In this issue of Blood, Alaterre et al investigate the molecular circuits that are crucial during plasma cell (PC) differentiation. By combining single-cell RNA-sequencing (RNA-seq) and single-cell assay for transposase-accessible chromatin using sequencing (ATAC-seq) in an in vitro model of human B cell to PC differentiation, the authors identify a complex and dynamic pattern of epigenetic and transcriptomic changes in the early stage of PC differentiation that dictate PC fate and prepare for PC functions.1
The differentiation of B cells to PCs is an essential process for humoral immunity and to protect the host against infections.2 Upon foreign antigen recognition, B cells proliferate, migrate to the border of T-cell zone or interfollicular region of secondary lymphoid organs, and get activated. A subset of these activated B cells migrates to extrafollicular foci and differentiates into short-lived plasmablasts (PBs) that secrete low-affinity antibodies, whereas some activated cells proliferate and return to the B-cell follicle to form germinal centers (GCs). Here the B cells increase their immunoglobulin affinity for antigen by undergoing somatic hypermutation and subsequent clonal selection, producing long-lived PC homing to the bone marrow and memory B cells.3 The transition of B cells into PCs is tightly regulated by a complex network of transcription factors (TFs).4 It involves the upregulation of PC-specific TFs such as IRF4, XBP1, and BLIMP1 and downregulation of B cell–specific TFs such as PAX5, IRF8, and BACH2. These transcriptional changes are followed by increased surface expression of specific cellular markers, such as CD27, CD38, CD138, and BCMA on PCs and simultaneous downregulation of the B-cell markers CD19 and CD20.5 Although these TFs associated with PC differentiation have been well studied, the differentiation trajectories converting naive B cells into PCs and the regulators that control PC fate and differentiation remain largely unclear. Bulk RNA sequencing studies performed on sorted B cells have identified differences between naive B cells, GC B cells, memory B cells, PBs, and PCs but could not delineate the molecular trajectories involved in PC differentiation at the different stages.6 The development of single-cell sequencing techniques has overcome this problem and allowed a better study of the different cellular subsets that are transitioning from one cellular differentiation state to another. As such, in a recent single-cell analysis Verstegen et al have identified in vitro novel B and antibody-secreting cell intermediates, rising from human naive B cells, that possibly progress through an alternative route of differentiation into memory B cells, recapitulating in vivo human GC reactions.7
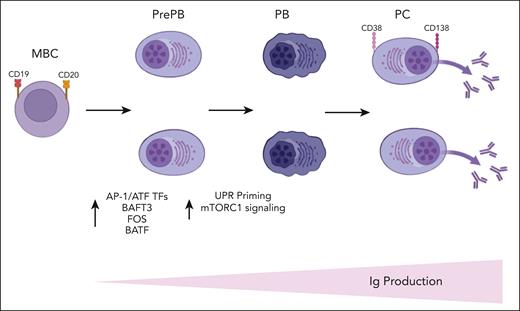
In this article the authors have combined single-cell RNA-seq and single-cell ATAC-seq to decipher the molecular trajectories involved in PC differentiation and to identify key regulators and pathways controlling this process. They have shown a strong transcriptional heterogeneity of the pre-PB and PB stages and a significant overexpression of TFs belonging to the AP-1/ATF superfamily of TFs such as BATF3, FOS, and BATF at pre-PB stage (see figure). Recent studies have demonstrated a role of the AP-1/ATF TFs in the transcriptional control of B-cell development8 and the interaction of BATF family members with elements of the interferon-regulatory factor family to control immune-regulatory networks.9 The increased expression of BATF3 was also validated by the authors at protein level in pre-PB cells, suggesting that it may represent a key transcriptional node that sustain PC fate and differentiation.
A schematic summary of Alaterre et al's findings is displayed here. Ig, immunoglobulin; MBC, memory B cells; PB, plasmablasts; PC, plasma cells; PrePB, preplasmablasts; TFs, transcriptional factors. The figure was created with BioRender.com.
A schematic summary of Alaterre et al's findings is displayed here. Ig, immunoglobulin; MBC, memory B cells; PB, plasmablasts; PC, plasma cells; PrePB, preplasmablasts; TFs, transcriptional factors. The figure was created with BioRender.com.
Of interest, integration of transcriptomic and epigenetic data at single-cell level revealed the presence of a more mature subpopulation of pre-PB cells characterized by an epigenetic profile of PCs together with unfolded protein response (UPR) activation. Of note, the first activation of UPR in pre-PB was associated with an overexpression of genes involved in mammalian target of rapamycin (mTORC1) signaling, and the second activation was associated with downregulation of these genes and overexpression of genes involved in protein secretion. The mTORC1 pathway is a key metabolic pathway known to positively regulate PC differentiation by promoting protein synthesis (see figure).10 Therefore, these findings are of central significance and suggest that PC fate is likely determined early during the differentiation of B cells. Some of the pre-PB cells are already committed to generate antibody-secreting cells and prime the UPR through the activation of mTORC1 pathway to prepare for PC function.
Future research including loss- or gain-of-function studies are required to further dissect the functional contribution of the identified pathways in PC differentiation.
In conclusion, the data presented in this article elegantly define the complex and dynamic pattern of epigenetic and transcriptomic changes occurring in the early stage of PC differentiation and dictating PCs’ fate. Furthermore, they identified major gene networks involved in PC development that can be ultimately targeted to optimize protective immune responses but also better understand PC disorders, caused by the failure to complete cell fate transition.
Conflict-of-interest disclosure: P.N. declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal