In this issue of Blood, Huffman et al1 present a multiethnic meta-analysis on fibrinogen combining whole genome-sequenced (WGS) and chip-typed individuals from 2 large consortia, Trans-Omics for Precision Medicine (TOPMed) and Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE). Fibrinogen plays a key part of the coagulation process, being the precursor of fibrin through enzymatic cleavage by thrombin. Its blood levels have been shown to correlate with various thrombotic conditions as well as inflammation. Due to the complexity of the coagulation process, and the multiple factors that affect fibrinogen levels, understanding the genetics affecting the regulation of fibrinogen can shed light on pathways involved and underlying causality between fibrinogen, inflammation, and downstream phenotypes such as thrombotic events or inflammatory conditions.
Genome-wide association studies (GWAS) have been very successful in identifying sequence variants that are associated with thousands of traits. This agnostic approach to link variations in DNA to phenotypes and diseases is particularly useful to generate hypotheses about biological processes and pathways. During these past 15 years of the GWAS era, researchers found thousands of previously unknown associations in almost all phenotypes studied, which increased our understanding of them.
TOPMed, with its over 30 000 WGS individuals, constitutes a more comprehensive reference panel for imputation of chip-typed individuals than older reference panels, such as 1000 Genomes. In turn, more variants can be imputed and with higher accuracy, allowing for identification of variants with lower frequencies. In fact, although the FGB locus is obvious and known from GWAS studies, it was first associated with a common variant in the region.2,3 The current study confirms that the rare missense variant rs148685782 (minor allele frequency [MAF] 0.4%), unavailable in early reference panels, is actually the strongest association in the region. And further, rare protein-altering variants, not included in genotype chips, when aggregated in gene-burden tests associate with every fibrinogen-producing gene.
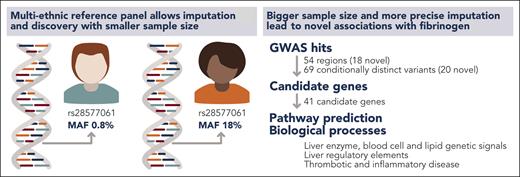
The study also shows the gains to be obtained by analyzing multiple ethnicities. The association between rs28577061 and fibrinogen could be found in the subset of individuals of African ancestry (∼11 000), because this variant is over 20 times more frequent among African subjects than among European subjects (see figure). More recent genomic cohorts, such as All of Us,4 recognize the advantages of recruiting individuals of diverse ethnical backgrounds, so we are likely to see an increase in associations coming from, up to this date, underrepresented ethnical groups.
The availability of multiethnic reference panels allows identification of sequence variants with varying MAFs among ethnicities. In turn, smaller sample sizes are needed to identify associations in ethnicities with a higher MAF. Huffman et al benefited from a multiethnic reference panel to improve imputation and identify novel loci, candidate genes, and biological processes related to fibrinogen. Professional illustration by Somersault18:24.
The availability of multiethnic reference panels allows identification of sequence variants with varying MAFs among ethnicities. In turn, smaller sample sizes are needed to identify associations in ethnicities with a higher MAF. Huffman et al benefited from a multiethnic reference panel to improve imputation and identify novel loci, candidate genes, and biological processes related to fibrinogen. Professional illustration by Somersault18:24.
An interesting and unexpected finding was the association of fibrinogen with gout. Even though genetically predicted fibrinogen seems to correlate with risk of gout, this correlation does not seem to be causal, according to the Mendelian randomization analysis run by the authors. We can trace a parallel to the association between elevated C-reactive protein levels and risk of cardiovascular disease, which has been largely documented in the literature5 but shown repeatedly not to be causal.6,7 It seems more likely, in light of current evidence, that elevated fibrinogen and gout are both downstream consequences of inflammatory processes and that many of the variants identified in this study have pleiotropic effects.
Finally, the study identified potentially novel coagulation pathway regulators. Three of these novel loci include missense variants in SERPINA1, ZFP36L2, and TLR10. SERPINA1 encodes the well-known protease inhibitor α-1 antitrypsin, TLR10 encodes a toll-like receptor, and ZFP36L2 encodes a less known gene family of AU-rich element RNA binding proteins. α-1 antitrypsin has previously been shown to be among the most abundant proteins in fibrin clots and might represent a link between the hemostatic and inflammatory systems.8 Less is known about the potential connection between ZFP36L2 and TLR10 and fibrinogen levels. This highlights the role of GWAS studies in identifying novel candidate pathway regulators.
Taken together, the findings by Huffman et al increase our understanding of fibrinogen genetics and underlying causality between fibrinogen levels and phenotypes. They also open new investigative avenues to be confirmed and increased in further studies.
Conflict-of-interest disclosure: V.T. and M.K.M. are employees of deCODE genetics/Amgen Inc.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal