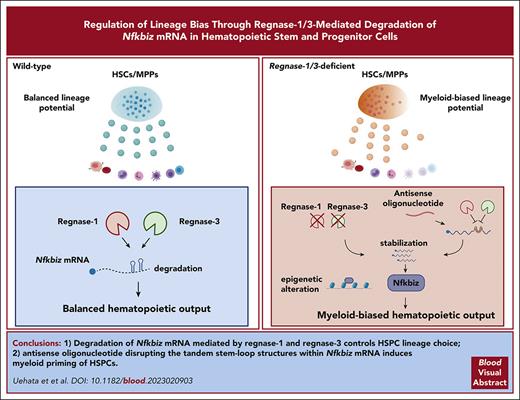
Reg1 and Reg3 are ribonucleases critical for determining lymphoid fate and restricting myeloid differentiation in HSPCs.
Reg1/Reg3-mediated decay of Nfkbiz, crucial for HSPC lineage choice, is potently manipulated by an antisense oligonucleotide.
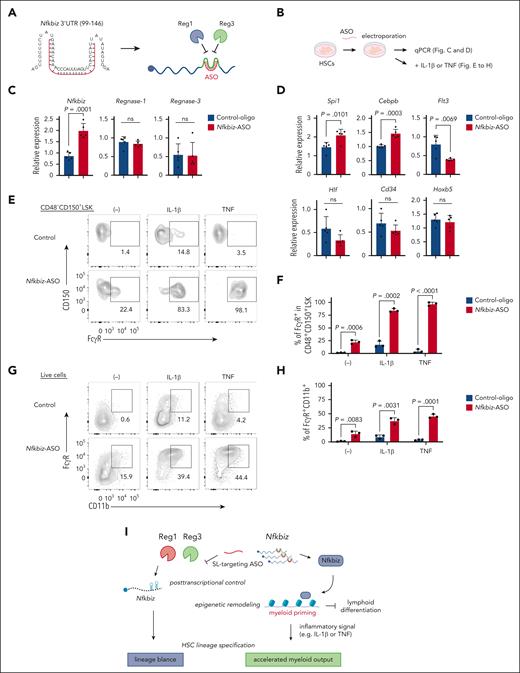
Visual Abstract
Regulation of lineage biases in hematopoietic stem and progenitor cells (HSPCs) is pivotal for balanced hematopoietic output. However, little is known about the mechanism behind lineage choice in HSPCs. Here, we show that messenger RNA (mRNA) decay factors regnase-1 (Reg1; Zc3h12a) and regnase-3 (Reg3; Zc3h12c) are essential for determining lymphoid fate and restricting myeloid differentiation in HSPCs. Loss of Reg1 and Reg3 resulted in severe impairment of lymphopoiesis and a mild increase in myelopoiesis in the bone marrow. Single-cell RNA sequencing analysis revealed that Reg1 and Reg3 regulate lineage directions in HSPCs via the control of a set of myeloid-related genes. Reg1- and Reg3-mediated control of mRNA encoding Nfkbiz, a transcriptional and epigenetic regulator, was essential for balancing lymphoid/myeloid lineage output in HSPCs in vivo. Furthermore, single-cell assay for transposase-accessible chromatin sequencing analysis revealed that Reg1 and Reg3 control the epigenetic landscape on myeloid-related gene loci in early stage HSPCs via Nfkbiz. Consistently, an antisense oligonucleotide designed to inhibit Reg1- and Reg3-mediated Nfkbiz mRNA degradation primed hematopoietic stem cells toward myeloid lineages by enhancing Nfkbiz expression. Collectively, the collaboration between posttranscriptional control and chromatin remodeling by the Reg1/Reg3-Nfkbiz axis governs HSPC lineage biases, ultimately dictating the fate of lymphoid vs myeloid differentiation.
Introduction
Hematopoietic stem and progenitor cells (HSPCs) in the bone marrow (BM) generate multilineages that contribute to the production of all mature blood and immune cells. This process is tightly controlled to maintain the balance between lymphoid, myeloid, erythroid, and megakaryocytic cell populations under steady state conditions.1-3 Consistent with the recent lineage-biased output model of HSPCs,4-7 the pattern of gene expression in the progenitor reflects that of the mature cell type toward which it is biased, a phenomenon known as transcriptional lineage priming.1,8,9 This is initiated by cell-intrinsic and -extrinsic factors controlling lineage-determining transcription programs. Under microbial infection, inflammatory cytokines such as interleukin-1β (IL-1β) and tumor necrosis factor (TNF) trigger reprograming of transcriptional networks in HSPCs to prime myelopoiesis by inducing the lineage-determining transcription factors (TFs) PU.1 and C/EBPβ.10-17 In addition, alterations in epigenetic landscape can contribute to transcriptional lineage priming in a cell-intrinsic manner.18-20 However, molecular mechanisms that explain the link between lineage output and transcriptional status need to be clarified.
Regnase-1 (Reg1) is an endoribonuclease that degrades messenger RNAs (mRNAs) harboring stem-loop (SL) structures with a pyrimidine-purine-pyrimidine loop in the 3’ untranslated regions (UTRs).21-23 Reg1 is critical for the suppression of inflammatory responses in mature immune cells and for the maintenance of immune homeostasis.24,25 There are 4 regnase family members (Reg1 to Reg4) in mammals characterized by the presence of a ribonuclease (RNase) domain and a CCCH-type zinc finger (Zf) domain. However, it remains unclear to what extent the regnase family members play redundant roles in the immune system.
In this study, we clarified the roles of Reg1 and Reg3 in HSPC lineage choice through the control of Nfkbiz mRNA stability. Further, we developed a method that induces hematopoietic stem cell (HSC) myeloid priming by using an antisense oligonucleotide (ASO) that inhibits Reg1/Reg3-mediated Nfkbiz degradation.
Methods
scRNA-seq
Single-cell sorting for single-cell RNA sequencing (scRNA-seq) was performed as previously described.26,27 Briefly, lineage-negative (Lin–) cells were purified from mice competitively reconstituted with wild type (WT; CD45.1) and Reg1/Reg3-deficient (DKO; CD45.2) fetal liver (FL) cells. WT and DKO Lin–Sca1+Kit+ (LSK) cells were sc-sorted into 384-well cell capture plates containing lysis solution and barcoded poly(T) reverse-transcription (RT) primers. Immediately after sorting, each plate was spun down to ensure cell immersion into the lysis solution, snap frozen on dry ice, and stored at −80°C until processed.
Full details are provided in the supplemental Methods (available on the Blood website).
Results
Reg1 and Reg3 are essential for balanced lineage output in BM
Among regnase family members, Lin– hematopoietic progenitor highly express Reg1 and Reg3 (supplemental Figure 1A-C). Therefore, we investigated whether Reg1 and Reg3 regulate hematopoietic differentiation in the BM. Because mice lacking both Reg1 and Reg3 (DKO) but not Reg1 or Reg3 alone (supplemental Figure 1C-E) exhibited perinatal lethality, accompanied by marked infiltration of neutrophils in the periportal region of the liver and the parenchyma of the pancreas (supplemental Figure 1F). To investigate hematopoiesis, we performed fetal liver (FL) transplantation using E15.5 fetuses of Reg1–/–, Reg3–/–, or DKO donors and euthanized them 4 to 6 weeks after transplantation. Interestingly, DKO FL-transplanted mice displayed a profound loss of B220+CD19+ B cells and a reciprocal increase in CD11b+ myeloid cells in the peripheral blood compared with control (supplemental Figure 1G-H). In contrast, deficiency of Reg1 or Reg3 alone did not lead to profound changes in immune cells. Consistently, E15.5 DKO FL cells exhibited decreased B cells and increased myeloid cells (supplemental Figure 1I). These results suggest the redundant roles of Reg1 and Reg3 in hematopoiesis.
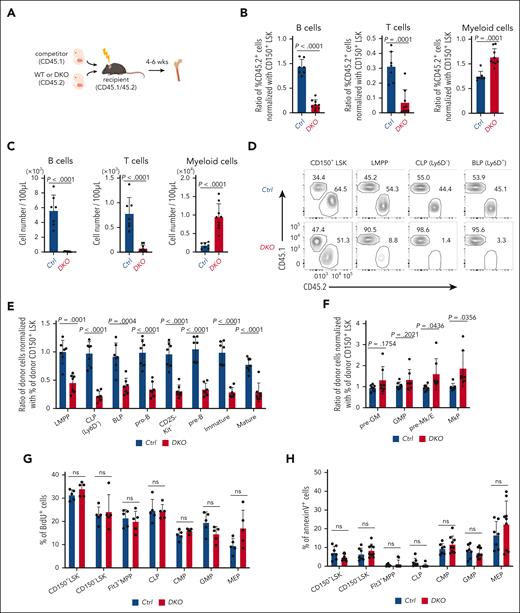
To investigate whether the influence of Reg1/Reg3 deficiency on BM hematopoiesis is cell-intrinsic, we performed competitive FL transplants. We engrafted equal numbers of control or DKO FL cells (CD45.2) alongside competitor cells (CD45.1) into lethally irradiated recipient mice (CD45.1/45.2) (Figure 1A). We observed comparable engraftment levels in mice receiving control and DKO FL cells (supplemental Figure 2A). DKO FL–transplanted mice showed a significant reduction of B/T-lymphocytes and a concomitant increase in myeloid cells in the peripheral blood in frequencies and numbers (Figure 1B-C). A similar trend was observed in the BM, marked by reduced B-cell numbers and an increase in Gr-1+CD11b+ myeloid cells (supplemental Figure 2B-C). Consistently, thymic T cell populations were reduced in DKO mice compared with that of controls (supplemental Figure 2D). The analysis of BM hematopoietic progenitors in mixed chimera revealed that DKO cells failed to give rise to all B cell lineage cells and showed unimpaired or mildly increased proportion of megakaryocytic-erythroid and myeloid lineages (Figure 1D-F).
Reg1 and Reg3 are essential for early stage lymphopoiesis in a cell-intrinsic manner. (A-F) Competitive FL transplantation. (A) Experimental workflow of competitive transplantation using FL cells (E15.5). (B-C) Relative frequencies and cell numbers of donor immune cells in the peripheral blood. (B) Each dot represents the ratio of the percentage of CD45.2 in each population to the percentage of CD45.2 in SLAM+ Lin–Sca1+Kit+ (LSK) from recipient mice. Donor B cells, T cells, and myeloid cells were defined as CD45.2+ B220+CD19+ cells, CD4/8+TCRβ+ cells, and CD11b+ cells, respectively. (D) Representative flow cytometric plots of CD45.1 and CD45.2 expression gated on CD150+ LSK, LMPP (Flt3hiLSK), CLP (Ly6D–Flt3+IL-7Rα+Sca1dullKitdull), and BLP (Ly6D+Flt3+IL-7Rα+Sca1dullKitdull). Numbers in plots indicate the percentage of CD45.1+ or CD45.2+ cells. (E-F) Relative frequencies of each population in the BM. Each dot represents the ratio of the percentage of CD45.2 in each population to the percentage of CD45.2 in CD150+ LSK from recipient mice. (G-H) in vivo BrdU incorporation and annexin V staining in CD150+ LSK, CD150– LSK, Flt3+ MPP, CLP, CMP, GMP, and MEP in the BM of mice competitively transplanted with FL cells, as in panel A. (E-F) Data are a composite of 3 independent experiments. (G-H) Data are representative from 3 independent experiments. Data are presented as mean ± standard deviation (SD) (B-C,E-H). Statistical significance was calculated by unpaired 2-tailed Student t test (B-C,E-H). BrdU, Bromodeoxyuridine; BLP, B-cell-biased lymphoid progenitor; CLP, common lymphoid progenitor; CMP, granulocyte-monocyte progenitor; GMP, granulocyte-monocyte progenitor; MEP, megakaryocyte–erythroid progenitor; LMPP, lymphoid primed multipotent progenitor; ns, not significant.
Reg1 and Reg3 are essential for early stage lymphopoiesis in a cell-intrinsic manner. (A-F) Competitive FL transplantation. (A) Experimental workflow of competitive transplantation using FL cells (E15.5). (B-C) Relative frequencies and cell numbers of donor immune cells in the peripheral blood. (B) Each dot represents the ratio of the percentage of CD45.2 in each population to the percentage of CD45.2 in SLAM+ Lin–Sca1+Kit+ (LSK) from recipient mice. Donor B cells, T cells, and myeloid cells were defined as CD45.2+ B220+CD19+ cells, CD4/8+TCRβ+ cells, and CD11b+ cells, respectively. (D) Representative flow cytometric plots of CD45.1 and CD45.2 expression gated on CD150+ LSK, LMPP (Flt3hiLSK), CLP (Ly6D–Flt3+IL-7Rα+Sca1dullKitdull), and BLP (Ly6D+Flt3+IL-7Rα+Sca1dullKitdull). Numbers in plots indicate the percentage of CD45.1+ or CD45.2+ cells. (E-F) Relative frequencies of each population in the BM. Each dot represents the ratio of the percentage of CD45.2 in each population to the percentage of CD45.2 in CD150+ LSK from recipient mice. (G-H) in vivo BrdU incorporation and annexin V staining in CD150+ LSK, CD150– LSK, Flt3+ MPP, CLP, CMP, GMP, and MEP in the BM of mice competitively transplanted with FL cells, as in panel A. (E-F) Data are a composite of 3 independent experiments. (G-H) Data are representative from 3 independent experiments. Data are presented as mean ± standard deviation (SD) (B-C,E-H). Statistical significance was calculated by unpaired 2-tailed Student t test (B-C,E-H). BrdU, Bromodeoxyuridine; BLP, B-cell-biased lymphoid progenitor; CLP, common lymphoid progenitor; CMP, granulocyte-monocyte progenitor; GMP, granulocyte-monocyte progenitor; MEP, megakaryocyte–erythroid progenitor; LMPP, lymphoid primed multipotent progenitor; ns, not significant.
We then asked whether Reg1 and Reg3 were required for B-lymphopoiesis in uncommitted or lymphoid lineage-committed progenitor stages. To address this, we used Il7r-cre+ROSA-yellow fluorescent protein (YFP)+Reg1fl/flReg3–/– mice (DKOIl7r-cre), in which Reg1 gene was deleted via Il7r-driven cre recombination. We confirmed Il7r-cre–mediated recombination at the pro-B stage and, to a lesser extent, the common lymphoid progenitor (CLP) based on RT polymerase chain reaction (RT-PCR) for Reg1 and YFP expression (supplemental Figure 2E-F). Through competitive FL transplantation experiments, we observed that B-cell differentiation remained unchanged in YFP-expressing DKOIl7r-cre cells compared with their control counterparts (supplemental Figure 2G). These results indicate that Reg1 and Reg3 function at uncommitted stages of B-lymphopoiesis and become dispensable once the cells undergo lineage commitment.
To understand how Reg1 and Reg3 regulate BM hematopoiesis, we examined the proliferative activity and cell viability of uncommitted progenitors. In vivo bromodeoxyuridine (BrdU) incorporation experiments using mixed chimera receiving control and DKO FL cells showed no difference in proliferative activity of CD150+ or CD150– LSK, Flt3+ multipotent progenitor (MPP), CLP, or myeloid progenitors such as common myeloid progenitor (CMP), granulocyte-monocyte progenitor (GMP), and megakaryocyte–erythroid progenitor (MEP) between these genotypes (Figure 1G). In addition, annexin V–positive apoptotic cell frequencies were comparable between the control and DKO (Figure 1H). These results suggest that Reg1 and Reg3 regulate lymphoid lineage differentiation, rather than affecting cell proliferation or viability.
To evaluate the roles of Reg1 and Reg3 in steady-state hematopoiesis, we generated Reg1 conditional knockout mice crossed with Reg3–/– animals, using tamoxifen-inducible cre recombinase system (supplemental Figure 3A-B). CreERT2+Reg1fl/flReg3–/– (DKOCreERT2) mice also exhibited a reduction in B220+CD19+ B cells and an increase in CD11b+ myeloid cells both in frequencies and numbers, at day 14 after tamoxifen injection (supplemental Figure 3C-D). We next examined HSPC subpopulations defined as long-term HSC (LT-HSC) (Flt3–CD150+CD48–LSK), short-term HSC (ST-HSC) (Flt3–CD150–CD48–LSK), MPP2 (Flt3–CD150+CD48+LSK), MPP3 (Flt3–CD150–CD48+LSK), MPP4(Flt3+CD150–CD48+LSK), and CLPs (Flt3+IL-7Rα+Lin–Sca1dullKitdull) (supplemental Figure 3E). DKOCreERT2 mice displayed a significant reduction in CLPs, whereas ST-HSC, MPP2, and MPP3 showed increased numbers compared with that of control mice (supplemental Figure 3F-G).
Taken together, our findings suggest that Reg1 and Reg3 are important for balancing lymphoid-myeloid differentiation in HSPCs.
Reg1 and Reg3 are required for the priming of lymphoid-biased HSPCs
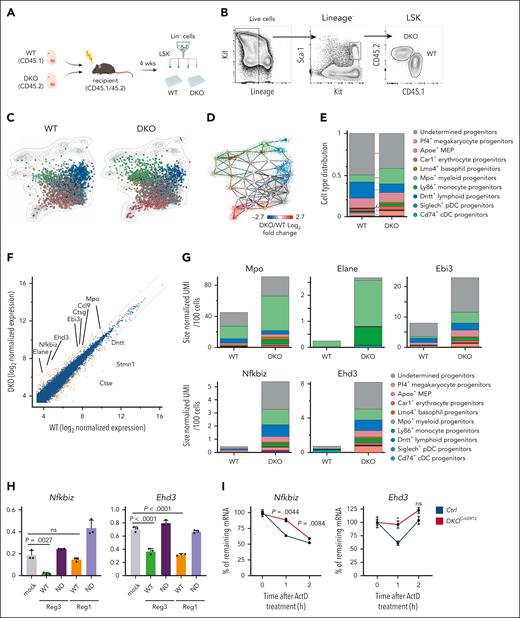
Recent scRNA-seq approaches suggest that transcriptional lineage priming is initiated in HSPCs and is correlated with their functional lineage biases.1,15,26,28 To investigate whether Reg1 and Reg3 contribute to shaping lineage biases in HSPCs, we performed scRNA-seq analysis of WT and DKO LSK cells containing HSCs and MPPs from competitively FL-transferred mice (Figure 2A). WT and DKO LSK populations were single cell–sorted into 384 well plates, and 2241 and 2242 cells were analyzed from each background, respectively (Figure 2B; supplemental Figure 4A). We used the MetaCell approach by which homologous populations were grouped based on similar transcriptional profiles to distinguish between closely related cell types (supplemental Figure 4B).26 When WT and DKO cells were projected onto the hematopoietic core map,26DKO cells were enriched in the direction of myeloid and megakaryocytic-erythroid progenitors compared with control, whereas lymphoid lineages were underrepresented in DKO cells (Figure 2C). Consistently, meta cells representing myeloid- and megakaryocytic/erythroid-biased populations increased in DKO samples compared with that of WT, whereas those representing lymphoid populations decreased in DKO samples (Figure 2C-D; supplemental Figure 4C; supplemental Table 1). The expression levels of phenotypic markers for myeloid-biased HSCs, such as CD41, did not significantly increase in DKO cells (supplemental Figure 4D-F). Thus, our results support the notion that Reg1 and Reg3 are critical for the generation of B-lymphoid lineages from HSCs to maintain balanced hematopoiesis.
Reg1 and Reg3 control lineage biases within HSPCs. (A) Schematic representation of experimental strategy for single-cell sorting. (B) Gating strategy for WT and Reg1–/–Reg3–/– (DKO) LSK cells sorted from the identical mouse receiving WT (CD45.1) and DKO (CD45.2) FL cells. (C) A 2-dimensional projection of 2241 single cells sorted from WT (CD45.1) LSK and 2242 single cells from DKO (CD45.2) LSK onto the reference model. Cells are colored according to assignment to reference model meta cells (see “Methods”). (D) Difference in meta-cell abundance between single cells sorted from DKO and WT mice. Each circle represents a reference meta cell. Circle color represents log2 (fold change) in meta-cell abundance between the 2 genetic backgrounds. (E) Cell type distribution of single cells sorted from WT and DKO mice. ∗P < .05; ∗∗P < .001; ∗∗∗P < .00001. (F) Differential gene expression between pooled cells from DKO cells compared with WT cells. Gold dots indicate differentially expressed genes (FDR corrected P < .001 under χ2 test) with fold change >2. Diagonal lines show log2 (fold change) = −1 or 1. Values represent log2 size–normalized expression. (G) Total expression of specific genes in different genetic backgrounds. Values represent size-normalized total transcripts per 100 cells. Colors represent relative contribution from the different hematopoietic core subsets. (H) Luciferase activity was assessed by using pGL3 plasmids containing the 3’ UTR of Nfkbiz and Ehd3, together with mock or plasmids for WT or nuclease-dead mutant for Reg1 and Reg3 (n = 3 each). Data are representative of at least 3 independent experiments. (I) mRNA stability of Nfkbiz and Ehd3 in Lin– cells from control and DKOCreERT2 mice. Cells were stimulated with IL-1β for 2 hours, followed by addition of actinomycin D (ActD). The remaining transcripts relative to the 0-hour time point were measured by quantitative RT-PCR. Data are representative of 2 independent experiments (H-I). Data are presented as mean ± SD (H) or ± standard error of the mean (I). Statistical significance was calculated by 2-tailed Fisher exact test (E), 1-way analysis of variance (ANOVA) with Holm-Sidak multiple comparisons test (H), or unpaired 2-tailed Student t test (I). ND, nuclease dead.
Reg1 and Reg3 control lineage biases within HSPCs. (A) Schematic representation of experimental strategy for single-cell sorting. (B) Gating strategy for WT and Reg1–/–Reg3–/– (DKO) LSK cells sorted from the identical mouse receiving WT (CD45.1) and DKO (CD45.2) FL cells. (C) A 2-dimensional projection of 2241 single cells sorted from WT (CD45.1) LSK and 2242 single cells from DKO (CD45.2) LSK onto the reference model. Cells are colored according to assignment to reference model meta cells (see “Methods”). (D) Difference in meta-cell abundance between single cells sorted from DKO and WT mice. Each circle represents a reference meta cell. Circle color represents log2 (fold change) in meta-cell abundance between the 2 genetic backgrounds. (E) Cell type distribution of single cells sorted from WT and DKO mice. ∗P < .05; ∗∗P < .001; ∗∗∗P < .00001. (F) Differential gene expression between pooled cells from DKO cells compared with WT cells. Gold dots indicate differentially expressed genes (FDR corrected P < .001 under χ2 test) with fold change >2. Diagonal lines show log2 (fold change) = −1 or 1. Values represent log2 size–normalized expression. (G) Total expression of specific genes in different genetic backgrounds. Values represent size-normalized total transcripts per 100 cells. Colors represent relative contribution from the different hematopoietic core subsets. (H) Luciferase activity was assessed by using pGL3 plasmids containing the 3’ UTR of Nfkbiz and Ehd3, together with mock or plasmids for WT or nuclease-dead mutant for Reg1 and Reg3 (n = 3 each). Data are representative of at least 3 independent experiments. (I) mRNA stability of Nfkbiz and Ehd3 in Lin– cells from control and DKOCreERT2 mice. Cells were stimulated with IL-1β for 2 hours, followed by addition of actinomycin D (ActD). The remaining transcripts relative to the 0-hour time point were measured by quantitative RT-PCR. Data are representative of 2 independent experiments (H-I). Data are presented as mean ± SD (H) or ± standard error of the mean (I). Statistical significance was calculated by 2-tailed Fisher exact test (E), 1-way analysis of variance (ANOVA) with Holm-Sidak multiple comparisons test (H), or unpaired 2-tailed Student t test (I). ND, nuclease dead.
DKO HSCs shift toward myeloid-biased hematopoiesis upon serial transplantation
We further assessed the long-term maintenance of lineage-biased hematopoiesis in DKO HSCs by transplanting LT-HSCs isolated from CreERT2+ control or CreERT2+Reg1fl/flReg3–/– mice (DKOCreERT2) along with supportive BM cells into lethally irradiated recipients (supplemental Figure 5A). DKOCreERT2 LT-HSCs gave rise to a higher percentage of granulocytes and monocytes and a lower percentage of B/T-lymphocytes than control after primary transplantation (supplemental Figure 5B). In secondary recipients, DKOCreERT2 LT-HSC–derived cells maintained skewed lineage outputs toward granulocytes and monocytes and away from B/T-lymphocytes, compared with that of control LT-HSC–derived cells (supplemental Figure 5C). Notably, the number of LT-HSCs was not altered 3 months after primary and secondary transplantation (supplemental Figure 5D-E). These finding indicate that myeloid-biased hematopoiesis persisted in the absence of Reg1 and Reg3 even after secondary transplantation.
Reg1 and Reg3 cooperatively regulate a set of mRNAs including Nfkbiz and Ehd3
We next explored how degradation of a common set of mRNAs in HSCs allows for Reg1 and Reg3 to prime and maintain B-lymphoid lineage–biased MPPs. We compared differentially expressed genes between WT and DKO HPSCs by reanalyzing the scRNA-seq data (supplemental Table 2). Remarkably, most of the upregulated genes in DKO cells were myeloid-lineage–associated genes, such as Mpo, Ccl9, Ctsg, Ebi3, and Elane (Figure 2F). In contrast, downregulated genes included Dntt and Stmn1, which are required for lymphoid development and suppression of megakaryocytic differentiation, respectively.29,30 Consistently, Mpo, Elane, and Ebi3 were highly expressed in myeloid and monocyte progenitors of DKO cells compared with WT counterparts (Figure 2G; supplemental Figure 6A). In addition to myeloid-associated genes, Nfkbiz and Ehd3 were found to be upregulated in DKO cells (Figure 2F-G; supplemental Figure 6A). Interestingly, these genes were increased in DKO cells even in undetermined progenitors, which potentially contain HSCs, as well as lymphoid- and myeloid-biased progenitors (Figure 2G; supplemental Figure 6A). Quantitative PCR analysis using mixed chimera mice receiving WT and DKO FL cells confirmed a significant elevation of Nfkbiz and Ehd3 in DKO CD150+LSK and lymphoid primed multipotent progenitor (LMPP), compared with WT cells (supplemental Figure 6B). On the contrary, lineage-related TFs that can skew HSPC differentiation toward myeloid or megakaryocytic-erythroid lineages, including Cebpa, Cebpb, Spi1, and Gata1, were not altered between WT and DKO cells (supplemental Figure 6B).
Then, we examined whether the genes increased in DKO HSPCs were directly controlled by these RNases. Luciferase reporter experiments showed that both Reg1 and Reg3 inhibited the luciferase activity in cells expressing luciferase constructs harboring 3’ UTRs of Nfkbiz and Ehd3 (Figure 2H). In contrast, nuclease-dead mutants of Reg1 (D141N) and Reg3 (D271N) failed to suppress the reporter activity, suggesting that Reg1 and Reg3 inhibited Nfkbiz and Ehd3 in a manner dependent on their RNase enzymatic activities. The 3’ UTRs of Irf8 and Cebpd were also responsive to the overexpression of Reg1 or Reg3, but no significant change in their gene expressions was observed in DKO progenitors (supplemental Figure 6B-C). However, Reg1/Reg3-doubly–deficient Lin– cells showed enhanced stability of Nfkbiz transcript and had only a partial effect on Ehd3, whereas the stability of Irf8 or Cebpd mRNAs were not influenced by the double-deficiency of Reg1 and Reg3 (Figure 2I; supplemental Figure 6D), confirming that both Reg1 and Reg3 are required to downregulate or suppress Nfkbiz and Ehd3 mRNA levels.
Reg1 and Reg3 control lymphoid-myeloid lineage balance via Nfkbiz in HSPCs
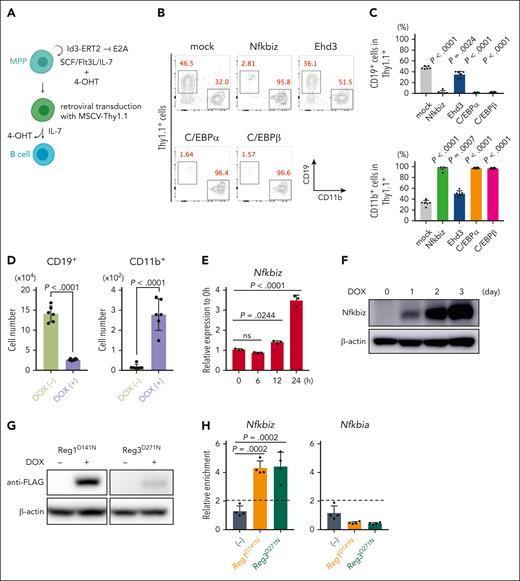
To investigate whether Nfkbiz or Ehd3 influences lymphoid and/or myeloid differentiation in HSPCs, we used a culture system of mouse primary MPPs maintained by overexpressing human Id3-ERT2 fusion protein (MPPhId3-ERT2) in the presence of 4-hydroxytamoxifen (Figure 3A).31 We retrovirally expressed Nfkbiz, Ehd3, and TFs implicated in myeloid differentiation as controls in MPPhId3-ERT2 cells. As expected, expression of myeloid master regulators, C/EBPα and C/EBPβ, strongly blocked B-cell differentiation and induced myeloid differentiation (Figure 3B-C). Interestingly, Nfkbiz expression in MPPhId3-ERT2 cells resulted in a profound defect in lymphoid differentiation and promoted myeloid-lineage differentiation to a degree similar to the effects of C/EBPα and C/EBPβ, whereas Ehd3 showed only a modest effect on myeloid-lymphoid differentiation in MPPhId3-ERT2 cells (Figure 3B-C). We then established MPPhId3-ERT2 cells from Reg1fl/flReg3–/– BM cells in which cre recombinase could be expressed by doxycycline (DOX) treatment in vitro (tetO-cre-Reg1fl/flReg3–/– MPPhId3-ERT2) (supplemental Figure 7A-C). Consistent with the hematopoietic changes in the BM caused by double-deficiency of Reg1 and Reg3, tetO-cre-Reg1fl/flReg3–/– MPPhId3-ERT2 cells failed to induce CD19+ cells but reciprocally increased CD11b+ myeloid cells under B-cell differentiating condition in the presence of DOX (Figure 3D). The Nfkbiz expression was strikingly elevated after Reg1 depletion both at mRNA and protein levels (Figure 3E-F). RNA-immunoprecipitation (IP) experiments showed that nuclease-dead mutants of Reg1 or Reg3 coprecipitated mRNA for Nfkbiz but not a Reg1 nontarget gene Nfkbia (Figure 3G-H), indicating that both Reg1 and Reg3 directly interact with Nfkbiz mRNA.
Nfkbiz is a direct target gene for Reg1 and Reg3 that is potent to promote myelopoiesis. (A) Schematic representation for B-cell differentiation of multipotent progenitor (MPPhId3-ERT2) cells retrovirally transduced with MSCV-Thy1.1 vector. Cells were then cultured under B-cell–differentiation condition. (B-C) In vitro B-cell differentiation in MPPhId3-ERT2 cells retrovirally overexpressing Nfkbiz and Ehd3 as well as myeloid-lineage TFs. At day 7, cells were analyzed by flow cytometry. Flow cytometry plots of CD19 and CD11b after gated on Thy1.1+ cells (B) and percentages of CD19+ and CD11b+ Thy1.1+ cells (C) were shown (n = 6 each). (D) Numbers of CD19+CD11b– and CD19–CD11b+ cells in tetO-cre-Reg1fl/flReg3–/– MPPhId3-ERT2 cells cultured under B-cell–differentiating condition in the presence or absence of DOX at day 6 (n = 6 each). (E-F) Quantitative RT-PCR (E) and immunoblot analysis (F) of Nfkbiz in tetO-cre-Reg1fl/flReg3–/– MPPhId3-ERT2 cells at the indicated time points after DOX treatment (n = 3 each in E). Cells were cultured under stem-cell condition for the indicated time. (G) DOX-inducible overexpression of FLAG-tagged Reg1 (D141N) or Reg3 (D271N) in tetO-cre-Reg1fl/flReg3–/– MPPhId3-ERT2 cells. (H) RNA immunoprecipitation quantitative PCR (RIP-qPCR) analysis of a set of genes from lysates of tetO-cre-Reg1fl/flReg3–/– MPPhId3-ERT2 cells expressing FLAG-tagged Reg1 or Reg3 as in (G) (n = 4 each). Data are representative of 2 (B-D, G-H) or at least 3 (E-F) independent experiments. Data are presented as mean ± SD (C-E,H). Statistical significance was calculated by 1-way ANOVA with Holm-Sidak multiple comparisons test (C,E,H) or unpaired 2-tailed Student t test (D).
Nfkbiz is a direct target gene for Reg1 and Reg3 that is potent to promote myelopoiesis. (A) Schematic representation for B-cell differentiation of multipotent progenitor (MPPhId3-ERT2) cells retrovirally transduced with MSCV-Thy1.1 vector. Cells were then cultured under B-cell–differentiation condition. (B-C) In vitro B-cell differentiation in MPPhId3-ERT2 cells retrovirally overexpressing Nfkbiz and Ehd3 as well as myeloid-lineage TFs. At day 7, cells were analyzed by flow cytometry. Flow cytometry plots of CD19 and CD11b after gated on Thy1.1+ cells (B) and percentages of CD19+ and CD11b+ Thy1.1+ cells (C) were shown (n = 6 each). (D) Numbers of CD19+CD11b– and CD19–CD11b+ cells in tetO-cre-Reg1fl/flReg3–/– MPPhId3-ERT2 cells cultured under B-cell–differentiating condition in the presence or absence of DOX at day 6 (n = 6 each). (E-F) Quantitative RT-PCR (E) and immunoblot analysis (F) of Nfkbiz in tetO-cre-Reg1fl/flReg3–/– MPPhId3-ERT2 cells at the indicated time points after DOX treatment (n = 3 each in E). Cells were cultured under stem-cell condition for the indicated time. (G) DOX-inducible overexpression of FLAG-tagged Reg1 (D141N) or Reg3 (D271N) in tetO-cre-Reg1fl/flReg3–/– MPPhId3-ERT2 cells. (H) RNA immunoprecipitation quantitative PCR (RIP-qPCR) analysis of a set of genes from lysates of tetO-cre-Reg1fl/flReg3–/– MPPhId3-ERT2 cells expressing FLAG-tagged Reg1 or Reg3 as in (G) (n = 4 each). Data are representative of 2 (B-D, G-H) or at least 3 (E-F) independent experiments. Data are presented as mean ± SD (C-E,H). Statistical significance was calculated by 1-way ANOVA with Holm-Sidak multiple comparisons test (C,E,H) or unpaired 2-tailed Student t test (D).
We then asked whether the lineage bias in hematopoiesis in the absence of Reg1 and Reg3 depends on in vivo Nfkbiz expression. For this purpose, we crossed DKOCreERT2 mice, which showed elevated Nfkbiz expression in Lin– BM cells (supplemental Figure 7D), with Nfkbizfl/fl (TKOCreERT2) mice and analyzed them at day14 after tamoxifen injection. Intriguingly, TKOCreERT2 mice showed a dramatic improvement in the frequencies and numbers of HSPC subpopulations compared with that of DKOCreERT2 mice (Figure 4A-C). In parallel, the frequency and number of CD19+B220+ B cells, as well as CD11b+ myeloid cells, were fully restored in TKOCreERT2 mice (Figure 4D-I). Moreover, B-cell differentiation potential was also restored in TKOCreERT2 cells in an in vitro B-cell differentiation assay (Figure 4J-L), indicating that the upregulation of Nfkbiz accounts for the alteration in HSC lineage determination under the Reg1/Reg3 double deficiency. Taken together, these findings demonstrate that the posttranscriptional control of Nfkbiz expression by Reg1 and Reg3 licenses lymphopoiesis by suppressing myeloid differentiation in HSPCs.
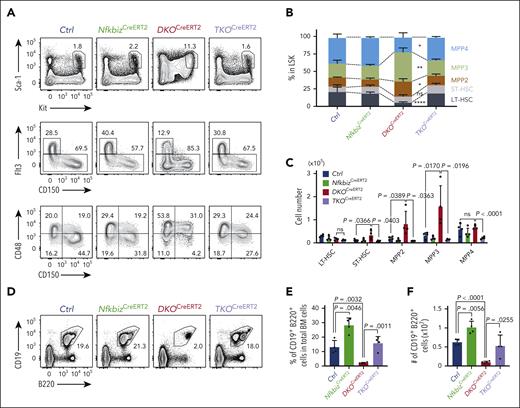
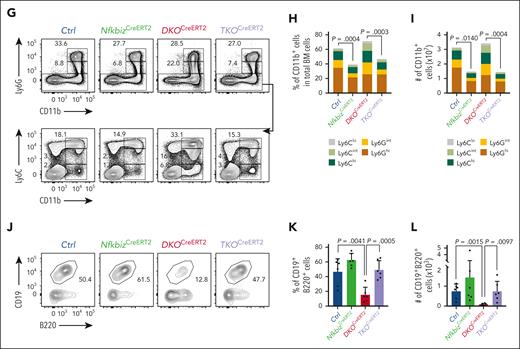
The Reg1/Reg3-Nfkbiz axis controls lymphoid-myeloid fate decision. (A-C) Representative flow cytometry plots (A), frequencies (B) and cell numbers (C) of HSPC subpopulations in mice of the indicated genotypes at day 14 after tamoxifen treatment. Ctrl, CreERT2+Reg1fl/+Reg3+/+Nfkbizfl/+; NfkbizCreERT2, CreERT2+Reg1fl/+Reg3+/+Nfkbizfl/fl; DKOCreERT2, CreERT2+Reg1fl/flReg3–/–Nfkbiz+/+; TKOCreERT2, CreERT2+Reg1fl/flReg3–/–Nfkbizfl/fl (n = 4 each). Asterisks represents P value (∗P < .05; ∗∗P < .01; ∗∗∗∗P < .0001). (D-F) Representative flow cytometry plots (D), and frequency (E) and cell number (F) of B cells in the indicated mice (n = 4 each). (G-I) Representative flow cytometry plots (G) and frequencies (H) and cell numbers (I) of myeloid cells in the indicated mice (n = 4 each). (J-L) In vitro B-cell differentiation using Lin–Kit+IL-7Rα+ FL cells derived from the indicated genotypes. Progenitor cells were differentiated under B-cell–promoting condition in the presence of 4-OHT. Ctrl, CreERT2+Reg1fl/+Reg3+/+Nfkbizfl/+; NfkbizCreERT2, CreERT2+Reg1fl/+Reg3+/+Nfkbizfl/fl; DKOCreERT2, CreERT2+Reg1fl/flReg3–/–Nfkbiz+/+; TKOCreERT2, CreERT2+Reg1fl/flReg3–/–Nfkbizfl/fl. Representative flow cytometry plots (J), frequency (K), and absolute number (L) of CD19+B220+ cells are shown. n = 6. Data are a composite of 2 independent experiments (A-I) or a representative of 2 independent experiments (J-L). Data are presented as mean ± SD (C,E-F,H-I,K-L). Statistical significance was calculated by 1-way ANOVA with Holm-Sidak multiple comparisons test. 4-OHT, 4-hydroxytamoxifen.
The Reg1/Reg3-Nfkbiz axis controls lymphoid-myeloid fate decision. (A-C) Representative flow cytometry plots (A), frequencies (B) and cell numbers (C) of HSPC subpopulations in mice of the indicated genotypes at day 14 after tamoxifen treatment. Ctrl, CreERT2+Reg1fl/+Reg3+/+Nfkbizfl/+; NfkbizCreERT2, CreERT2+Reg1fl/+Reg3+/+Nfkbizfl/fl; DKOCreERT2, CreERT2+Reg1fl/flReg3–/–Nfkbiz+/+; TKOCreERT2, CreERT2+Reg1fl/flReg3–/–Nfkbizfl/fl (n = 4 each). Asterisks represents P value (∗P < .05; ∗∗P < .01; ∗∗∗∗P < .0001). (D-F) Representative flow cytometry plots (D), and frequency (E) and cell number (F) of B cells in the indicated mice (n = 4 each). (G-I) Representative flow cytometry plots (G) and frequencies (H) and cell numbers (I) of myeloid cells in the indicated mice (n = 4 each). (J-L) In vitro B-cell differentiation using Lin–Kit+IL-7Rα+ FL cells derived from the indicated genotypes. Progenitor cells were differentiated under B-cell–promoting condition in the presence of 4-OHT. Ctrl, CreERT2+Reg1fl/+Reg3+/+Nfkbizfl/+; NfkbizCreERT2, CreERT2+Reg1fl/+Reg3+/+Nfkbizfl/fl; DKOCreERT2, CreERT2+Reg1fl/flReg3–/–Nfkbiz+/+; TKOCreERT2, CreERT2+Reg1fl/flReg3–/–Nfkbizfl/fl. Representative flow cytometry plots (J), frequency (K), and absolute number (L) of CD19+B220+ cells are shown. n = 6. Data are a composite of 2 independent experiments (A-I) or a representative of 2 independent experiments (J-L). Data are presented as mean ± SD (C,E-F,H-I,K-L). Statistical significance was calculated by 1-way ANOVA with Holm-Sidak multiple comparisons test. 4-OHT, 4-hydroxytamoxifen.
The Reg1/Reg3-Nfkbiz axis shapes epigenetic lineage priming in HSC
Because Nfkbiz acts as a transcriptional modulator inducing chromatin remodeling,32,33 we hypothesized that the increased Nfkbiz expression modified epigenetic landscape in HSCs. To address this possibility, we examined the single-cell chromatin accessibility landscape using single cell-assay for transposase-accessible chromatin sequencing (scATAC-seq) of Flt3–CD48– LSK population, which contains HSCs and the intermediate cells between HSCs and CD48+ lineage-biased MPPs (Figure 5A).34,35 We sequenced 13 859, 9936, 10 991, and 6902 cells for control, NfkbizCreERT2, DKOCreERT2, and TKOCreERT2, respectively. Uniform Manifold approximation and projection (UMAP) plot for total cells showed that clustering based on the differential chromatin accessibilities generated 18 clusters (C1-C18) (Figure 5B). To dissect the chromatin accessibility profiles across the clusters, we examined the selected markers for HSCs and mature cell lineages.26,36 The chromatin of stemness-related genes (Hoxa9, Hoxa10, Gata2, Mllt3, Hlf, Meis1, and Msi2) was more accessible in clusters C1 to C8 than in the rest of the clusters (Figure 5C; supplemental Figure 8A-C). In contrast, clusters C10 to C14 displayed distinct patterns of accessibilities in a set of myeloid-related genes (Spi1, Csf1r, Csf2rb, Csf3r, F13a1, and Fcer1g) and lymphoid-related genes (Flt3 and Satb1) (Figure 5C; supplemental Figure 8A-C), suggesting that cells in these clusters harbor myeloid/lymphoid epigenetic features reminiscent of MPP5 and MPP6 that are phenotypically defined as CD34– and CD34+ Flt3–CD150–CD48– LSK, respectively.35 Among clusters C1 to C8, clusters C1 to C5 were consolidated in the UMAP plot and showed common features in the increased accessibility of lymphoid- and myeloid-related genes compared with that of clusters C6 to C8 (Figure 5B-C). In contrast, erythroid-related genes (Gata1, Epor, and Car1) were accessible in clusters C6 to C8 (Figure 5C).
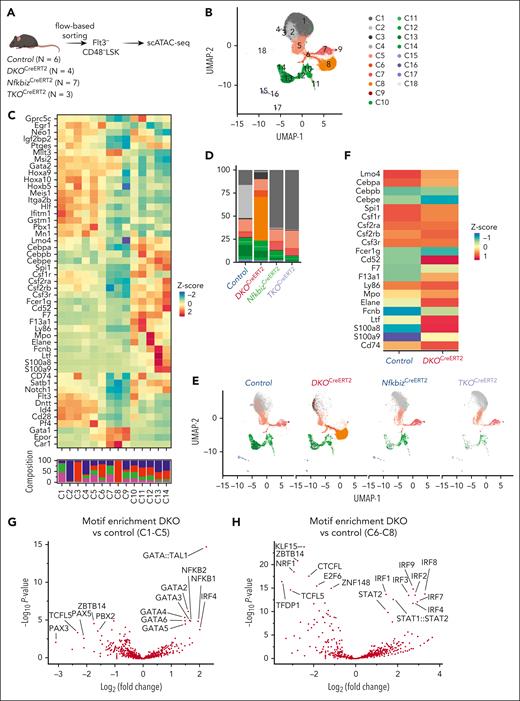
The Reg1/Reg3-Nfkbiz axis regulates lineage priming of HSCs/early MPPs via chromatin remodeling. (A) Schematic representation of scATAC-seq using Flt3–CD48– LSK. In each genotype, samples are pooled from multiple mice to create a single sample for sequencing, as indicated by the number of mice represented. (B) UMAP visualization of all cells obtained from scATAC-seq data. Colors represent the cluster identity. (C) Heatmap showing chromatin accessibility for stemness-related and lineage-associated genes. (D) Cluster distribution of single cells across the indicated genotypes. Colors represent the cluster identity as in (B). (E) A 2-dimensional projection of 13859, 10991, 9936, and 6902 single cells from control, DKOCreERT2, NfkbizCreERT2, and TKOCreERT2, respectively, onto the UMAP plot (B). (F) Heatmap showing differential chromatin accessibilities of the selected myeloid-related gene loci between control and DKOCreERT2 cells in C1 to C5. (G-H) Volcano plot showing the comparison of TF motif enrichment between control vs DKOCreERT2 in C1 to C5 (G) and C6 to C8 (H).
The Reg1/Reg3-Nfkbiz axis regulates lineage priming of HSCs/early MPPs via chromatin remodeling. (A) Schematic representation of scATAC-seq using Flt3–CD48– LSK. In each genotype, samples are pooled from multiple mice to create a single sample for sequencing, as indicated by the number of mice represented. (B) UMAP visualization of all cells obtained from scATAC-seq data. Colors represent the cluster identity. (C) Heatmap showing chromatin accessibility for stemness-related and lineage-associated genes. (D) Cluster distribution of single cells across the indicated genotypes. Colors represent the cluster identity as in (B). (E) A 2-dimensional projection of 13859, 10991, 9936, and 6902 single cells from control, DKOCreERT2, NfkbizCreERT2, and TKOCreERT2, respectively, onto the UMAP plot (B). (F) Heatmap showing differential chromatin accessibilities of the selected myeloid-related gene loci between control and DKOCreERT2 cells in C1 to C5. (G-H) Volcano plot showing the comparison of TF motif enrichment between control vs DKOCreERT2 in C1 to C5 (G) and C6 to C8 (H).
The chromatin accessibility landscape was dramatically altered in DKOCreERT2 cells characterized by their dominance in C3 and C8 together with the underrepresentation of C1, C2, C4, C5, and C7 (Figure 5D-E). In contrast, DKOCreERT2 cells were evenly observed in clusters C10 to C14. Interestingly, DKOCreERT2 cells showed more accessibility for myeloid-related genes such as Cd74 and Csf2rb in clusters C1 to C8 than control (Figure 5C,F). Notably, the changes in chromatin accessibility in DKO cells were drastically restored by further deletion of Nfkbiz, implying that Reg1 and Reg3 regulate epigenetic homeostasis in HSCs by suppressing the expression of Nfkbiz (Figure 5D-F).
Consistent with the role of Nfkbiz in controlling gene expression via interaction with TFs such as NF-κB, NF-κB–binding motifs were enriched in regions more accessible in DKOCreERT2 cells than in control cells in clusters C1 to C5 (Figure 5G). Furthermore, GATA and IRF family–binding motifs were also more frequently observed in regions highly accessible in DKOCreERT2 cells in clusters C1 to C5 and C6 to C8, respectively (Figure 5G-H). These findings suggest that the activation of Nfkbiz in DKOCreERT2 cells can facilitate the recruitment of TFs involved in lineage priming via chromatin remodeling.
Reg3 and Reg1 recognize and destabilize mRNAs with common signatures
Next, we analyzed the mechanisms of how Reg1 and Reg3 recognize target mRNAs. Because Reg1-interacting stem-loop motifs were identified by high-throughput sequencing of crosslinking and IP (HITS-CLIP) analyses,23 we analyzed the transcriptome-wide binding mode of Reg3 with the same technique. To this end, HEK293T cells stably expressing DOX-inducible FLAG-tagged Reg3 were generated and UV crosslinked. IP of FLAG-Reg3 yielded Reg3-RNA complexes after induction of Reg3 by DOX treatment (Figure 6A). Recovered RNA fragments from 2 independent HITS-CLIP experiments were deep sequenced, and genome-aligned reads were clustered, allowing for the identification of 6639 and 7678 putative Reg3-binding sites in 2 biological duplicate libraries, respectively (supplemental Table 3). We found that the uniquely mapped reads of Reg3-binding sites were enriched in 3’ UTRs (Figure 6B). When the secondary structures of these sequences were analyzed by RNAfold,37 SL structures with 3 to 7 nucleotide stems and trinucleotide loops were significantly enriched (Figure 6C). In addition, motif analyses of the SL sequences revealed that the uridine-adenine-uridine (UAU) triplet was overrepresented in the hairpin sequences of Reg3-binding motifs (Figure 6E). This is reminiscent of the Reg1-binding motifs revealed by the HITS-CLIP analysis (Figure 6D,F).23 Interestingly, Reg3- and Reg1-binding sequences overlapped significantly (supplemental Table 4), indicating that Reg3 and Reg1 target an overlapping set of mRNAs by recognizing a common binding motif.
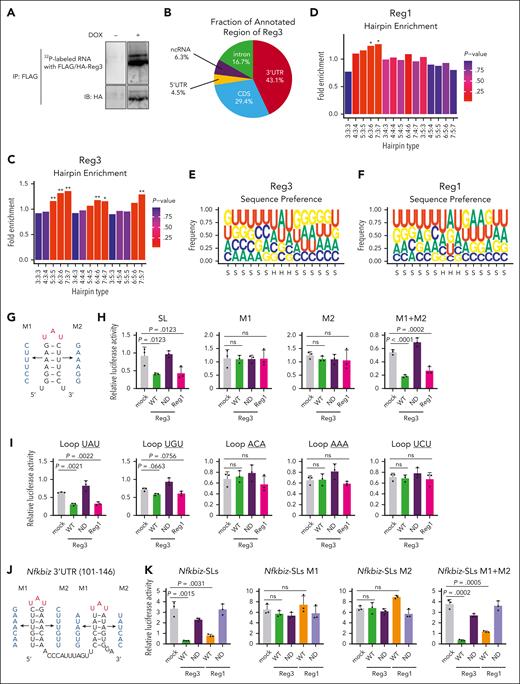
Reg1 and Reg3 recognize the tandem UAU-SL structures present in the Nfkbiz 3’ UTR. (A) Phosphoimage of SDS-PAGE of radiolabelled Reg3-RNA complexes obtained by immunoprecipitation with anti-FLAG antibody. (B) Pie chart deciphering the distribution of Reg3-binding sites obtained by the intersection of 2 biological HITS-CLIP replicates. (C-D) SL enrichments in Reg3- and Reg1-bound CLIP tags of the 3’ UTR compared with 1000 individual sequence permutations. X-axis indicates hairpin types in which the numbers represent the lengths of stem and loop. ∗P < .01; ∗∗P < .001. (E-F) Sequence logo representations of the recognition motives for Reg3 and Reg1. Stem and hairpin regions are indicated as S and H, respectively, below the x-axis. (G) Schematic representation of the artificial SL structure and its mutant forms (M1 and M2) in which the SL structure is disrupted. (H) Luciferase activity was assessed by using pGL3-βGlo plasmids containing the 3’ UTR with SL, M1, M2, and M1+ M2 as shown in (G). HeLa cells were transfected with pGL3 plasmids together with expression plasmids for WT Reg1, WT, or ND mutant of Reg3 (n = 3 each). (I) Luciferase activity was assessed by using pGL3-βGlo plasmids containing the 3’ UTR with a set of loop sequences. The loop sequences tested are shown in the top of figures. Transfection was performed as in (H) (n = 3 each). (J) Schematic representation of mouse Nfkbiz SL structures targeted by Reg1 and Reg3. M1 and M2 represent mutant sequences that no longer form SL structures. (K) Luciferase activity was assessed by using pGL3-βGlo plasmids containing either WT or mutated SL sequences (M1, M2, or M1+ M2) as shown in (J). HeLa cells were transfected with pGL3 plasmids together with expression plasmids for WT or ND mutant of Reg1 and Reg3 (n = 3 each). Data are representative of 2 to 3 independent experiments (H-I,K). Data are presented as mean ± SD (H-I,K). Statistical significance was calculated by 1-way ANOVA with Holm-Sidak multiple comparisons test (H-I,K).
Reg1 and Reg3 recognize the tandem UAU-SL structures present in the Nfkbiz 3’ UTR. (A) Phosphoimage of SDS-PAGE of radiolabelled Reg3-RNA complexes obtained by immunoprecipitation with anti-FLAG antibody. (B) Pie chart deciphering the distribution of Reg3-binding sites obtained by the intersection of 2 biological HITS-CLIP replicates. (C-D) SL enrichments in Reg3- and Reg1-bound CLIP tags of the 3’ UTR compared with 1000 individual sequence permutations. X-axis indicates hairpin types in which the numbers represent the lengths of stem and loop. ∗P < .01; ∗∗P < .001. (E-F) Sequence logo representations of the recognition motives for Reg3 and Reg1. Stem and hairpin regions are indicated as S and H, respectively, below the x-axis. (G) Schematic representation of the artificial SL structure and its mutant forms (M1 and M2) in which the SL structure is disrupted. (H) Luciferase activity was assessed by using pGL3-βGlo plasmids containing the 3’ UTR with SL, M1, M2, and M1+ M2 as shown in (G). HeLa cells were transfected with pGL3 plasmids together with expression plasmids for WT Reg1, WT, or ND mutant of Reg3 (n = 3 each). (I) Luciferase activity was assessed by using pGL3-βGlo plasmids containing the 3’ UTR with a set of loop sequences. The loop sequences tested are shown in the top of figures. Transfection was performed as in (H) (n = 3 each). (J) Schematic representation of mouse Nfkbiz SL structures targeted by Reg1 and Reg3. M1 and M2 represent mutant sequences that no longer form SL structures. (K) Luciferase activity was assessed by using pGL3-βGlo plasmids containing either WT or mutated SL sequences (M1, M2, or M1+ M2) as shown in (J). HeLa cells were transfected with pGL3 plasmids together with expression plasmids for WT or ND mutant of Reg1 and Reg3 (n = 3 each). Data are representative of 2 to 3 independent experiments (H-I,K). Data are presented as mean ± SD (H-I,K). Statistical significance was calculated by 1-way ANOVA with Holm-Sidak multiple comparisons test (H-I,K).
Consistently, a luciferase reporter RNA harboring the SL-containing sequence with an UAU-loop sequence in the 3’ UTR was suppressed by the overexpression of either RNase-competent Reg3 or Reg1 (Figure 6G-H). Disruption of the SL structure by mutating either side of the stem sequence abrogated the Reg3-mediated suppression of the luciferase activity (Figure 6G-H). Restoration of the SL structure by mutating both sides of the stem sequences restored Reg3 responsiveness (Figure 6H). SLs harboring UAU and, to a lesser extent, UGU, but not ACA, AAA or UCU, were suppressed by Reg3, suggesting that the pyrimidine-purine-pyrimidine loop sequences are required for Reg3 suppression, similar to Reg1 (Figure 6I). These results demonstrate that Reg3 and Reg1 recognize overlapping sets of mRNAs by recognizing common SL sequences present in the mRNA 3’ UTR.
Next, we investigated SL structure(s) in the Nfkbiz 3’ UTR responsible for the Reg1/Reg3-mediated decay. Luciferase reporter assays revealed that the Nfkbiz instability element was located within the 1 to 200 region of its 3’ UTR and contains 4 SL structures (SL1-SL4) (supplemental Figure 9A-B). Among these, SL3 and SL4, but not SL1 and SL2, were responsible for the suppression by both Reg1 and Reg3 (supplemental Figure 9C). These SLs harbor UAU-loop sequences (supplemental Figure 9B), a consensus motif for Reg1 and Reg3 recognition (Figure 6E-F). Consistently, a reporter containing either SL3 or SL4 was suppressed by the overexpression of Reg1 or Reg3 (supplemental Figure 9D). The Reg1/Reg3 responsiveness of SL3 and SL4 was abrogated by mutation of SL structures and was rescued by restoration of SL structures by introducing mutations in both sides of the stem sequences (Figure 6J-K), indicating that Reg1 and Reg3 commonly recognize SL structures in SL3 and SL4 for degrading Nfkbiz.
Enhancement of Nfkbiz expression via its 3’ UTR leads to myeloid priming in HSCs
The aforementioned results prompted us to hypothesize that enhancement of Nfkbiz expression by inhibiting Reg1/Reg3-mediated decay could control HSC lineage bias. We achieved an increase in Reg1 expression by targeting its self-regulation through the use of ASOs against SL structures located in the Reg1 3’ UTR.38 To inhibit Reg1/Reg3-mediated Nfkbiz mRNA decay, we designed morpholino ASOs to target and disrupt the SL structures of SL1, SL2, andSL3-4 (supplemental Figure 9B). Because SL3 and SL4 aligned tandemly with a short interval, we designed an ASO targeting both SL3 and SL4 (Figure 7A). When we introduced ASOs together with the reporter harboring the full-length Nfkbiz 3’ UTR, the SL3-4–targeting ASO highly increased luciferase activity compared with control-oligo or ASOs against SL1 or SL2 (supplemental Figure 10A). The results suggest that the SL3-4–targeting ASO (hereafter Nfkbiz-ASO) is potent to stabilize Nfkbiz mRNA by altering the SL structure. Similarly, in BM-derived macrophages, the Nfkbiz-ASO stabilized Nfkbiz mRNA and enhanced Nfkbiz expression both at mRNA and protein levels (supplemental Figure 10B-D). Consistent with the proinflammatory function of Nfkbiz in macrophages, the Nfkbiz-ASO augmented IL-6 production after lipopolysaccharide (LPS) treatment (supplemental Figure 10E). In contrast, Nfkbiz-ASO failed to increase Nfkbiz expression in Reg1/Reg3 DKO macrophages (supplemental Figure 10F), suggesting that the ASO enhanced Nfkbiz expression by inhibiting Reg1/Reg3-mediated mRNA decay.
Manipulation of Nfkbiz expression by ASO alters HSC lineage priming toward myeloid lineage. (A) Schematic representation of ASO (shown in red) for Reg1/Reg3-targeting SL structures in the mouse Nfkbiz 3’ UTR. (B) Experimental design for ASO-transduced HSCs. HSCs were electroporated with control oligo or Nfkbiz-targeting ASO. Cells were used for subsequent analyses in (C-H). (C-D) qRT-PCR analysis of Nfkbiz, Reg1, Reg3 and HSC/lineage-related genes in cultured HSCs at day 5 after electroporation (n = 5 each). (E-F) Representative flow cytometry plots and frequency of FcγR expression in CD48–CD150+ LSK from cultured HSCs stimulated with IL-1β or TNF for 24 hours (n = 3 each). (G-H) Representative flow cytometry plots and frequency of FcγR+CD11b+ cells from cultured HSCs stimulated with IL-1β or TNF for 24 hours (n = 3 each). (I) Model of the Reg1/Reg3-Nfkbiz axis in the control of HSC lineage priming. Data are representative of 3 independent experiments (C-H). Data are presented as mean ± SD (C-D,F,H) Statistical significance was calculated by unpaired 2-tailed Student t test (C-D,F,H).
Manipulation of Nfkbiz expression by ASO alters HSC lineage priming toward myeloid lineage. (A) Schematic representation of ASO (shown in red) for Reg1/Reg3-targeting SL structures in the mouse Nfkbiz 3’ UTR. (B) Experimental design for ASO-transduced HSCs. HSCs were electroporated with control oligo or Nfkbiz-targeting ASO. Cells were used for subsequent analyses in (C-H). (C-D) qRT-PCR analysis of Nfkbiz, Reg1, Reg3 and HSC/lineage-related genes in cultured HSCs at day 5 after electroporation (n = 5 each). (E-F) Representative flow cytometry plots and frequency of FcγR expression in CD48–CD150+ LSK from cultured HSCs stimulated with IL-1β or TNF for 24 hours (n = 3 each). (G-H) Representative flow cytometry plots and frequency of FcγR+CD11b+ cells from cultured HSCs stimulated with IL-1β or TNF for 24 hours (n = 3 each). (I) Model of the Reg1/Reg3-Nfkbiz axis in the control of HSC lineage priming. Data are representative of 3 independent experiments (C-H). Data are presented as mean ± SD (C-D,F,H) Statistical significance was calculated by unpaired 2-tailed Student t test (C-D,F,H).
Next, we investigated whether the Nfkbiz-ASO is also potent to increase the Nfkbiz expression in HSC by using a serum-free HSC culture system that enables efficient enrichment of CD150+CD48–CD201+ LSK population (Figure 7B; supplemental Figure 10G-H).39 We found that a Fluorescein isothiocyanate (FITC)-conjugated ASO was efficiently introduced into cultured HSCs (cHSCs) by electroporation and persisted at least for 5 days (supplemental Figure 10I-J). As expected, the introduction of the Nfkbiz-ASO enhanced the expression of Nfkbiz in cHSCs compared with SL1- or SL2-targeting ASOs (Figure 7C; supplemental Figure 10K). In contrast, the Nfkbiz-ASO did not influence the expression of Reg1 or Reg3, suggesting that the ASO specifically acts on Nfkbiz transcripts. Interestingly, Nfkbiz-ASO treatment for 5 days mildly but significantly enhanced the expression of myeloid-related genes Spi1 and Cebpb, whereas the treatment reduced Flt3 expression in cHSCs (Figure 7D). Consistently, cell surface expression of FcγR, a myeloid marker, was slightly augmented in CD150+CD48– LSK population by treatment with Nfkbiz-ASO (Figure 7E-F). In contrast, the Nfkbiz-ASO did not alter the expression of HSC-related genes including Hlf, Cd34, and Hoxb5 (Figure 7D) or the frequency of LSK population (supplemental Figure 10L-M). Although Nfkbiz-ASO treatment alone slightly induced myeloid differentiation, simultaneous treatment with TNF or IL-1β drastically promoted myeloid differentiation of cultured HSCs (Figure 7G-H). Interestingly, TNF or IL-1β synergized with Nfkbiz-ASO and dramatically enhanced FcγR expression in CD150+CD48– LSK population of cHSCs (Figure 7E-F). Taken together, the Nfkbiz-ASO is potent to induce myeloid-lineage priming in HSCs by enhancing the expression of Nfkbiz.
Discussion
In this study, we discovered that the Reg1/Reg3-Nfkbiz axis acts as the novel regulatory mechanism governing lineage bias in HSPCs, identifying it as a promising target for manipulating hematopoiesis (Figure 7I). It was demonstrated that epigenetic regulation is critical for the lineage specification of HSPCs, and it is difficult to separate these features of stem cells by known epigenetic factors.40-43 In this regard, we believe that Reg1 and Reg3 are novel cellular factors to be identified that determine myeloid-lymphoid cell fate decision by controlling HSPCs.
Despite the multiple functions of Nfkbiz in various mature immune cells,32,44-46 its role in HSCs has not yet been discussed. The IκB-ζ protein, encoded by the Nfkbiz gene, interacts with NF-κBp50 to reinforce NF-κB activation32 and also participates in chromatin remodeling under inflammatory conditions.44,46 NF-κB has been shown to be involved in emergency myelopoiesis by TNF,16 and the cooperation between NF-κBp50 and C/EBPα in the granulocytic progenitors for promoting myelopoiesis has been demonstrated.47 Therefore, Nfkbiz expressed by the suppression of Reg1/Reg3 may initiate the myeloid program in cooperation with the NF-κB and/or by recruiting the chromatin remodelers to induce epigenetic changes.
The Reg1/Reg3-Nfkbiz regulatory axis is a novel therapeutic target to manipulate myeloid- and lymphoid-biased outputs in hematopoiesis. We found that an ASO disrupting SL structures in the Nfkbiz 3’ UTR is potent to increase Nfkbiz expression and induce myeloid-biased lineage priming in HSCs. Given that this ASO specifically targets Nfkbiz expression, this strategy might be beneficial compared with the treatment with cytokines/TLR ligands, which can induce broad responses. Indeed, the SL structures targeted by Reg1 and Reg3 are conserved in human NFKBIZ. Nevertheless, further studies are required to optimize the sequences and nucleic acids modification as well as drug delivery to improve the efficacy of the therapeutic potential.
In summary, we demonstrate here that Reg1 and Reg3 act as molecular switch to control lineage biases in HSPCs by shaping transcriptional lineage priming. These 2 RNases control multipotent cell lineage choice by targeting Nfkbiz for degradation. Nfkbiz acts as a key regulator to promote myeloid-lineage commitment. Our findings provide insights into how HSC-lineage priming is regulated.
Acknowledgments
The authors thank H. Miyachi, S. Kitano, R. Takeda, and S. Nagai for animal technical service; T. Maruyama for providing Nfkbiz flox mice; Y. Agata for plasmid construction of retroviral expression of human Id3, S. Yamazaki for Soluplus, T. Nagasawa, N. Chevrier, and all members of the Takeuchi laboratory for insights and suggestions. Flow cytometric analysis and cell sorting using LSRFortessa (BD Biosciences) and AriaIII (BD Biosciences) were performed at the Medical Research Support Center, Graduate School of Medicine, Kyoto University, which was supported by Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research) from Japan Agency for Medical Research and Development (AMED) (grant number JP20am0101092). Figures were created using BioRender.com.
This study was supported by Japan Society for the Promotion of Science KAKENHI (18H05278, 23H00402) (O.T.) and (18K15185) (T.U.) and Core-to-Core Program (O.T.) and grant-in-aid for Scientific Research on Innovative Areas ‘Genome Science’ (221S0002, 16H06279) (T.M.). This study was also supported by AMED (grant numbers JP20gm4010002, JP21ae0121030, 23ama221327h0001) (O.T.) and the Cooperative Research Program (Joint Usage/Research Center program) of Institute for Frontier Life and Medical Sciences, Kyoto University (O.T.) and a grant-in-aid for Transformative Research Areas (A) “Material Symbiosis” (grant 20H05874) (T.U.) and ISHIZUE 2017 of Kyoto University Research Development Program (T.U.), an Office of Directors’ Research Grant provided by the Institute for Frontier Life and Medical Sciences, Kyoto University (A.V.), and the Fujiwara Memorial Foundation (T.U.).
Authorship
Contribution: T.U. and O.T. were in charge of experimental designs, analyses, interpretation of data, and drafting of the manuscript; S.Y., D.O., K. Toratani, and M.M. participated in the acquisition, analysis, and interpretation of data; A.J., A.G., and I.A. handled single cell RNA-sequencing (scRNA-seq) samples and performed analyses on scRNA-seq data; A.V. and K. Takeuchi. performed analysis of scRNA-seq and scATAC-seq (single-cell sequencing assay for transposase-accessible chromatin) data; Y.M. and M.L. performed HITS-CLIP (high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation) of Reg3; H. Kiryu analyzed HITS-CLIP of Reg1 and Reg3; H.W. and G.K. contributed to genetic engineering to generate multiple mouse lines; T.T. performed histological examination; T.I. and R.Y. contributed to in vitro culture of multipotent progenitors and hematopoietic stem cells, respectively; D.M.S. carried out structural modeling; T.M., M.M., R.Y., H.-R.R., and H. Kawamoto provided critical discussions and suggestions; and O.T. supervised the overall study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for D.O. is Laboratory of Molecular Immunobiology, Division of Biological Science, Graduate School of Science and Technology, Nara Institute of Science and Technology, Ikoma, Nara, Japan.
Correspondence: Osamu Takeuchi, Department of Medical Chemistry, Graduate School of Medicine, Kyoto University, Yoshida-Konoe-cho, Sakyo-ku, Kyoto 606-8501, Japan; email: otake@mfour.med.kyoto-u.ac.jp.
References
Author notes
∗T.U. and S.Y. contributed equally to this study.
Data for single cell RNA sequencing and single-cell sequencing assay for transposase-accessible chromatin have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus under accession codes GSE228923 and GSE229065, respectively.
Mouse strains generated in this work are available for noncommercial research purposes following a reasonable request. Source data are provided with this article. Data are available on request from the corresponding author, Osamu Takeuchi (otake@mfour.med.kyoto-u.ac.jp).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal