IP-sj1 and 2; lane 5, HL60 cells treated with the mixture of hFES-AS-sj1 and 2; lane 6, HL60 cells treated with the mixture of hFES-S1, 2 and 3; lane 7, HL60 cells treated with a mixture of hFES-IP1, 2 and 3; lane 8, HL60 cells treated with hFES-AS1, 2 and 3; lane 9, negative control performed with HL60 cells genomic DNA; lane 10, negative control performed without the cDNA template.
Detection by RT-PCR of the c-fes mRNA in HL60 cells treated with c-fes ODN. Total cellular RNA was extracted after 5 days of c-fes AS ODN treatment as described in Materials and Methods. The 500-bp amplified c-fes fragment was obtained using the oligonucleotide primers hFES-DP (5′-ACTATGGGCTTCTCTTCCGAGCTG-3′ ) and hFES-RP (5′-TCACGGTCCTTGTCTTTGCTGGCCT-3′ ). The 162-bp human β2-microglobulin fragment, used as a quantitative control, was obtained using the oligonucleotide primers hβ2m-DP (5′-CTCGCGCTACTCTCTCTTTCT-3′ ) and hβ2m-RP (5′-TCCATTCTTCAGTAAGTCAACT-3′ ); Lane 1, DNA mol wt marker VIII (Boehringer); lane 2, untreated HL60 cells; lane 3, HL60 cells treated with the mixture of hFES-S-sj1 and 2; lane 4, HL60 cells treated with the mixture of hFES-IP-sj1 and 2; lane 5, HL60 cells treated with the mixture of hFES-AS-sj1 and 2; lane 6, HL60 cells treated with the mixture of hFES-S1, 2 and 3; lane 7, HL60 cells treated with a mixture of hFES-IP1, 2 and 3; lane 8, HL60 cells treated with hFES-AS1, 2 and 3; lane 9, negative control performed with HL60 cells genomic DNA; lane 10, negative control performed without the cDNA template.
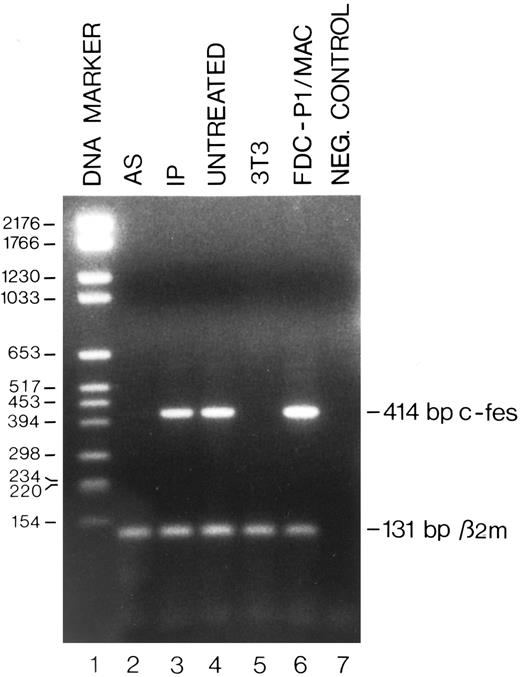
Detection by RT-PCR of the c-fes mRNA in FDC-P1/MAC-11 cells treated with c-fes ODN. Total cellular RNA was extracted after 5 days of murine c-fes AS ODN treatment as described in Materials and Methods. The 414-bp amplified murine c-fes fragment was obtained using the oligonucleotide primers mFES-DP (5′-CAGAGCTGGAGCAGCGGCCCCGACA-3′ ) and mFES-RP (5′-GGTGCAGCTGTGCGGCCCTCACACC-3′ ). The 131-bp murine β2-microglobulin fragment, used as a quantitative control, was obtained using the oligonucleotide primers mβ2m-DP (5′-GGTGCTTGTCTCACTGACCGGCTT-3′ ) and mβ2m-RP (5′-GAGGCGGGTGGAACTGTGTTACGT -3′ ); lane 1, DNA mol wt marker VI (Boehringer); lane 2, FDC-P1/MAC-11 cells treated with the mixture of mFES-AS1, 2, and 3; lane 3, FDC-P1/MAC-11 cells treated with the mixture of mFES-IP1, 2 and 3; lane 4, ODN untreated FDC-P1/MAC-11 cells; lane 5, negative control performed with NIH-3T3 cells not expressing the c-fes gene; lane 6, proliferating FDC-P1/MAC-11 cells; lane 7, negative control performed without the cDNA template.
Detection by RT-PCR of the c-fes mRNA in FDC-P1/MAC-11 cells treated with c-fes ODN. Total cellular RNA was extracted after 5 days of murine c-fes AS ODN treatment as described in Materials and Methods. The 414-bp amplified murine c-fes fragment was obtained using the oligonucleotide primers mFES-DP (5′-CAGAGCTGGAGCAGCGGCCCCGACA-3′ ) and mFES-RP (5′-GGTGCAGCTGTGCGGCCCTCACACC-3′ ). The 131-bp murine β2-microglobulin fragment, used as a quantitative control, was obtained using the oligonucleotide primers mβ2m-DP (5′-GGTGCTTGTCTCACTGACCGGCTT-3′ ) and mβ2m-RP (5′-GAGGCGGGTGGAACTGTGTTACGT -3′ ); lane 1, DNA mol wt marker VI (Boehringer); lane 2, FDC-P1/MAC-11 cells treated with the mixture of mFES-AS1, 2, and 3; lane 3, FDC-P1/MAC-11 cells treated with the mixture of mFES-IP1, 2 and 3; lane 4, ODN untreated FDC-P1/MAC-11 cells; lane 5, negative control performed with NIH-3T3 cells not expressing the c-fes gene; lane 6, proliferating FDC-P1/MAC-11 cells; lane 7, negative control performed without the cDNA template.
Thus, a 500-bp fragment corresponding to the human c-fes coding region, nt 73 to nt 572 of the HSCFES locus (EMBL, Accession No. X52192), was generated during 35 cycles of PCR (denaturation at 94°C for 40 seconds, annealing at 68°C for 2 minutes, and extension at 72°C for 4 minutes); a 414-bp fragment corresponding to the murine c-fes coding region, nt 234 to nt 648 of MMFESCR (GenBank, Accession No. X12616), was generated during 30 cycles of PCR (denaturation at 94°C for 40 seconds, annealing and extension at 72°C for 2 minutes). Ten microliters of PCR reaction was then electrophoresed on a 2% agarose gel ethidium bromide stained. ODN specific for the human36 and mouse37 β2-microglobulin cDNA (see legend of Figs 1 and 2, respectively) were used, in a parallel amplification reaction, to monitor the amount of RNA in each sample.
Antibodies.For Western blotting analysis we used the following Antibodies (Ab): anti v-fes monoclonal Ab (Ab-1) (Oncogene Sciences Inc, Uniondale, NY) which recognizes the mammalian p92c-fes. Anti-hck polyclonal Ab (Santa Cruz Biotechnology Inc, Santa Cruz, CA), specific for the mammalian p56 and p59 hck.
The Ab used in biparametric cell cycle analysis is a mouse monoclonal antibody (MoAb) anti-Bromodeoxyuridine (Becton Dickinson, San Jose, CA).
Flow cytometric analysis to study integrin and matrix protein expression was performed with the following human-specific MoAb: anti-CD49d (VLA-4, α-chain of α4β1 fibronectin receptor) (AMAC Inc, Westbrook, ME); anti-CD49e (VLA-5, α-chain of α5β1 fibronectin receptor) (Telios Inc, San Diego, CA); anti-CD49f (VLA-6, α-chain of α6β1 laminin receptor) (Immunotech S.A., Marseille, France); anti-CD51 (α-chain of the αvβ3 vitronectin receptor) (AMAC Inc); anti-CD11a (Serotec, Oxford, UK), anti-CD11b and anti-CD11c (Becton Dickinson, San Jose, CA); anti-fibronectin (UBI, Lake Placid, NY), anti-laminin (Immunotech S.A.) and anti-vitronectin (Biogenesis LTD, Bournemouth, UK).
MoAb to Mac-3 were obtained from American Type Culture Collection (ATCC, Rockville, MD).
Preparation of cell lysates and Western blot analysis.5 ×10 5 HL60 cells were washed twice in ice-cold phosphate-buffered saline (PBS) and lysed in ice-cold lysis buffer (20 mmol/L Tris pH 8, 150 mmol/L NaCl, 10% glycerol, 1% NP-40, 1 mmol/L phenyl-methyl-sulphonil fluoride, 1 μg/mL aprotinin, 1 μg/mL leupeptin, 1 μg/mL pepstatin, 10 mmol/L EDTA, 1 mmol/L Na orthovanadate), and sonicated. The protein content was normalized by Lowry procedure.38 Fifty micrograms of cell extract was dissolved in 50 mmol/L Tris pH 8, 5% β-mercaptoethanol, 2% sodium dodecyl sulphate (SDS), 0.1% bromophenol blue, 10% glycerol, loaded onto 7.5% to 15% SDS-polyacrylamide gel electrophoresis and electrophoresed in TGS buffer (25 mmol/L Tris pH 8.3, 250 mmol/L glycine, 0.1% SDS). The separated proteins were transferred at 4°C onto a nitrocellulose sheet by electroblotting procedure39 in TGM buffer (25 mmol/L Tris pH 8.3, 250 mmol/L glycine, 20% methanol) for 2 hours at 0.9 A. To monitor the electroblotting efficiency the membrane was stained in 0.2% Ponceau S/0.3% trichloracetic acid and destained in 0.3% trichoracetic acid. Membranes were pretreated with 4 mg/mL goat serum in TBST (10 mmol/L Tris pH 8, 150 mmol/L NaCl, 0.05% Tween 20) for 40 minutes, incubated for 1 hour with 0.5 μg/mL primary antibody, washed twice with TBST, incubated for 30 minutes with secondary antibody horseradish peroxidase (HRP)-conjugated and then washed twice with TBST.40 41 The detection was performed out by BM Chemiluminescence Western blotting kit (Boehringer).
Flow cytometric analysis.Flow cytometric analysis was performed with a FACScan (Becton Dickinson & Co, Mountain View, CA) equipped with a single 488-nm argon laser and Lysis II software. The cell cycle analysis of untreated and ODN treated cells was determined by flow cytometry according to Dolbeare et al.42 Briefly, 106 cells were incubated in cell culture medium with 10 μmol/L BrdU (Sigma Chemical Co) at 37°C for 10 minutes. A mouse MoAb against BrdU (anti-BrdU, Becton Dickinson, NJ) was bound to BrdU incorporated into neosynthesized DNA. The complex was detected by rabbit fluorescein isothiocyanate (FITC)-conjugated antimouse Ab (Dako A/S, Glostrup, Denmark). Cells were stained with propidium iodide (50 μg/mL) and FACS analyzed. The indirect immunofluorescence has been used to evaluate CD49d, CD49e, CD49f, CD51, fibronectin, laminin, and vitronectin expression in HL60 cells and Mac-3 expression in FDC-P1/MAC-11 cells. Briefly, 5 × 105 cells were incubated with the corresponding Ab for 1 hour at 4°C. After 2 washes with PBS the cells were incubated with the corresponding secondary Ab (rabbit antimouse FITC-conjugated or rabbit antirat FITC-conjugated; Dako A/S) for 20 minutes at 4°C. The cells were washed twice with PBS and then subjected to cytofluorimetric analysis. The direct immunofluorescence has been used to evaluate the CD14, CD11b, and CD45 expression. Briefly, 5 ×105 cells were incubated with the corresponding fluorochrome-conjugated Ab for 20 minutes at 4°C. After 2 washes with PBS the cells were subjected to cytofluorimetric analysis. The analysis was performed on 2 × 104 cells for each sample.
RESULTS
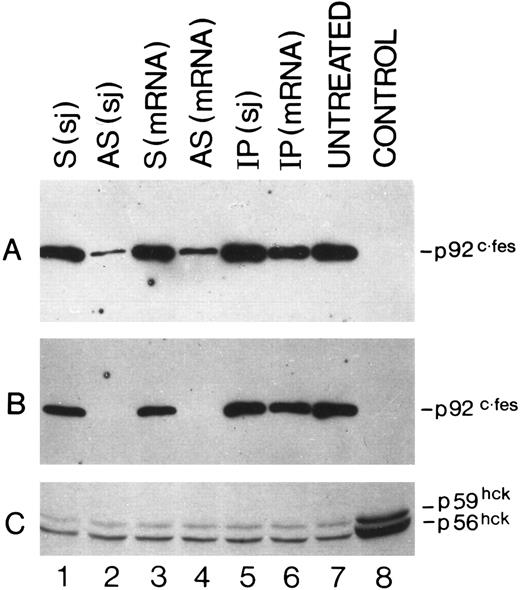
Specific inhibition of c-fes expression in HL60 and FDC-P1/MAC-11 cells.We used several c-fes AS and control ODN, whose sequences are reported in Tables 1 and 2. As shown in Fig 1, after 5 days of HL60 cells treatment with the mixture of hFES-AS1, 2 and 3 ODN, specific for c-fes mRNA, a dramatic decrease of mRNA abundance is evident even if a faint band is still detectable (Fig 1, lane 8), whereas no detectable c-fes mRNA is observed using the mixture of hFES-AS-sj1 and 2 AS ODN, specific for the human c-fes primary transcript (Fig 1, lane 5). The abundance of the β2-microglobulin mRNA, used as a quantitative control, is superimposable in all the samples studied (Fig 1, lanes 2 to 8). No detectable murine c-fes mRNA is observed in FDC-P1/MAC-11 cells after 5 days of AS-ODN treatment using the mixture of mFES-AS1, 2 and 3 (Fig 2, lane 2), whereas c-fes mRNA is clearly detectable in IP-ODN treated and untreated cells (Fig 2, lanes 3 and 4, respectively). In these RT-PCR experiments, the abundance of the murine β2-microglobulin mRNA, used as a quantitative control, is superimposable in all the samples studied (Fig 2, lanes 2 to 6). The Western blot analysis has clearly shown in HL60 cells, with both the antisense strategies, that p92c-fes protein is still detectable after 3 days (Fig 3A, lanes 2 and 4) but not after 5 days of ODN treatment (Fig 3B, lanes 2 and 4). On the contrary the p56-59 hck tyrosine kinase, belonging to the src family,43 is clearly expressed in all the samples studied (Fig 3C, lanes 1 to 8), demonstrating the specificity of c-fes inactivation. It has to be pointed out that the AS strategy based on the inactivation of c-fes primary transcript seems to be more efficient than that on c-fes mRNA in reducing the p92c-fes protein abundance (Fig 3A, compare lane 2 and 4). As shown in Figs 1 and 3, no inhibition of the expression of c-fes mRNA and corresponding protein is evident after treatment of HL60 cells with c-fes S and IP ODN when compared with untreated cells (Fig 3A and B, lanes 1, 3, 5, 6, and 7). No inhibition of cell proliferation is observed after 5 days of c-fes AS ODN treatment, since the fraction of HL60 and FDC-P1/MAC-11 cells in each phase of the cell cycle, as evaluated by flow cytometric analysis, remains unmodified as compared with untreated or control ODN treated cells (data not shown). These experiments confirm our previous results obtained in HL60 cells with a different c-fes AS strategy.8 10
Detection by Western blot analysis of p92c-fes protein in HL60 cells treated with c-fes ODN. p92c-fes was monitored in HL60 cells after 3 days (A) and 5 days (B) of c-fes ODN treatment, whereas the level of expression of hck protein was evaluated only after 5 days of treatment (C). Lane 1, HL60 cells treated with the mixture of hFES-S-sj1 and 2; lane 2, HL60 cells treated with the mixture of hFES-AS-sj1 and 2; lane 3, HL60 cells treated with the mixture of the hFES-S1, 2, and 3; lane 4, HL60 cells treated with the mixture of hFES-AS1, 2, and 3; lane 5, HL60 cells treated with the mixture of hFES-IP-sj1 and 2; lane 6, HL60 cells treated with the mixture of the hFES-IP1, 2, and 3; lane 7, ODN untreated HL60 cells; lane 8, negative control performed with K562 cells not expressing the p92c-fes (A and B); (C) shows the hck expression in HL60 cells treated with all-trans retinoic acid (positive control) as already described.8
Detection by Western blot analysis of p92c-fes protein in HL60 cells treated with c-fes ODN. p92c-fes was monitored in HL60 cells after 3 days (A) and 5 days (B) of c-fes ODN treatment, whereas the level of expression of hck protein was evaluated only after 5 days of treatment (C). Lane 1, HL60 cells treated with the mixture of hFES-S-sj1 and 2; lane 2, HL60 cells treated with the mixture of hFES-AS-sj1 and 2; lane 3, HL60 cells treated with the mixture of the hFES-S1, 2, and 3; lane 4, HL60 cells treated with the mixture of hFES-AS1, 2, and 3; lane 5, HL60 cells treated with the mixture of hFES-IP-sj1 and 2; lane 6, HL60 cells treated with the mixture of the hFES-IP1, 2, and 3; lane 7, ODN untreated HL60 cells; lane 8, negative control performed with K562 cells not expressing the p92c-fes (A and B); (C) shows the hck expression in HL60 cells treated with all-trans retinoic acid (positive control) as already described.8
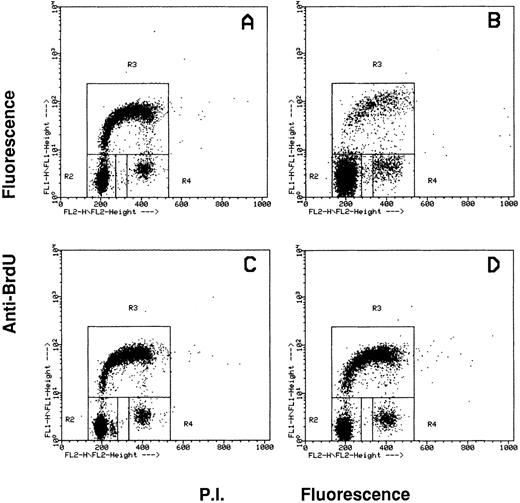
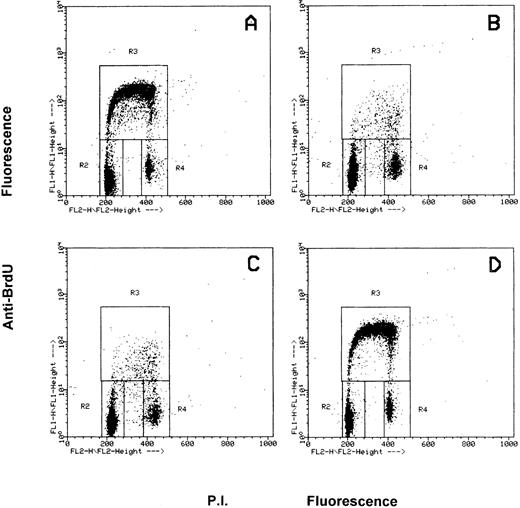
Effects of the inhibition of c-fes gene expression on HL60 and FDC-P1/MAC-11 cells induced to macrophage differentiation with PMA.The fraction of HL60 cells in the different phases of the cell cycle after c-fes ODN treatment and PMA induction is shown in Fig 4. No perturbation in the cell cycle progression is evident in HL60 cells treated for 5 days with hFES-IP-sj1 and 2, being 43% of the cells in G1, 49% in S and 8% in G2-M (Fig 4A). Superimposable results are obtained in untreated and control ODN treated HL60 cells, ie, a mixture of hFES-S1, 2, 3, or hFES-IP1, 2, 3, or hFES-S-sj1 and 2 (data not shown). When hFES-IP-sj ODN treated cells are induced to macrophage differentiation by PMA treatment for 48 hours, a considerable accumulation (about 80%) of HL60 cells in the G1 phase of the cell cycle occurs, and only 12% and 8% of the cells are respectively in S and G2-M phases (Fig 4B). Superimposable results are obtained with the other control cultures (data not shown). On the contrary, when HL60 cells are treated for 5 days with the mixture of hFES-AS-sj1 and 2 (Fig 4C) or with the mixture of hFES-AS 1, 2 and 3 (Fig 4D), and then induced with PMA, the distribution of the cells in the cycle is very similar to that of control cultures before PMA induction (Fig 4, compare C and D with A), showing an unmodified proliferative capacity. In fact with both inactivation strategies, the percentage of PMA-treated c-fes− HL60 cells in each phase of the cycle is about 50% in G1, 42% in S, and 8% in G2-M.
Flow cytometric cell cycle analysis of HL60 cells treated with c-fes ODN before and after induction for 48 hours with PMA. In each panel, the rectangles marked R2, R3, and R4 represent the cells in the Go/G1, S and G2/M phases of the cell cycle, respectively. (A) hFES-IP-sj-treated HL60 cells: the percentage of the cells in each phase of the cycle is 42% (Go/G1), 49% (S) and 9% (G2/M), respectively. Superimposable percentages were obtained in untreated, AS and control ODN treated cells, in several different experiments (data not shown). (B) hFES-IP-sj-treated HL60 cells induced with PMA; the percentage of the cells in the G1 phase of the cycle is 79%, in S phase 14% and in G2/M phases 7%. (C) Distribution of HL60 cells in the different phases of the cycle after 5 days of hFES-AS-sj1 and 2 ODN and PMA treatment. The percentage of the cells in the G1 phase is 50%, in S is 42% and in G2-M is 8%. (D) Superimposable results to that described in (C) were obtained when HL60 cells were treated with the mixture of hFES-AS1, 2, and 3 and induced to macrophage differentiation.
Flow cytometric cell cycle analysis of HL60 cells treated with c-fes ODN before and after induction for 48 hours with PMA. In each panel, the rectangles marked R2, R3, and R4 represent the cells in the Go/G1, S and G2/M phases of the cell cycle, respectively. (A) hFES-IP-sj-treated HL60 cells: the percentage of the cells in each phase of the cycle is 42% (Go/G1), 49% (S) and 9% (G2/M), respectively. Superimposable percentages were obtained in untreated, AS and control ODN treated cells, in several different experiments (data not shown). (B) hFES-IP-sj-treated HL60 cells induced with PMA; the percentage of the cells in the G1 phase of the cycle is 79%, in S phase 14% and in G2/M phases 7%. (C) Distribution of HL60 cells in the different phases of the cycle after 5 days of hFES-AS-sj1 and 2 ODN and PMA treatment. The percentage of the cells in the G1 phase is 50%, in S is 42% and in G2-M is 8%. (D) Superimposable results to that described in (C) were obtained when HL60 cells were treated with the mixture of hFES-AS1, 2, and 3 and induced to macrophage differentiation.
Figure 5 shows the distribution of FDC-P1/MAC-11 cells in the different phases of the cell cycle before and after c-fes ODN and PMA treatment. Figure 5A shows that in proliferating, untreated FDCP-1/MAC-11 cells 47% are in the G1 phase of the cycle, 44% in S phase and 9% in G2-M. After 48 hours of PMA induction, untreated or IP-ODN treated FDC-P1/MAC-11 cells (Fig 5B and C) accumulate in the G1 phase of the cycle (79%), whereas a drastic decrease of S phase cells occurs (10% left) and 11% of the cells are in G2-M. AS-ODN and PMA treated FDC-P1/MAC-11 cells continue to proliferate being their distribution in the cycle quite similar to that of untreated cells, ie, 49% in G1, 43% in S, and 8% in G2-M (Fig 5D, compare also with 5A).
Flow cytometric cell cycle analysis of FDC-P1/MAC-11 cells treated with c-fes ODN before and after induction for 48 hours with PMA. In each panel, the rectangles marked R2, R3, and R4 represent the cells in the Go/G1, S and G2/M phases of the cell cycle, respectively. (A) ODN untreated FDC-P1/MAC-11 cells: the percentage of the cells in each phase of the cycle is 47% (Go/G1), 44% (S) and 9% (G2/M), respectively. Superimposable percentages were obtained in c-fes mIP1, 2, and 3 ODN treated cells, in several different experiments (data not shown). (B) ODN untreated FDC-P1/MAC-11 cells induced with PMA; the percentage of the cells in the G1 phase of the cycle is 79%, in S phase 10%, and in G2/M phases 11%. (C) Distribution in the cell cycle phases of mIP ODN treated FDC-P1/MAC-11 cells after PMA treatment. The percentage of the cells in the G1 phase is 79%, in S is 11%, and in G2-M is 10%. (D) Superimposable results to that described in (A) were obtained when FDC-P1/MAC-11 cells were treated with the mixture of mFES-AS1, 2, and 3 and induced to macrophage differentiation with TPA (49% in G1, 43% in S, and 8% in G2-M).
Flow cytometric cell cycle analysis of FDC-P1/MAC-11 cells treated with c-fes ODN before and after induction for 48 hours with PMA. In each panel, the rectangles marked R2, R3, and R4 represent the cells in the Go/G1, S and G2/M phases of the cell cycle, respectively. (A) ODN untreated FDC-P1/MAC-11 cells: the percentage of the cells in each phase of the cycle is 47% (Go/G1), 44% (S) and 9% (G2/M), respectively. Superimposable percentages were obtained in c-fes mIP1, 2, and 3 ODN treated cells, in several different experiments (data not shown). (B) ODN untreated FDC-P1/MAC-11 cells induced with PMA; the percentage of the cells in the G1 phase of the cycle is 79%, in S phase 10%, and in G2/M phases 11%. (C) Distribution in the cell cycle phases of mIP ODN treated FDC-P1/MAC-11 cells after PMA treatment. The percentage of the cells in the G1 phase is 79%, in S is 11%, and in G2-M is 10%. (D) Superimposable results to that described in (A) were obtained when FDC-P1/MAC-11 cells were treated with the mixture of mFES-AS1, 2, and 3 and induced to macrophage differentiation with TPA (49% in G1, 43% in S, and 8% in G2-M).
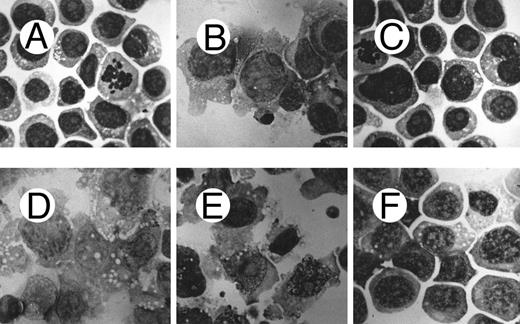
Morphological analysis of PMA-treated c-fes− HL60 cells shows a high degree of cellular immaturity. In Fig 6 the morphology of the ODN untreated HL60 cells (Fig 6A) is shown in comparison with proliferating c-fes− HL60 cells (Fig 6C) and with c-fes− HL60 cells after PMA treatment (Fig 6F ). Untreated, S or IP ODN treated HL60 cells undergo macrophage differentiation when induced with PMA (Fig 6B, D, and E). Thus, it is evident that the morphology of c-fes− HL60 cells after PMA treatment is that of immature cells, very similar to the morphology of ODN untreated (Fig 6A) and AS ODN treated HL60 cells before PMA induction (Fig 6C). The same biological behavior was observed in FDC-P1/MAC-11 murine myeloid precursor cells because only untreated or IP ODN treated cells can differentiate to macrophages when induced for 48 hours with PMA as evaluated by morphological analysis (Fig 7B and C). c-fes AS-ODN treated FDC-P1/MAC-11 cells induced for 48 hours with PMA shows a high degree of immaturity and a morphology very similar to that of untreated cells (Fig 7D, compare with 7A).
Morphological characteristics of HL60 cells treated with c-fes ODN before and after induction to macrophage differentiation by 48 hours of PMA treatment. (A) ODN untreated HL60 cells; (B) ODN untreated HL60 cells induced to macrophage differentiation with PMA; (C) HL60 cells after 5 days of treatment with hFES-AS-sj1 and 2 ODN; (D) macrophage morphology observed when HL60 cells, pretreated for 5 days with hFES-S-sj1 and 2 ODN, are induced to differentiate with PMA; (E) macrophage morphology observed when HL60 cells, pretreated for 5 days with hFES-IP-sj1 and 2 ODN, are induced to differentiate with PMA; (F ) morphology of HL60 cells pretreated with hFES-AS-sj1 and 2 ODN and PMA induced. This panel is representative of the results obtained by several different experiments. Superimposable results are obtained with both the antisense strategies. Original magnification ×1,000.
Morphological characteristics of HL60 cells treated with c-fes ODN before and after induction to macrophage differentiation by 48 hours of PMA treatment. (A) ODN untreated HL60 cells; (B) ODN untreated HL60 cells induced to macrophage differentiation with PMA; (C) HL60 cells after 5 days of treatment with hFES-AS-sj1 and 2 ODN; (D) macrophage morphology observed when HL60 cells, pretreated for 5 days with hFES-S-sj1 and 2 ODN, are induced to differentiate with PMA; (E) macrophage morphology observed when HL60 cells, pretreated for 5 days with hFES-IP-sj1 and 2 ODN, are induced to differentiate with PMA; (F ) morphology of HL60 cells pretreated with hFES-AS-sj1 and 2 ODN and PMA induced. This panel is representative of the results obtained by several different experiments. Superimposable results are obtained with both the antisense strategies. Original magnification ×1,000.
Morphological characteristics of FDC-P1/MAC-11 cells treated with c-fes ODN before and after induction to macrophage differentiation by 48 hours of PMA treatment. (A) ODN untreated FDC-P1/MAC-11 cells; (B) ODN untreated FDC-P1/MAC-11 cells induced to macrophage differentiation with PMA; (C) macrophage morphology observed when FDC-P1/MAC-11 cells, pretreated for 5 days with the mFES-IP1, 2, and 3 ODN, are induced to differentiate with PMA; (D) morphology of FDC-P1/MAC-11 cells pretreated with mFES-AS1, 2, and 3 ODN and PMA induced. This panel is representative of the results obtained by several different experiments. Original magnification ×1,000.
Morphological characteristics of FDC-P1/MAC-11 cells treated with c-fes ODN before and after induction to macrophage differentiation by 48 hours of PMA treatment. (A) ODN untreated FDC-P1/MAC-11 cells; (B) ODN untreated FDC-P1/MAC-11 cells induced to macrophage differentiation with PMA; (C) macrophage morphology observed when FDC-P1/MAC-11 cells, pretreated for 5 days with the mFES-IP1, 2, and 3 ODN, are induced to differentiate with PMA; (D) morphology of FDC-P1/MAC-11 cells pretreated with mFES-AS1, 2, and 3 ODN and PMA induced. This panel is representative of the results obtained by several different experiments. Original magnification ×1,000.
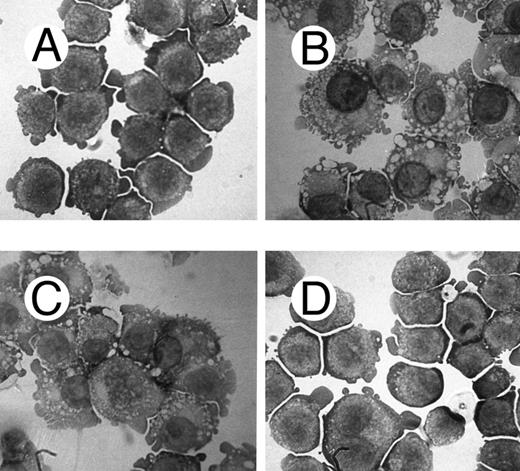
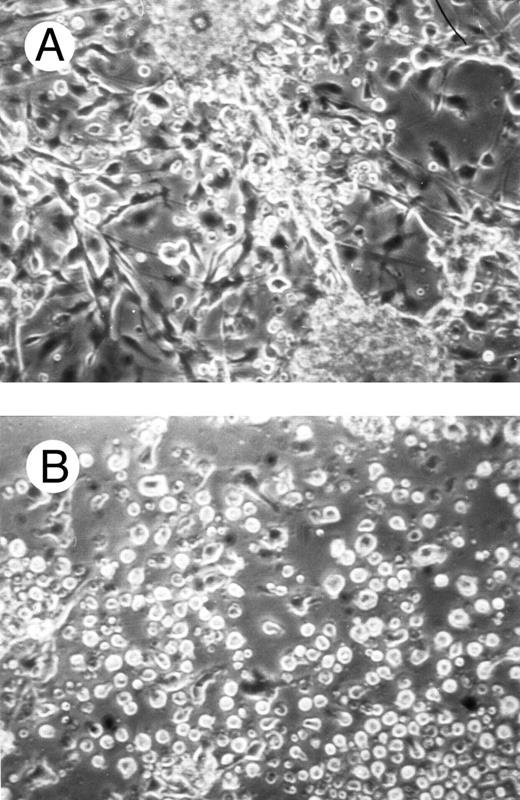
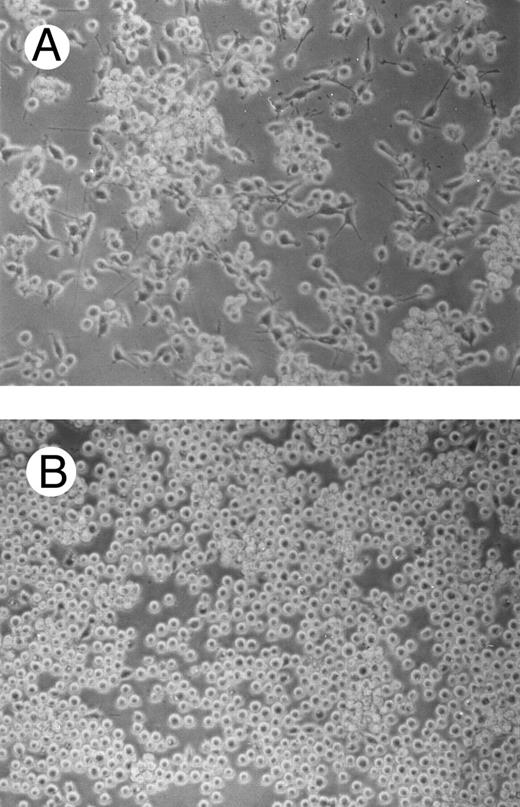
One of the main features of c-fes− HL60 and c-fes− FDC-P1/MAC-11 cells induced to macrophage differentiation with PMA is the almost complete loss of the adherence capability to the substrate (Figs 8B and 9B) when compared to the control cultures (Figs 8A and 9A).
Effects of c-fes AS ODN on the adhesion capacity of HL60 cells after 48 hours of PMA induction. Micrographs of HL60 cells treated with hFES-IP-sj1 and 2 (A) and treated with hFES-AS-sj1 and 2 (B). Original magnification ×200.
Effects of c-fes AS ODN on the adhesion capacity of HL60 cells after 48 hours of PMA induction. Micrographs of HL60 cells treated with hFES-IP-sj1 and 2 (A) and treated with hFES-AS-sj1 and 2 (B). Original magnification ×200.
Effects of c-fes AS ODN on the adhesion capacity of FDC-P1/MAC-11 cells after 48 hours of PMA induction. Micrographs of FDC-P1/MAC-11 cells treated with mFES-IP1, 2, and 3 (A) and treated with mFES-AS1, 2, and 3 (B). Original magnification ×200.
Effects of c-fes AS ODN on the adhesion capacity of FDC-P1/MAC-11 cells after 48 hours of PMA induction. Micrographs of FDC-P1/MAC-11 cells treated with mFES-IP1, 2, and 3 (A) and treated with mFES-AS1, 2, and 3 (B). Original magnification ×200.
Moreover, PMA-treated c-fes− HL60 are characterized by a low level of expression of α-naphtil-butyrate esterase and a high level of myeloperoxydase; this pattern is very similar to that obtained in proliferating HL60 cells, whereas an opposite pattern of expression is obtained in terminally differentiated macrophages,19 see Table 3.
Myeloid Differentiation Marker Expression in Oligomer- and PMA-Treated HL60 Cells
| Marker . | Untreated . | AS-Treated . | S-Treated . | IP-Treated . |
|---|---|---|---|---|
| α-Naphthylbutyrate esterase3-150 | 65 | 7 | 67 | 69 |
| Naphthol-AS-D-cloroacetate esterase3-150 | 10 | 7 | 6 | 6 |
| Myeloperoxydase3-150 | 10 | 97 | 8 | 14 |
| CD11c† | 90.7 | 12.4 | 91.7 | 94.3 |
| CD14† | 2.1 | 4.3 | 2.7 | 5.2 |
| CD45† | 95.7 | 94.8 | 97.3 | 90.9 |
| Marker . | Untreated . | AS-Treated . | S-Treated . | IP-Treated . |
|---|---|---|---|---|
| α-Naphthylbutyrate esterase3-150 | 65 | 7 | 67 | 69 |
| Naphthol-AS-D-cloroacetate esterase3-150 | 10 | 7 | 6 | 6 |
| Myeloperoxydase3-150 | 10 | 97 | 8 | 14 |
| CD11c† | 90.7 | 12.4 | 91.7 | 94.3 |
| CD14† | 2.1 | 4.3 | 2.7 | 5.2 |
| CD45† | 95.7 | 94.8 | 97.3 | 90.9 |
Values represent the average of three separate experiments. The variability was less than 10%.
Percent of positive cells determined by cytochemistry, and † by flow cytometry.
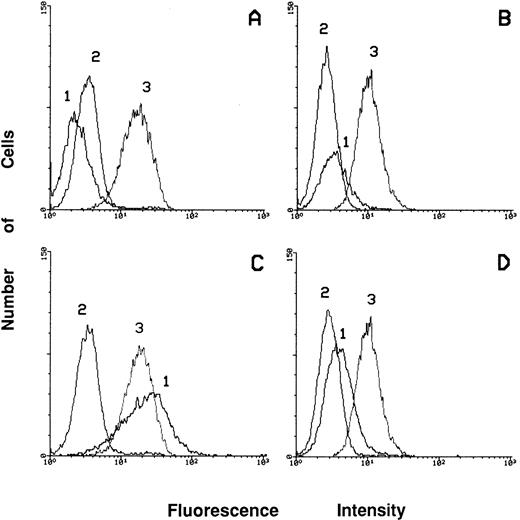
To better evaluate the differentiation level of PMA-treated c-fes− HL60 cells we studied the expression of surface markers such as CD45, CD14, and CD11c by flow cytometric analysis (Fig 10). The level of expression of the CD45 Antigen in these cells (Fig 10D, peak 3), is very similar to that of proliferating c-fes− HL60 cells (Fig 10B, peak 3) and to that of untreated, S or IP ODN treated cells before and after PMA induction (Fig 10A and C, peak 3). These results indicate a high degree of viability as shown also by the morphological analysis (Fig 6). The lack of expression of the macrophage differentiation marker CD11c indicates that PMA-treated c-fes− HL60 cells cannot reach terminal macrophage differentiation. In fact, the induction of CD11c integrin is evident only in the control cultures after PMA treatment (Fig 10C, peak 1) but not in PMA-treated c-fes− HL60 cells (Fig 10D, peak 1). As expected, no CD14 expression is ever evident in both the control and AS ODN treated HL60 cells before and after PMA treatment (Fig 10A to D, peak 2).
Flow cytometric analysis showing the expression of CD14, CD11c, and CD45 markers in ODN treated HL60 cells before and after 48 hours of PMA induction. Peak 1 represents the expression of the adhesion molecule CD11c; peak 2 shows the level of expression of the CD14 antigen and peak 3 of CD45 antigen. The expression of the above listed markers has been evaluated in HL60 cells treated with the mixture of hFES-IP-sj1 and 2 (A); in HL60 cells treated with the mixture of hFES-AS-sj1 and 2 (B); in HL60 cells treated as described in (A) and induced to macrophage differentiation in the presence of control ODN (C); in HL60 cells treated as described in (B) and induced to macrophage differentiation (D).
Flow cytometric analysis showing the expression of CD14, CD11c, and CD45 markers in ODN treated HL60 cells before and after 48 hours of PMA induction. Peak 1 represents the expression of the adhesion molecule CD11c; peak 2 shows the level of expression of the CD14 antigen and peak 3 of CD45 antigen. The expression of the above listed markers has been evaluated in HL60 cells treated with the mixture of hFES-IP-sj1 and 2 (A); in HL60 cells treated with the mixture of hFES-AS-sj1 and 2 (B); in HL60 cells treated as described in (A) and induced to macrophage differentiation in the presence of control ODN (C); in HL60 cells treated as described in (B) and induced to macrophage differentiation (D).
The levels of expression of the mono-macrophagic marker Mac-3 show a slight increase after PMA induction only in untreated or IP-treated FDC-P1/MAC-11 cells, whereas its expression remains unmodified in AS-treated cells. On the contrary, no modulation of CD11b expression was detected in c-fes− FDC-P1/MAC-11 and in control cell populations before and after PMA treatment (data not shown).
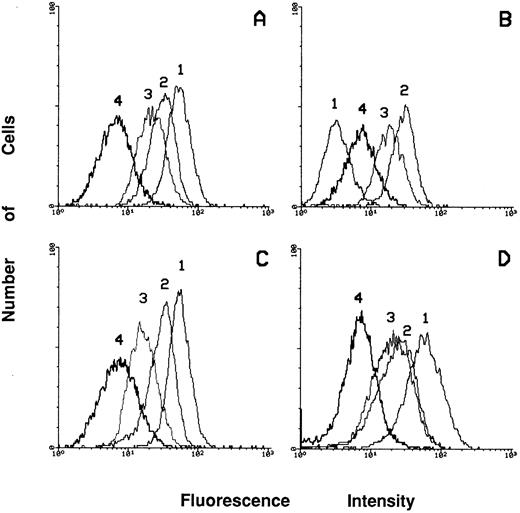
Expression of integrins in c-fes− HL60 cells before and after PMA treatment.To clarify if the loss of adherence capacity is simply the consequence of the differentiation arrest of PMA-treated c-fes− HL60 cells or also an impairment of the expression of adhesion molecules or extracellular matrix proteins we have studied the expression of these proteins in c-fes− HL60 cells before and after PMA treatment. As shown in Fig 11B, peak 1, an almost complete downregulation of the α4β1 fibronectin receptor, recognized by anti-CD49d MoAb, is evident in c-fes− HL60 cells. On the contrary, untreated, S or IP ODN treated HL60 cells express this receptor at high levels (Fig 11A, C, and D, respectively, peak 1).
Flow cytometric analysis of the expression of the α4β1 (peak 1), α5β1 (peak 2), α6β1 (peak 3) and αvβ3 (peak 4) receptors in HL60 cells after treatment with c-fes ODN. (A) Shows the receptors expression in ODN untreated HL60 cells; (B) shows the modulations of the expression of the receptors after treatment of HL60 cells with the mixture of hFES-AS-sj1 and 2; (C) shows the levels of expression of the receptors when HL60 cells are treated with the mixture of the hFES-S-sj1 and 2; (D) shows the receptors expression in HL60 cells treated with the mixture of the two hFES-IP-sj ODN.
Flow cytometric analysis of the expression of the α4β1 (peak 1), α5β1 (peak 2), α6β1 (peak 3) and αvβ3 (peak 4) receptors in HL60 cells after treatment with c-fes ODN. (A) Shows the receptors expression in ODN untreated HL60 cells; (B) shows the modulations of the expression of the receptors after treatment of HL60 cells with the mixture of hFES-AS-sj1 and 2; (C) shows the levels of expression of the receptors when HL60 cells are treated with the mixture of the hFES-S-sj1 and 2; (D) shows the receptors expression in HL60 cells treated with the mixture of the two hFES-IP-sj ODN.
Furthermore, the α5β1 fibronectin receptor (CD49e) and the α6β1 laminin receptor (CD49f ) are highly expressed in c-fes− HL60 cells (Fig 11B, peak 2 and 3, respectively) as well as in control cell cultures (Fig 11A, C, and D, peaks 2 and 3), whereas the level of expression of the αvβ3 vitronectin receptor (CD51) is weakly detectable in HL60 cells independently of the ODN treatment (Fig 11A, B, C, and D, peak 4).
The pattern of expression of the above listed integrins remains unmodified in c-fes− HL60 cells or in control cell cultures, after treatment with PMA. It is worth mentioning that in c-fes− HL60 cells, PMA was not capable to restore the α4β1 fibronectin receptor expression (data not shown).
Furthermore, it is rather evident that the levels of expression of CD11a and CD11b integrins are superimposable in the c-fes− HL60 cells when compared to the same cells treated with PMA. Control cell populations before and after induction with PMA, express superimposable levels of CD11a and CD11b (data not shown).
Expression of extracellular matrix proteins in PMA-treated c-fes− HL60 cells.We have studied, by flow cytometric analysis, the level of expression of fibronectin, vitronectin, and laminin in proliferating and PMA-treated HL60 cells in the presence or in the absence of the p92c-fes protein. Control ODN treated and AS ODN treated cells express almost at the same level these proteins. The levels of expression of fibronectin, laminin, and vitronectin are not modified by PMA treatment, independently from c-fes inhibition (data not shown).
DISCUSSION
In the work presented here we studied the involvement of c-fes protooncogene in PMA induced macrophage differentiation in two different systems: HL60 cells, a human leukemic myeloid cell line, and FDC-P1/MAC-11, a murine myeloid precursor cell line. These cell lines are particularly suitable for an in vitro study of myeloid differentiation. In fact HL60 cells (M2/M3 type of acute myeloid leukemia, growth factors independent and tumorigenic in mice) can differentiate to granulocytes when treated with retinoic acid, to monocytes with Vit. D3 and to macrophages with phorbol esters.44,20 FDC-P1/MAC-11 cells are not tumorigenic and depend on either IL-3 or granulocyte-macrophage colony-stimulating factor for survival and proliferation. These myeloid precursor cells express the c-fms proto-oncogene and differentiate to monocytes with M-CSF.17 Furthermore, our data clearly indicate that PMA treatment of these cells induces macrophage differentiation.
The study was carried on using the methodology of AS ODN to create a dominant negative inactivation of the c-fes proto-oncogene in these two cell lines. C-fes− HL60 and FDC-P1/MAC-11 cells were obtained by treatment with Phosphodiesters AS ODN, using different AS strategies. The first strategy was performed using a mixture of three AS ODN complementary to the 5′ region of human and mouse c-fes mRNA. This strategy has been selected because the N-terminal region to the SH2 domain of c-fes mRNA has no obvious counterpart in other tyrosine kinases, such as src and abl, and can be therefore considered a specific sequence domain.25,27 The second strategy, used only in HL60 cells where the genetic locus is characterized, was based on treatment with a mixture of two AS ODN complementary to two 5′ splice-junctions of c-fes primary transcript. With both these strategies the disappearance of the c-fes mRNA and protein, evaluated by the RT-PCR and Western blotting, is evident after 5 days of AS ODN treatment. The specificity of c-fes inactivation is validated by the different control experiments performed with S or IP ODN. The FES-IP ODN has the advantage to maintain the same nucleotide composition of the AS ODN and therefore it is a good control to monitor the nonspecific effects sequence independent.23 HL60 cells treatment with these control ODN did not interfere with c-fes mRNA or protein expression. Furthermore, in HL60 cells, the different hFES-AS ODN did not inactivate other tyrosin kinases, and particularly hck.
No perturbation of HL60 and FDC-P1/MAC-11 cell growth is evident after 5 days of treatment with the different FES-AS ODN, since the fraction of the cells in each phase of the cell cycle remains unmodified as compared with untreated or control ODN treated cells.
Using these specific c-fes AS ODN we have shown that the PMA-treated c-fes− HL60 and FDC-P1/MAC-11 cells undergo a maturation arrest. In fact cytospins, prepared at 24, 36, and 48 hours of PMA treatment of c-fes− HL60 and FDC-P1/MAC-11, do not show cells with macrophage morphology. Moreover, cytochemical and immunological studies show no evidence of maturation since low level of expression of α-naphtil-butyrrate-esterase enzyme and CD11c integrin are detectable in c-fes− HL60 cells and no induction of Mac-3 antigen occurs in c-fes− FDC-P1/MAC-11 cells after 48 hours of PMA treatment. Furthermore, PMA-treated c-fes− HL60 and FDC-P1/MAC-11 cells are not capable to accumulate in the G1 phase of the cycle and therefore to decrease the percentage in the S phase, which is the biological behavior of differentiating cells.45 46 These morphological, cytochemical, and immunological studies and the cytofluorimetric cell cycle analysis confirm that HL60 and FDC-P1/MAC-11 cells, lacking the c-fes protein cannot proceed along the macrophage differentiation pathway when treated with PMA for 48 hours and continue to proliferate as uninduced cells.
The high levels of expression of the CD45 antigen in PMA-treated c-fes− HL60 indicate that there is not a significative percentage of cell death. This behavior is confirmed also by the morphological analysis since no apoptotic cells are detectable in both cell lines. Moreover PMA-treated c-fes− HL60 and FDC-P1/MAC-11 cells never show the plastic-adherent behavior characteristic of terminally differentiated macrophages.
No modulation in the expression of the integrins CD11a and CD11b is evident in c-fes− HL60 cells and in control cell populations after 48 hours of PMA treatment.
On the contrary, a complete downregulation of the α4β1 fibronectin receptor expression is observed in c-fes− HL60 cells in all the experiments performed, before and after PMA treatment, when compared with the control cultures. The αvβ3 vitronectin receptor is weakly expressed in proliferating or PMA-treated HL60 cells, independently from c-fes expression, whereas the α5β1 fibronectin receptor and the α6β1 laminin receptor are expressed at high levels, independently from c-fes expression and PMA treatment. The complete downregulation of the α4β1 fibronetin receptor in PMA-treated c-fes− HL60 cells can be therefore one of the contributory causes of the loss of adhesiveness to the substrate.
Furthermore, the lack of p92c-fes protein in HL60 cells, treated or not with PMA, cannot interfere with the expression of several extracellular matrix proteins such as fibronectin, laminin, and vitronectin.
As a whole, these data allow us to conclude that the lack of the c-fes proto-oncogene product in PMA treated HL60 and FDC-P1/MAC-11 cells leads to a maturation arrest and to a complete downregulation of the α4β1 fibronectin receptor in c-fes− HL60 cells. It has to be pointed out that several reports demonstrate that many effects of PMA are mediated by protein kinase C (PKC),47, 48 even if other observations suggest that PKC activation cannot, by itself, cause HL60 macrophage differentiation.49
Our data show strong evidence that c-fes proto-oncogene is involved in the signal transduction pathway underlying PMA-induced macrophage differentiation. Further experiments are needed to clarify the possible correlations between c-fes inhibition and PKC activation during PMA treatment of precursor myeloid cells. It remains also to identify which are the molecular mechanisms underlying the control of the α4β1 fibronectin receptor expression by the c-fes proto-oncogene and if the lack of phosphorylation of c-fes specific substrates50 can play a role in the macrophage differentiation arrest which occur in PMA-treated c-fes− HL60 and FDC-P1/MAC-11 cells.
Supported by grants from A.I.R.C. (Associazione Italiana per la Ricerca sul Cancro) and from P.F. ACRO C.N.R. (Consiglio Nazionale delle Ricerche).
Address reprint requests to Sergio Ferrari, MD, Dipartimento di Scienze Biomediche, Sez. di Chimica Biologica, Università degli Studi di Modena, Via Campi 287, 41100 Modena, Italy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal