Key Points
TP53 allelic state and VAF significantly correlates with outcomes in patients with MDS-del(5q).
Based on TP53 allelic status and a VAF cutoff of 20%, 2 distinct groups with clinical implications are identified among MDS-del(5q).
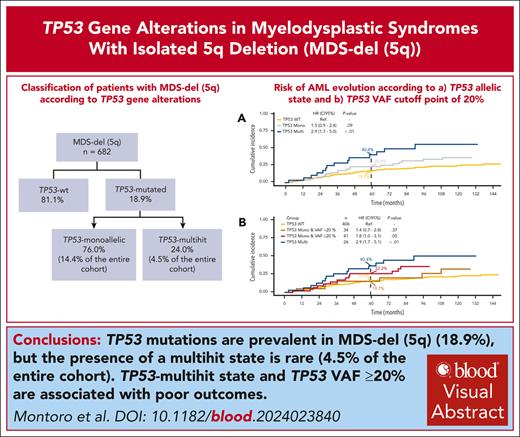
Visual Abstract
Mutations in the TP53 gene, particularly multihit alterations, have been associated with unfavorable clinical features and prognosis in patients diagnosed with myelodysplastic syndrome (MDS). Despite this, the role of TP53 gene aberrations in MDS with isolated deletion of chromosome 5 [MDS-del(5q)] remains unclear. This study aimed to assess the impact of TP53 gene mutations and their allelic state in patients with MDS-del(5q). To that end, a comprehensive analysis of TP53 abnormalities, examining both TP53 mutations and allelic imbalances, in 682 patients diagnosed with MDS-del(5q) was conducted. Twenty-four percent of TP53-mutated patients exhibited multihit alterations, whereas the remaining patients displayed monoallelic mutations. TP53-multihit alterations were predictive of an increased risk of leukemic transformation. The impact of monoallelic alterations was dependent on the variant allele frequency (VAF); patients with TP53-monoallelic mutations and VAF <20% exhibited behavior similar to TP53 wild type, and those with TP53-monoallelic mutations and VAF ≥20% presented outcomes equivalent to TP53-multihit patients. This study underscores the importance of considering TP53 allelic state and VAF in the risk stratification and treatment decision-making process for patients with MDS-del(5q).
Medscape Continuing Medical Education online

In support of improving patient care, this activity has been planned and implemented by Medscape, LLC and the American Society of Hematology. Medscape, LLC is jointly accredited with commendation by the Accreditation Council for Continuing Medical Education (ACCME), the Accreditation Council for Pharmacy Education (ACPE), and the American Nurses Credentialing Center (ANCC), to provide continuing education for the healthcare team.
Medscape, LLC designates this Journal-based CME activity for a maximum of 1.0 AMA PRA Category 1 Credit(s)™. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
Successful completion of this CME activity, which includes participation in the evaluation component, enables the participant to earn up to 1.0 MOC points in the American Board of Internal Medicine's (ABIM) Maintenance of Certification (MOC) program. Participants will earn MOC points equivalent to the amount of CME credits claimed for the activity. It is the CME activity provider's responsibility to submit participant completion information to ACCME for the purpose of granting ABIM MOC credit.
All other clinicians completing this activity will be issued a certificate of participation. To participate in this journal CME activity: (1) review the learning objectives; (2) study the education content; (3) take the post-test with a 75% minimum passing score and complete the evaluation at https://www.medscape.org/journal/blood; and (4) view/print certificate. For CME questions, see page 1753.
Disclosures
CME questions author Laurie Barclay, freelance writer and reviewer, Medscape, LLC, declares no competing financial interests.
Learning objectives
Upon completion of this activity, participants will:
Describe genetic findings in myelodysplastic syndrome and isolated deletion of chromosome 5 [MDS-del(5q)], based on a comprehensive analysis of TP53 abnormalities
Determine prognostic significance of clinical and genetic findings in MDS-del(5q), based on a comprehensive analysis of TP53 abnormalities
Identify clinical implications of genetic findings in MDS-del(5q), based on a comprehensive analysis of TP53 abnormalities
Release date: October 17, 2024; Expiration date: October 17, 2025
Introduction
Myelodysplastic syndromes (MDSs) characterized by the isolated deletion of chromosome 5q [MDS-del(5q)] constitute a unique entity of MDS. As per the 2017 World Health Organization (WHO) classification, this entity is defined by the presence of dysplasia in 1 to 3 lineages, bone marrow blasts <5%, circulating peripheral blasts <1%, and the deletion of chromosome 5q, either alone or with 1 additional chromosomal abnormality, except for monosomy 7 or 7q deletion.1 It is widely accepted that patients diagnosed with MDS-del(5q) exhibit a very good prognosis owing to the unique biology of the disease and its exceptional sensitivity to lenalidomide.2,3
The emergence of next-generation sequencing (NGS) has prompted the identification of recurrent somatic gene mutations with significant prognostic impact in patients with MDS.4,5 Mutations in the TP53 gene, present in 5% to 10% of patients diagnosed with de novo MDS, are associated with the poorest overall survival (OS) and the highest risk of transformation to acute myeloid leukemia (AML).6-8 Importantly, recent studies have demonstrated that the dismal prognosis associated with TP53 gene aberrations in MDS is linked to the biallelic inactivation of the TP53 gene through multiple alterations, as opposed to isolated mutations in which a wild-type allele remains intact.9,10
Notably, there is a robust association between TP53 mutations and del(5q). Specifically, 20% of patients with MDS-del(5q) and 70% to 90% of MDS with complex karyotype including del(5q) exhibit TP53 mutations.8 Despite the presence of various factors influencing the prognosis of patients with MDS-del(5q),11-13 the implications of TP53 mutations and its allelic state within this subgroup of MDS remain unclear, with inconsistent data reported so far.8,14-17
Against this background, we investigated the clinical and biological implications of TP53 gene mutations and their allelic state in, to our knowledge, the largest cohort of MDS-del(5q) assembled to date. To achieve this, we correlated genetic data, including a comprehensive analysis of TP53 gene and a panel of genes recurrently mutated in MDS, with the risk of transformation to AML and OS.
Methods
Patients
Patients diagnosed with de novo MDS-del(5q) according to the WHO 2017 classification and who underwent TP53 mutational analysis were included.1 Therapy-related MDS-del(5q) and those under disease modifying therapy before the genetic analysis were excluded. Clinical characteristics and laboratory data, encompassing sex, age, hemoglobin level, platelet counts, white blood cell counts, neutrophils and monocytes levels, and the percentage of peripheral blood and bone marrow blasts, were collected at the moment of diagnosis. Risk stratification was performed according to the Revised International Prognostic Scoring System (IPSS-R).18 Treatment details were compiled when available. Clinical outcomes included the date of death from any cause or last follow-up and the date of AML transformation. Patient information was collected in accordance with the Institutional Review Board–approved protocol of each center in which the research was conducted and by the tenets of the Declaration of Helsinki.
Genetic data
For a comprehensive characterization, cytogenetic and molecular data were gathered for all patients at the time of diagnosis or before initiating any treatment. Genetic profiling was performed at each institution and included conventional G-banding analyses in all cases. Additionally, and depending on the institution, the profiling included fluorescence in situ hybridization (FISH) of 17p (TP53), single nucleotide polymorphism microarrays (SNP-As), Sanger sequencing of TP53 gene, targeted NGS, whole-exome sequencing (WES), and whole-genome sequencing (WGS; supplemental data, available in the Blood website). A set of 27 genes recurrently mutated in MDS were common across all NGS analyses and included ASXL1, CALR, CBL, CEBPA, CSF3R, DNMT3A, ETV6, EZH2, FLT3, IDH1, IDH2, JAK2, KIT, KRAS, MPL, NPM1, NRAS, PTPN11, RUNX1, SETBP1, SF3B1, SRSF2, TET2, TP53, U2AF1, WT1, and ZRSR2. Variant filtering and categorization were performed as previously described,19 in which only variants with a variant allele frequency (VAF) ≥2% were considered. Allelic imbalances, defined as deletions and copy number–neutral loss of heterozygosity (cnLOH) of chromosome 17p affecting the TP53 locus, were evaluated by FISH, SNP-A, and various NGS techniques (WES, WGS, and targeted panels; supplemental Data).
Definition of TP53 status
Patients were categorized based on their TP53 mutational status into 2 groups: TP53 wild type (TP53-wt), indicating the absence of detected mutations, and TP53 mutated (TP53-mut), indicating the presence of ≥1 mutations. After integrating the mutational and allelic imbalance data, in accordance with the criteria defined by Bernard et al, and the 2022 WHO classification,9,20 2 TP53-mut subgroups were further defined: (1) TP53 monoallelic, when only 1 mutation with a VAF <50% and no allelic imbalances were detected; noteworthy, patients with a sole TP53 mutation (VAF <50%) and unavailable allelic imbalances information were also classified as TP53 monoallelic, in line with previous data9,20; and (2) TP53 multihit, in cases with multiple mutations, mutation(s) alongside concomitant allelic imbalance, or a single TP53 mutation with VAF ≥50% (because 17p deletion or cnLOH can be inferred in these cases).
Statistical analysis
Clinical variables were summarized using descriptive statistics. A logistic model was used to calculate P values and identify significant differences between groups in descriptive analysis. Kaplan-Meier method was used for OS analysis and reported with 95% confidence interval (95% CI). A Cox proportional hazards model was used to detect differences in survival end points; hazard ratios (HRs) with 95% CIs and P values were reported. The progression to AML was analyzed using the competing risks method, and Fine and Gray proportional subdistribution hazards model was used for comparison. This model facilitated the derivation of HRs and associated P values. Patients were categorized into 3 groups: progression to AML (the event of interest), death without prior progression (the competing event), or censoring in the absence of these events. Those who underwent allogeneic hematopoietic stem cell transplantation were censored at the time of transplant. Additionally, cumulative incidence data at 60 months are presented to offer a detailed understanding of progression over time. The maximum log-rank method was used to determine the optimum VAF percentage cutoff. P values <.05 were considered statistically significant. No missing data imputation was performed. All statistical analyses were performed using R (version 4.2.2).
Results
Clinical characteristics and outcome of patients with MDS-del(5q)
A total of 682 patients with MDS-del(5q) across 19 institutions were included. The median follow-up for the entire cohort was 68.5 months (95% CI, 63.0-81.1). The median age at diagnosis was 74 years (interquartile range [IQR], 66-80), and 73.3% (n = 500) were female. According to the IPSS-R criteria, data from 527 patients were analyzed. Among them, 476 patients (90.3%) were classified as very low or low risk, whereas 51 patients (9.7%) fell into the intermediate-risk category. Complete blood count information was missing for the remaining patients, making IPSS-R assessment not possible. The main features of the cohort are summarized in Table 1.
Main clinical characteristics of the study cohort (n = 682 patients)
| . | Median (IQR) . |
|---|---|
| Age at diagnosis, y | 74 (66-80) |
| Female sex, % | 73.3 |
| Hemoglobin, g/dL | 9.2 (8.1-10.3) |
| WBC count, × 109/L | 4.1 (3-5.5) |
| Neutrophils, × 109/L | 2.0 (1.3-3.1) |
| Platelet count, × 109/L | 249 (169-347) |
| Bone marrow blasts, % | 2 (1-3.5) |
| Cytogenetics, n (%) | |
| Isolated del(q) | 568 (83.3) |
| +1 additional abnormality | 114 (16.7) |
| IPSS-R, n (%) | |
| Very low | 184 (38.5) |
| Low | 243 (50.8) |
| Intermediate | 51 (10.7) |
| . | Median (IQR) . |
|---|---|
| Age at diagnosis, y | 74 (66-80) |
| Female sex, % | 73.3 |
| Hemoglobin, g/dL | 9.2 (8.1-10.3) |
| WBC count, × 109/L | 4.1 (3-5.5) |
| Neutrophils, × 109/L | 2.0 (1.3-3.1) |
| Platelet count, × 109/L | 249 (169-347) |
| Bone marrow blasts, % | 2 (1-3.5) |
| Cytogenetics, n (%) | |
| Isolated del(q) | 568 (83.3) |
| +1 additional abnormality | 114 (16.7) |
| IPSS-R, n (%) | |
| Very low | 184 (38.5) |
| Low | 243 (50.8) |
| Intermediate | 51 (10.7) |
WBC, white blood cell.
Survival and AML progression data were accessible in 539 (79.0%) and 507 patients (73.6%), respectively. The median OS for the entire cohort was 73.8 months (95% CI, 63.2-83.4). A total of 115 patients (22.7%) experienced progression to AML, with a mean time of 32 months (IQR, 18-57) after diagnosis. The risk of evolution to AML at 60 months was calculated at 21.8%.
Genetic and molecular characterization of patients with MDS-del(5q)
According to the conventional G-banding analyses findings, 568 patients (83.2%) presented an isolated del(5q), and 114 (16.7%) displayed 1 extra cytogenetic alteration. Interestingly, chromosome 17 was only affected in 1 patient, who showed an isochromosome 17q (supplemental Figure 1). FISH to detect 17p deletion was available in 30.1% of cases (n = 207), revealing the absence of TP53 deletion in all cases.
Analysis of mutations of the TP53 gene was carried out in 673 of 682 patients (98.7%) at diagnosis and in 9 of 682 (1.3%) before any treatment. Specifically, NGS was used in 91.8% of cases (n = 626), whereas Sanger sequencing in 8.2% of cases (n = 56). Data regarding TP53 allelic imbalances (deletions and cnLOH) were available for 411 patients (60%): 134 (33%) by SNP-A; 26 (6%) by targeted NGS; 11 (3%) by WES; and 240 (58%) by WGS.
Molecular landscape was assessed by NGS in 626 patients (91.8%): 375 (59.9%) by targeted NGS; 240 (38.3%) by WGS; and 11 (1.8%) by WES. A virtual panel of 133 genes associated with hematological neoplasms was analyzed in cases sequenced by WES or WGS. Intriguingly, 182 patients (29.1%) showed an absence of mutations in the analyzed genes. In addition, when focusing on the subset of 27 genes commonly assessed across all NGS analyses, it was observed that 222 patients (35.5%) exhibited no mutations, 223 (35.6%) harbored a single mutation, 107 (17.1%) carried 2 mutations, and 74 (11.8%) displayed ≥3 mutations. Notably, the most frequently mutated gene among this cohort was TP53 (20%), followed by SF3B1 (19%), DNMT3A (17%), TET2 (13%), ASXL1 (9%), and JAK2 (7%). Moreover, beyond these 27 core genes, only CSNK1A1 was mutated in ≥5% of patients, with an incidence of 7% (31/435; supplemental Figures 2 and 3; supplemental Table 1). Correlation analysis in these genes revealed that mutations in ASXL1 exhibited a mutually exclusive pattern with mutations in JAK2 and CSNK1A1, whereas no other significant associations were observed (supplemental Table 2). The remaining genes were mutated in ≤2% of the patients (supplemental Figures 2 and 3; supplemental Table 1).
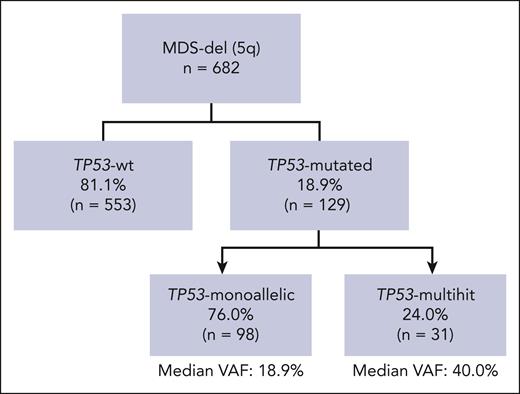
Classification of MDS-del(5q) according to TP53 gene alterations
Overall, 129 patients (18.9%) were categorized as TP53-mut, whereas 553 (81.1%) were designated as TP53-wt. Of note, among TP53-wt patients for whom allelic imbalances of 17p were analyzed (n = 357 [65%]), no deletions or cnLOH were detected. Upon the integration of all molecular data, TP53-mut patients were further categorized into TP53 monoallelic in 76% of cases (n = 98), 14.4% of the entire cohort, and TP53 multihit in 24% (n = 31), 4.5% of the entire cohort. Among TP53-monoallelic patients, 46.9% (n = 46) had a single TP53 mutation with a VAF of <50%, without deletion or cnLOH. In the remaining 53.1% (n = 52), data of deletions and/or cnLOH were unavailable, thus a monoallelic status was inferred. Importantly, 48% of these cases (n = 25) presented a VAF of <20%, strongly suggesting a monoallelic state. Among TP53-multihit patients, 16 (51.6%) presented multiple mutations (15 patients had 2, and 1 had 3 mutations); 13 (41.9%) showed 1 mutation with a VAF >50%; and 2 (6.5%) had 1 TP53 mutation plus cnLOH (Figure 1).
Classification of MDS-del(5q) according to TP53 gene alterations. A total of 129 patients (18.9%) were categorized as TP53-mut, whereas 553 (81.1%) were designated as TP53-wt. Upon the integration of all molecular data, TP53-mut patients were further categorized into TP53 monoallelic in 76% of cases (n = 98), 14.4% of the entire cohort, and TP53 multihit in 24% (n = 31), 4.5% of the entire cohort. Among TP53-monoallelic patients, 46 (46.9%) had a single TP53 mutation with a VAF of <50%, without deletion or cnLOH. In the remaining 53.1% (n = 52) data of deletions and/or cnLOH were unavailable. Among TP53-multihit patients, 51.6% (n = 16) presented multiple mutations (15 patients had 2 mutations, and 1 had 3 mutations); 41.9% (n = 13) showed 1 mutation with VAF >50%, and 6.5% (n = 2) showed 1 TP53 mutation plus cnLOH.
Classification of MDS-del(5q) according to TP53 gene alterations. A total of 129 patients (18.9%) were categorized as TP53-mut, whereas 553 (81.1%) were designated as TP53-wt. Upon the integration of all molecular data, TP53-mut patients were further categorized into TP53 monoallelic in 76% of cases (n = 98), 14.4% of the entire cohort, and TP53 multihit in 24% (n = 31), 4.5% of the entire cohort. Among TP53-monoallelic patients, 46 (46.9%) had a single TP53 mutation with a VAF of <50%, without deletion or cnLOH. In the remaining 53.1% (n = 52) data of deletions and/or cnLOH were unavailable. Among TP53-multihit patients, 51.6% (n = 16) presented multiple mutations (15 patients had 2 mutations, and 1 had 3 mutations); 41.9% (n = 13) showed 1 mutation with VAF >50%, and 6.5% (n = 2) showed 1 TP53 mutation plus cnLOH.
Clinical characteristics and prognostic impact of TP53 mutational status in patients with MDS-del(5q)
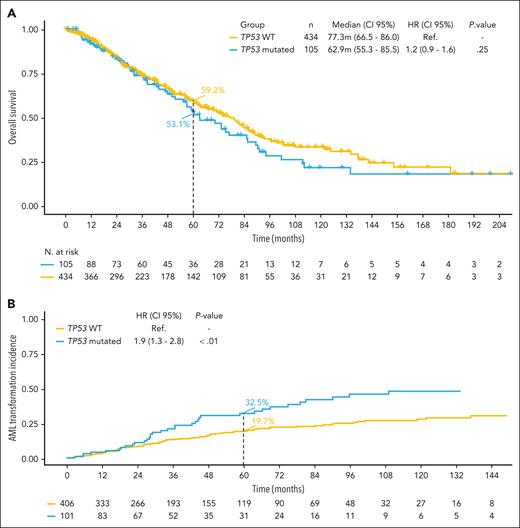
No significant differences were found in the main clinical variables between TP53-wt and TP53-mut patients (supplemental Table 3). The median OS was similar between both groups: 77.3 months (95% CI, 66.5-86.0) for TP53-wt and 62.9 months (95% CI, 55.3-85.5) for TP53-mut patients (P = .25). However, TP53-mut patients presented an increased risk of evolution to AML compared with TP53-wt patients (at 60 months, 32.5% vs 19.7%, respectively; HR, 1.9; 95% CI, 1.3-2.8; P < .01; Figure 2).
Kaplan-Meier curves for OS and risk of AML evolution according to TP53 mutational status. (A) The median OS was 77.3 months (95% CI, 66.5-85.3) for TP53-wt; and 62.9 months (95% CI, 55.3-85.5) for TP53-mut patients (P = .25). (B) The risk of evolution to AML at 60 months was 19.7% for TP53-wt and 32.5% for TP53-mut patients (HR, 1.9; 95% CI, 1.3-2.8; P < .01). Ref., reference.
Kaplan-Meier curves for OS and risk of AML evolution according to TP53 mutational status. (A) The median OS was 77.3 months (95% CI, 66.5-85.3) for TP53-wt; and 62.9 months (95% CI, 55.3-85.5) for TP53-mut patients (P = .25). (B) The risk of evolution to AML at 60 months was 19.7% for TP53-wt and 32.5% for TP53-mut patients (HR, 1.9; 95% CI, 1.3-2.8; P < .01). Ref., reference.
Clinical characteristics and prognostic impact of TP53 allelic state in patients with MDS-del(5q)
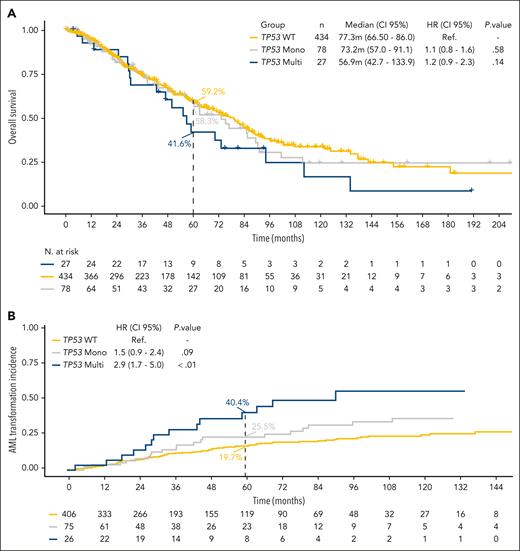
We then analyzed the clinical differences between the TP53-monoallelic and TP53-multihit groups. Clinical variables were also similar between both groups except for female sex, which was more prevalent in the TP53-monoallelic group than in the TP53-multihit group (81.6% vs 54.8%, respectively; P < .01). The median TP53 VAF of the TP53-mut cohort was 20.4% (IQR, 8.9-34.0). As expected, in TP53-monoallelic patients, it was 18.9% (IQR, 6.9-31.3), whereas it was 40.0% (IQR, 12.8%-70.2%) for TP53-multihit patients (P < .01; supplemental Table 3). Survival was superior in TP53-monoallelic patients (median OS, 73.2 months; 95% CI, 57.0-91.1) compared with TP53-multihit patients (median OS, 56.9 months; 95% CI, 42.7-133.9), although the difference did not reach statistical significance (P = .33). In contrast, MDS-del(5q) patients with TP53 multihit presented an increased risk of evolution to AML compared with TP53-monoallelic MDS-del(5q) (at 60 months, 40.4% vs 25.5%, respectively; HR, 1.9; 95% CI, 1.0-3.7; P = .04; Figure 3).
Kaplan-Meier curves for OS and risk of AML evolution according to TP53 allelic state. (A) The median OS was 73.2 months (95% CI, 57.0-91.1) for TP53-monoallelic and 56.9 (95% CI, 42.7-133.9) for TP53-multihit patients (P = .33). (B) The risk of evolution to AML at 60 months was 25.5% for TP53-monoallelic and 40.4% for TP53-multihit patients (HR, 1.9; 95% CI, 1.0-3.7; P = .04).
Kaplan-Meier curves for OS and risk of AML evolution according to TP53 allelic state. (A) The median OS was 73.2 months (95% CI, 57.0-91.1) for TP53-monoallelic and 56.9 (95% CI, 42.7-133.9) for TP53-multihit patients (P = .33). (B) The risk of evolution to AML at 60 months was 25.5% for TP53-monoallelic and 40.4% for TP53-multihit patients (HR, 1.9; 95% CI, 1.0-3.7; P = .04).
Considering the reported prognostic relevance of TP53 VAF in MDS,9 we investigated its impact within our cohort of MDS-del(5q) patients. Cutoff point analysis unveiled that TP53-monoallelic patients with a VAF ≥20% (n = 48 [50%]) showed a risk of death and AML evolution comparable with that of TP53-multihit MDS-del(5q). Specifically, their median OS was 51.5 and 56.9 months, respectively (P = .46), and AML evolution at 60 months was 32.2% and 40.4%, respectively (P = .17). Similarly, there were no differences in survival or AML progression risk between TP53-monoallelic patients with VAF <20% and TP53-wt patients. The median OS was 91.1 and 77.3 months, respectively (P = .22), and the risk of AML evolution at 60 months was 19.7% and 19.7%, respectively (P = .35; Figure 4). Moreover, there were no significant differences in the clinical characteristics according to the cutoff point VAF of 20% (supplemental Table 4).
Kaplan-Meier curves for OS risk of AML evolution according to TP53 VAF cutoff point of 20%. (A) TP53-monoallelic patients with a VAF ≥20% (n = 48 [50%]) presented similar OS to TP53-multihit patients (median OS was 51.5 and 56.9 months, respectively; P = .46). TP53-monoallelic patients with a VAF <20% (n = 48 [50%]) presented similar OS to TP53-wt patients (median OS was 91.1 and 77.3 months, respectively; P = .22). (B) TP53-monoallelic patients with a VAF ≥20% (n = 48 [50%]) presented similar risk of AML evolution at 60 months to TP53-multihit patients (32.2% and 40.4%, respectively; P = .17). TP53-monoallelic patients with a VAF <20% (n = 48 [50%]) presented similar risk of AML evolution at 60 months to TP53-wt patients (19.7% and 19.7%, respectively; P = .35).
Kaplan-Meier curves for OS risk of AML evolution according to TP53 VAF cutoff point of 20%. (A) TP53-monoallelic patients with a VAF ≥20% (n = 48 [50%]) presented similar OS to TP53-multihit patients (median OS was 51.5 and 56.9 months, respectively; P = .46). TP53-monoallelic patients with a VAF <20% (n = 48 [50%]) presented similar OS to TP53-wt patients (median OS was 91.1 and 77.3 months, respectively; P = .22). (B) TP53-monoallelic patients with a VAF ≥20% (n = 48 [50%]) presented similar risk of AML evolution at 60 months to TP53-multihit patients (32.2% and 40.4%, respectively; P = .17). TP53-monoallelic patients with a VAF <20% (n = 48 [50%]) presented similar risk of AML evolution at 60 months to TP53-wt patients (19.7% and 19.7%, respectively; P = .35).
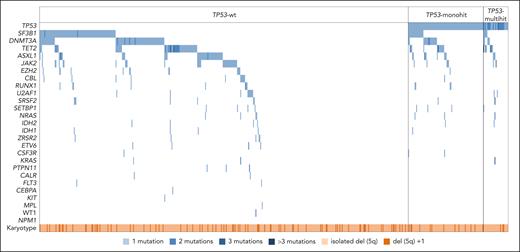
Finally, the mutational landscape and genetic complexity were compared across TP53-wt, TP53-monoallelic, and TP53-multihit patients. Mutational patterns and number of mutations did not exhibit significant differences among the 3 groups. No specific gene mutations showed significant enrichment, and in each subgroup, the median number of mutations was consistently 1 (Figure 5; supplemental Figures 4 and 5).
Molecular profile of patients with MDS-del(5q) according to TP53 allelic state. Oncoplot showing the distribution of gene mutations in the 27 genes analyzed across all patients with NGS data (n = 626) in the different groups according to TP53 allelic state (TP53-wt, n = 501; TP53-mono, n = 94; TP53 multihit, n = 31). Mutational patterns did not exhibit significant differences among TP53-wt, TP53-monoallelic, and TP53-multihit patients. The most frequently mutated gene among this cohort was TP53 (20%), followed by SF3B1 (19%), DNMT3A (17%), TET2 (13%), ASXL1 (9%), and JAK2 (7%). The median number of mutations was 1 in each subgroup. Each column represents 1 patient. All 27 analyzed genes as well as the occurrence of del(5q) as a sole aberration or with 1 additional cytogenetic aberration are given for each patient and represented in rows. White represents wild type; blue, mutated (number of mutations are depicted in different shades of blue); light orange, isolated del(5q); and dark orange, del(5q) and 1 additional lesion.
Molecular profile of patients with MDS-del(5q) according to TP53 allelic state. Oncoplot showing the distribution of gene mutations in the 27 genes analyzed across all patients with NGS data (n = 626) in the different groups according to TP53 allelic state (TP53-wt, n = 501; TP53-mono, n = 94; TP53 multihit, n = 31). Mutational patterns did not exhibit significant differences among TP53-wt, TP53-monoallelic, and TP53-multihit patients. The most frequently mutated gene among this cohort was TP53 (20%), followed by SF3B1 (19%), DNMT3A (17%), TET2 (13%), ASXL1 (9%), and JAK2 (7%). The median number of mutations was 1 in each subgroup. Each column represents 1 patient. All 27 analyzed genes as well as the occurrence of del(5q) as a sole aberration or with 1 additional cytogenetic aberration are given for each patient and represented in rows. White represents wild type; blue, mutated (number of mutations are depicted in different shades of blue); light orange, isolated del(5q); and dark orange, del(5q) and 1 additional lesion.
Predictive value of TP53 gene abnormalities in MDS-del(5q)
Treatment data were available for 64.4% (n = 439) of our cohort of patients with MDS-del(5q). Among these, 294 (67%) received treatment with lenalidomide at some point during the disease, whereas 48 (11%) were exclusively treated with erythropoietic stimulating agents. Furthermore, 63 (14.3%) received either red blood cell transfusions or no treatment, and 34 (7.7%) received other treatments. A total of 25 patients (5.7%) underwent allogeneic hematopoietic stem cell transplantation. Data on response to treatment with lenalidomide were available in 260 patients (88.4%): 186 (71.5%) showed response (either complete or partial response), and 74 (28.5%) did not attain a response (stable disease or progression). Interestingly, a trend toward higher responses was observed in TP53-wt patients than TP53-mut patients (response rate of 74.3% vs 61% for TP53-wt and TP53-mut patients, respectively; P = .07). When considering the allelic status, no significant differences were found between TP53 monoallelic and TP53 multihit (response rate of 56.7% vs 75%, respectively; P = .3).
Prognostic significance of clinical and biological variables in patients with MDS-del(5q)
Finally, we delved into the influence of the different clinical and biological variables in our entire cohort of patients with MDS-del(5q). In the multivariate analysis for OS, female sex, hemoglobin >10 g/dL, and age <70 years were associated with better survival. Conversely, mutations in RUNX1 were associated with an inferior OS. Interestingly, TP53 multihit and TP53 monoallelic with VAF ≥20%, showed a trend for a worse OS (P = .09). In the multivariable analysis for AML progression, platelet count >100 × 109/L was linked with a reduced risk of progression, whereas age <70 years, blasts >2%, mutations in SF3B1, and TP53 multihit and TP53 monoallelic with VAF ≥20% were associated with an increased risk of progression to AML (supplemental Figure 6).
Discussion
This international multicenter cooperative study, to our knowledge, stands as the largest cohort to date investigating patients with MDS-del(5q) and provides a comprehensive analysis of TP53 mutations and their allelic state. Our findings underscore that although TP53 mutations are prevalent in MDS-del(5q), the presence of multihit state is rare. Notably, the occurrence of TP53 multihit, along with a VAF ≥20%, significantly correlates with poor outcomes.
In our MDS-del(5q) cohort, TP53 mutations were observed in 18.9% of patients. However, only one-fourth of these patients showed TP53-multihit alterations. When taking into account the complete molecular landscape, the majority of patients in our series displayed either no mutations or a single mutation, which aligns with observations from 2 other MDS-del(5q) series.15,16 This is in contrast with the mutational profile of the general MDS population, in which TP53 mutations are observed in 5% to 10% of patients, with the majority characterized by TP53-multihit alterations and complex karyotypes.6,9,10 Furthermore, overall MDS typically exhibits a median of 3 mutations per patient,4,5 as opposed to the median of 1 mutation observed in our series. This low genetic complexity in MDS-del(5q) emphasizes the unique pathophysiology and clinical behavior of this entity, in which the sole deletion of chromosome 5 is likely a sufficient genetic alteration to drive the disease. Correlation analysis revealed a previously unreported phenomenon of mutual exclusivity between ASXL1 mutations with JAK2 and CSNK1A. Notably, in our cohort, the co-occurrence of CSNK1A1 and TP53 mutations, reported as mutually exclusive, was observed infrequently (n = 2).21 This finding further strengthens the hypothesis of shared pathways in MDS-del(5q). It must be noted that the frequency of double-hit events caused by allelic imbalances was low: only 6.5% of TP53-multihit patients presented a mutation plus a loss of heterozygosity, and no deletions in chromosome 17 have been detected. This can be partially explained by the inherent definition of the MDS-del(5q) entity (deletion of chromosome 5q, either alone or with 1 additional chromosomal abnormality, besides −7/7q), together with the notion that large deletions of chromosome 17p are usually observed in the context of complex karyotypes. Consistent with our findings, Sebaa et al did not observe deletions in 17p by FISH in the cohort of MDS-del(5q).22
Intriguingly, our series of patients with MDS-del(5q) harboring multihit abnormalities in the TP53 gene did not exhibit adverse risk factors such as heightened cytopenia or increased blast counts compared with TP53-monoallelic and TP53-wt subgroups. This is also in contrast with what has been observed in general MDS populations, in which the presence of TP53-multihit status is usually associated with adverse clinical characteristics.9,10 Furthermore, besides a higher proportion of male patients observed within the TP53-multihit subgroup, no other significant differences were identified when compared with the TP53-monoallelic subgroup. As mentioned earlier, the specific definition of MDS-del(5q) precludes the inclusion of high-risk variables (ie, blasts and adverse cytogenetics). Consequently, the prognostic utility of IPSS-R score is constrained within the MDS-del(5q), because it did not show impact in the multivariable analysis. Therefore, a comprehensive genetic assessment becomes paramount for precise prognostic evaluations in patients with MDS-del(5q).
The prognostic significance of TP53 mutations in MDS-del(5q) have been explored by several research groups with inconsistent results. Initial studies with limited number of patients indicated that TP53 mutations were associated with an increased risk of leukemic evolution and poor survival.8,14 In contrast, recent studies with significantly larger patient cohorts did not reproduce these findings.15-17 Our analysis in 682 patients confirmed twofold increased risk for progression to AML in patients with TP53-monoallelic mutations compared with TP53-wt, attributable to the presence of TP53-multihit alterations. In the general MDS population, TP53-multihit abnormalities are associated with both worse survival and risk of transformation to AML.9,10 It is intriguing that TP53 mutations do not significantly influence survival in MDS-del(5q). However, considering that our cohort presents a median age at diagnosis of 74 years along with the observation that AML transformation is a late event, the effect of TP53 mutations on OS is probably nuanced, particularly in this older population who likely succumb to age-related comorbidities. Nevertheless, the presence of TP53 multihit and TP53 monoallelic with VAF >20% showed a trend for a worse OS in the multivariable analysis.
In our cohort, optimal TP53 VAF threshold discriminating TP53-monoallelic patients into 2 groups with different prognosis was 20%. Indeed, TP53-monoallelic patients with VAF ≥20% (occurring in 50% of patients) shared similar prognosis to TP53-multihit patients. In contrast, patients with TP53 monoallelic with VAF <20% showed a prognosis similar to that of TP53-wt patients. These findings closely correspond to those reported by Bernard et al for overall MDS, indicating a VAF of 22%, and by Fleti et al and Bahaj et al, with a VAF of 23% identified as the optimal cutoff for MDS-del(5q).9,17,23
Recently, the 2022 WHO classification and the International Consensus Classification recognized TP53-mut cases as a distinct disease entity. In both classifications, the presence of TP53 mutations takes precedence over the definition of MDS-del(5q).20,24 In this setting, our data further reveal that TP53-multihit status and VAF significantly influence the prognosis of these patients, supporting the recommendations of the WHO classification and International Consensus Classification on the inclusion of the new entity “TP53-mutated myeloid neoplasms” (TP53-MN). However, based on our data regarding the outcomes of these patients, we propose a refinement of the defining criteria for MDS-del(5q) by incorporating those cases with TP53-monoallelic mutations with VAF <20% under the MDS-del(5q) subgroup, whereas patients with TP53 multihit or TP53 monoallelic with VAF ≥20% (in contrast to the VAF of 10%) under the TP53-mutated myeloid neoplasm subgroup.
In line with previous studies,14 we observed a higher prevalence of TP53 mutations in patients with no response to lenalidomide. However, this difference did not achieve statistical significance. Owing to the retrospective nature of the study and the lack of data for a significant proportion of patients, prospective studies are needed to correctly evaluate the impact of TP53 mutational state in the response to lenalidomide.
The primary limitation of our study lies in its retrospective and multicenter design, which could introduce methodological heterogeneity. Nevertheless, TP53 mutational status was analyzed by NGS methods in most of the patients, and the mutational profile was focused on the 27 genes commonly assessed across all NGS studies, yielding results consistent with prior research.15,16 Furthermore, because TP53 allelic status in MDS has only recently gained recognition,9 its analysis is not commonly included in routine diagnostics or in previous studies.8,14-17 In contrast, our study examined the allelic state in nearly half of the patients. Notably, among cases in which allelic analysis was absent, 48% displayed a VAF ≤20%, strongly suggesting a monoallelic state.23
In conclusion, our findings reveal that a meticulous diagnosis and prognostication of patients with MDS-del(5q) requires a thorough assessment of TP53 alterations. From a pragmatic standpoint, our study recommends the inclusion of the TP53 allelic status in this assessment and a VAF cutoff of 20%. Accordingly, we propose the categorization of 2 relevant subgroups with clinical implications within MDS-del(5q): the first includes MDS-del(5q) with TP53-wt or with TP53-monoallelic status with VAF <20%; the second, MDS-del(5q) with TP53-multihit status or TP53 monoallelic with VAF ≥20%. In the latter group, close monitoring is highly warranted to identify patients who might benefit from more intensive treatments.
Acknowledgments
V.S. was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC; grant IG-26537-2021). M.D.P. was supported by the Horizon 2020/2023 program of the European Union (GenoMed4All, 101017549; Synthema project, 1101095530); the TRANSCAN 7 Horizon 2020 program (EuroMDS 20180424); European Union–Next Generation EU, NRRP M6C2 (Investment 2.1 Enhancement and strengthening of biomedical research in the NHS [project PNRR-MAD-2022-12376695]); AIRC Foundation for Cancer Research (29483); and 5 × 1000 MYeloid NEoplasms Research Venture AIRC (project, 21267). F.S. was supported by the Instituto de Salud Carlos III (PI20/0053); TRANSCAN (AECC AC 18/000002); the Generalitat de Catalunya (2017 SGR288 and 2021 SGR00560); and the CERCA Programme/Generalitat de Catalunya, Fundación Internacional Josep Carreras. F.B. was supported by the Instituto de Salud Carlos III (grants PI17/00943 and PI20/01274). D.V. was supported by the Instituto de Salud Carlos III (grants PI17/01088 and PI20/00881).
Authorship
Contribution: M.J.M. and L.P. designed the study and wrote the manuscript; V.N. performed statistical analyses; M.J.M., L.P., P.A., Y.K., F.S., M.M., R.B., N.A.A., F.L.-C., T.G., L.N.E., A.J., Y.-H.W., A.C., E.S., and N.D.V. collected clinical and molecular data; M.J.M., L.P., A.J., D.V., and F.B. contributed to interpretation of the data and report design; and all authors contributed to manuscript preparation and approved its content.
Conflict-of-interest disclosure: U.P. received honoraria and research funding from Bristol Myers Squibb. C.H. declares equity ownership of the MLL Munich Leukemia Laboratory. A.J. received consulting fees from Novartis and Bristol Myers Squibb; and research funding from AstraZeneca. M.D.-C. received honoraria from Bristol Myers Squibb, Novartis, and Keros; and consulting fees from Novartis, Bristol Myers Squibb, Blueprint Medicines, Agios, Hemavan, Syros, Keros, Curis, and GlaxoSmithKline. The remaining authors declare no competing financial interests.
Correspondence: Maria Julia Montoro, Department of Hematology, Vall d’Hebron Barcelona Hospital Campus, Vall d’Hebron Institute of Oncology, Paseo Vall Hebron 119-129, 08006 Barcelona, Spain; email: jmontoro@vhio.net.
References
Author notes
Presented in part at the 65th Annual Meeting of the American Society of Hematology, San Diego, CA, 7-10 December 2023.
Data are available on request from the corresponding author, Maria Julia Montoro (jmontoro@vhio.net).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![Kaplan-Meier curves for OS risk of AML evolution according to TP53 VAF cutoff point of 20%. (A) TP53-monoallelic patients with a VAF ≥20% (n = 48 [50%]) presented similar OS to TP53-multihit patients (median OS was 51.5 and 56.9 months, respectively; P = .46). TP53-monoallelic patients with a VAF <20% (n = 48 [50%]) presented similar OS to TP53-wt patients (median OS was 91.1 and 77.3 months, respectively; P = .22). (B) TP53-monoallelic patients with a VAF ≥20% (n = 48 [50%]) presented similar risk of AML evolution at 60 months to TP53-multihit patients (32.2% and 40.4%, respectively; P = .17). TP53-monoallelic patients with a VAF <20% (n = 48 [50%]) presented similar risk of AML evolution at 60 months to TP53-wt patients (19.7% and 19.7%, respectively; P = .35).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/144/16/10.1182_blood.2024023840/3/m_blood_bld-2024-023840-gr4.jpeg?Expires=1764959405&Signature=yrnwZ5sb575IuUVx0~7~lAzpWKgB36hDcTZePe-xZ~i0Xymp4QegErggmMG3OvwuyDe7apXfy8s1BvQw-vw4C9~bi0WAuUWF0LJK9-qe0Dl8lTCnuSzoSsToOOI-Akjdk8mlDm~0zuKd4m686z1IsqtyxghJcG35rl1PP0QGxd4uDDB2FeKSm0P0zj1kWK4OPF5F0mrA3~lQwjmJgwAEZTJtnEV~VoTrSJmENAX6sdBhTShUVkG7gldbACJLO74yo-CobQyAcBusYYiRsP07quks5rqIhPI9lh2f99ULqeP0qnSW4iFg9I1YuRSZ0kPL4vzSCgNjcOecHaUhB-lvOw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal