In this issue of Blood, Chohan et al1 report a multiomic analysis of immunoglobulin M (IgM) gammopathies identifying molecular subtypes with distinct modes of transcriptional and epigenetic regulation.
Monoclonal gammopathies of underdetermined significance (MGUS) occur in at least 3% of the population aged 50 years or older and transform into plasma-cell dyscrasias and certain lymphomas at approximately 1% per annum.2 Long-term follow-up studies show that Waldenström macroglobulinemia (WM) is preceded by IgM expressing MGUS, and the more common class-switched MGUS proceeds to multiple myeloma.2 Multiple myeloma is more common and defined by distinct structural genetic events that often define subtypes that confer prognostic and therapeutic implications. In contrast, WM is less common and almost uniformly driven by MYD88 L265P mutations.3 Thus, it is unsurprising that less is known about the range of biological diseases in WM. Nonetheless, the disease has long been known to present with a spectrum of phenotypes ranging from more lymphoblastic to more plasmacytic, present not only between patients but also within an individual patient.
In the current study Chohan et al integrate layers of high-throughput data to identify the gene expression and DNA methylation programs of IgM gammopathies.1 This builds on previous work from the same group that identified genetic, metabolic, and immune alterations in the same cohort.4 Comparing healthy controls to IgM MGUS and WM, the authors report a progressive loss of DNA methylation, which mirrors proliferation-linked changes that occur during normal B-cell to plasma-cell differentiation.5,6 By integrating gene expression and DNA methylation, the authors identify concordant changes in cell cycle, immune, and signal transduction pathways between IgM gammopathies and controls. Notably, the authors report concordant promoter hypomethylation and upregulation of both CXCR4 and CD79B as well as downstream PI3K and MAPK components of these pathways. Given that CXCR4-activating mutations are found in nearly one-third of patients with WM7 and CD79B is altered in 10% to 15% of patients, this suggests that these pathways may be epigenetically configured for activation as compared with healthy B cells even in cases without activating mutations. Another notable observation was concordant promoter hypomethylation and upregulation of CCND1. This may parallel certain subtypes of multiple myeloma driven by CCND1 that present with a partial B-cell phenotype and distinct epigenetic program.8 Cumulatively, these data indicate that epigenetic reprogramming occurs at oncogenic pathways during the genesis of WM.
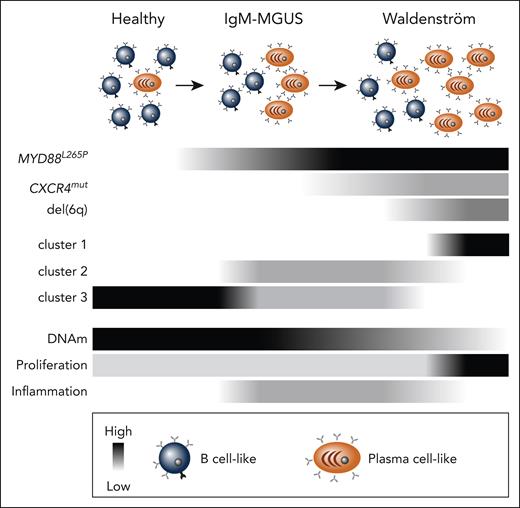
Beyond comparing healthy and IgM gammopathies, the authors compared the DNA methylation program in subtypes that they previously defined by gene expression.4 Here, cluster 1 is exclusively WM and believed to be a more aggressive, proliferative disease; cluster 2 is a mix of WM and MGUS with more immune cytokine and inflammatory signaling; and cluster 3 includes WM, MGUS, and healthy controls (see figure). It is important to note that previous work from Roos-Weil et al9 elegantly grouped WM into memory B cell–like and plasma cell–like subtypes based on a bifurcation in their DNA methylation profile, with the plasma cell–like subtype having less DNA methylation-mirroring plasma cells.5,6 In the current work, cluster 1 appears to have less DNA methylation as compared with clusters 2 and 3, aligning it with the plasma cell–like WM subtype reported by Roos-Weil et al.9 Consistent with this, both cluster 1 and the plasma cell–like WM subtype are enriched for deletions of chromosome 6q.4,9 Clusters 2 and 3 likely correspond with the previously reported memory B cell–like WM subtype,9 although it is hard to know for certain. Interestingly, Chohan et al found a pronounced correspondence of CpG island and promoter hypomethylation with transcription of cytokine signaling pathways in cluster 2. This is consistent with the group’s previous analyses indicating this subset has an inflammatory milieu.4 Indeed, immune dysfunction is a known hallmark of WM10; however, the current study indicates these hallmarks may not be a universal feature, as cells in cluster 1 did not show the same intrinsic immune signaling. Cumulatively, by integrating multiple data sets on the same samples, a richer picture is painted of the distinct biologies in IgM gammopathies.
Model of IgM gammopathies composed of B cell–like and plasma cell–like cells and the prevalence of common mutations denoted in black (key at bottom). Clusters 1 to 3 are IgM gammopathy subtypes described by Mondello et al.4 Global DNA methylation (DNAm) levels are inferred from previous studies,5,9 and B cell–intrinsic inflammation and proliferation correlated with DNA methylation are reported by Chohan et al.
Model of IgM gammopathies composed of B cell–like and plasma cell–like cells and the prevalence of common mutations denoted in black (key at bottom). Clusters 1 to 3 are IgM gammopathy subtypes described by Mondello et al.4 Global DNA methylation (DNAm) levels are inferred from previous studies,5,9 and B cell–intrinsic inflammation and proliferation correlated with DNA methylation are reported by Chohan et al.
Taken together these studies4,9 indicate IgM gammopathies are composed of distinct epigenetic subtypes that reflect disease biology and will likely have implications for clinical management. To translate these observations into actionable information, it will be critical to (1) identify markers that denote WM subtypes that can be easily assayed and (2) develop a consensus nomenclature to describe WM subtypes. These are especially important given the relatively rare prevalence of IgM gammopathies. Additionally, further detailed characterization and mechanistic studies are needed. DNA methylation often helps “lock in” a gene expression state due to its stability, but often epigenetic reprogramming is initiated by transcription factors and chromatin remodelers. Further detailed studies are needed to identify both the transcriptional regulators and epigenetic mechanisms underlying the distinct phenotypes seen in IgM gammopathies.
Conflict-of-interest disclosure: B.G.B. declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal