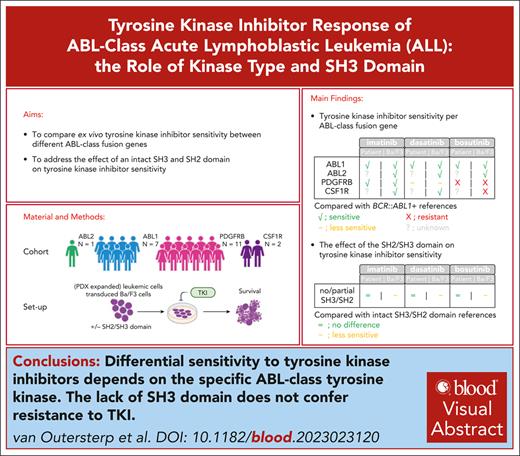
Differential sensitivity to TKIs depends on the specific ABL-class tyrosine kinase.
Lack of the SH3 domain does not confer resistance to TKI.
Visual Abstract
Acute lymphoblastic leukemia (ALL) with fusions of ABL-class tyrosine kinase genes other than BCR::ABL1 occurs in ∼3% of children with ALL. The tyrosine kinase genes involved in this BCR::ABL1-like (Ph-like) subtype include ABL1, PDGFRB, ABL2, and CSF1R, each of which has up to 10 described partner genes. ABL-class ALL resembles BCR::ABL1-positive ALL with a similar gene expression profile, poor response to chemotherapy, and sensitivity to tyrosine kinase inhibitors (TKIs). There is a lack of comprehensive data regarding TKI sensitivity in the heterogeneous group of ABL-class ALL. We observed variability in TKI sensitivity within and among each ABL-class tyrosine kinase gene subgroup. We showed that ALL samples with fusions for any of the 4 tyrosine kinase genes were relatively sensitive to imatinib. In contrast, the PDGFRB-fused ALL samples were less sensitive to dasatinib and bosutinib. Variation in ex vivo TKI response within the subset of samples with the same ABL-class tyrosine kinase gene was not associated with the ALL immunophenotype, 5′ fusion partner, presence or absence of Src-homology-2/3 domains, or deletions of IKZF1, PAX5, or CDKN2A/B. In conclusion, the tyrosine kinase gene involved in ABL-class ALL is the main determinant of TKI sensitivity and relevant for specific TKI selection.
Introduction
Acute lymphoblastic leukemia (ALL) is a heterogeneous malignancy driven by diverse genetic alterations. The BCR::ABL1-positive subtype and ABL-class subtype, each accounting for ∼3% of newly diagnosed pediatric ALL cases, involve fusions with ABL family tyrosine kinase genes.1 In this study, the ABL-class subtype was defined as fusions involving tyrosine kinases ABL1, ABL2, PDGFRB, or CSF1R fused to various partner genes, excluding the sentinel BCR::ABL1 fusion. In the context of B-cell precursor ALL (BCP-ALL), ABL-class fusions act as driving lesions and comprise ∼50% of PDGFRB fusions and ∼30% of ABL1 fusions, and the remaining cases involve fusions with either CSF1R or ABL2.1-3 In T-ALL, the most common ABL-class fusion is NUP214::ABL1, which is present in ∼3% of cases,3 is often amplified, and can be present subclonally.3
Although ABL-class ALL has been defined as one entity, the underlying biology may vary depending on the involved tyrosine kinase gene. In a retrospective study, it was shown that MRD at the end of induction was associated with the involved tyrosine kinase gene for patients treated with chemotherapy alone.2 In addition, there was a trend toward a differential outcome between the different involved tyrosine kinase genes.2 Patients with ABL2 and PDGFRB fusions had the highest MRD levels and worst event-free survival. This suggests that there are underlying differences between the involved ABL-class tyrosine kinase genes that affect sensitivity to chemotherapy. All 4 ABL-class tyrosine kinase genes activate STAT5, whereas only ABL1 and ABL2 activate CRKL.4,5 Moreover, although in BCR::ABL1-positive ALL, the Src-homology-3 (SH3) and SH2 domains of ABL1 are generally retained, some ABL1- and ABL2-fused leukemias lack these domains, resulting in differential downstream signaling.6 Wild-type PDGFRB and CSF1R do not have SH2 or SH3 domains. The effect of these biological differences on sensitivity to tyrosine kinase inhibitors (TKIs) remains to be established.
The addition of TKIs in combination with intensive chemotherapy has substantially improved outcomes in children with newly diagnosed BCR::ABL1-positive ALL.7-12 Imatinib, the first approved inhibitor of BCR::ABL1-positive ALL, is a type II TKI that primarily binds to the inactive conformation of ABL1. Upon the discovery of resistance caused by ABL1 kinase domain mutations hindering the binding of imatinib, the next-generation TKIs dasatinib, bosutinib, nilotinib, and ponatinib were developed.13 In pro-B murine model cell lines, these next-generation TKIs exhibited higher activity toward BCR::ABL1 than imatinib and were able to inhibit clinically relevant imatinib-resistant forms of BCR::ABL1.14-18 Preclinical studies using pro-B murine model cell lines expressing ABL-class fusions have shown that imatinib, dasatinib, nilotinib, and ponatinib target all 4 ABL-class tyrosine kinases,4,5,19 whereas the inhibitory activity of bosutinib appears to be limited to ABL1 and ABL2.4 Moreover, studies using patient–derived xenograft (PDX) models have shown sensitivity after in vivo or ex vivo exposure to imatinib or dasatinib for ABL1-, ABL2-, PDGFRB-, and CSF1R-fused ALL.4,20-22 Based on these preclinical studies and several case studies,4,5,19-50 current ALL treatment protocols include either imatinib (EsPhALL2017/COG AALL1631/ALLTogether-1) or dasatinib (COG AALL1131/DFCI ALL 16-001/Total Therapy XVII) as first-line treatment for pediatric patients with ABL-class ALL given the approval of these TKIs for patients with BCR::ABL1-positive ALL.51
Although the initial findings appear promising, there is currently a lack of comprehensive and comparable data concerning the response of ABL-class ALL in relation to the specific tyrosine kinase gene involved. Studies using transduced Ba/F3 cell line models have not recapitulated the heterogeneous genetic background of ALL samples. The in vivo and ex vivo assessments of TKI sensitivity in the primary material or PDX samples were conducted in limited-sized cohorts (≤6 samples per study) or reported as a case study.4,20-22,47,50 Moreover, comparability across these studies is hindered by the diverse techniques used for TKI sensitivity measurement and the variable TKI concentrations used. The combined preclinical and clinical data suggest that patients with ABL-class ALL are responsive to TKI therapy in general. However, the response to different TKIs in patients with ABL-class ALL has not been studied systematically.
In our study, the testing of TKIs on patient material was constrained to 3 TKIs due to a limited number of patient cells per sample. We assessed sensitivity to imatinib and dasatinib, given their current inclusion in clinical trials. Furthermore, we included bosutinib in our study, which was in clinical trials for pediatric chronic myeloid leukemia (CML) at the outset of the study and has recently gained approval for treating children with newly diagnosed or TKI-resistant/intolerant CML based on results from the BCHILD clinical trial (NCT04258943).52 An alternative TKI could be ponatinib, which demonstrated a more durable response in adults with BCR::ABL1-positive ALL than in those treated with imatinib.53,54 However, ponatinib is not approved for pediatric patients with leukemia yet. Moreover, in our study, we decided to use bosutinib because of the serious side effects associated with ponatinib reported in clinical trials at that moment.55 Furthermore, we chose bosutinib over nilotinib because of its distinct target profile compared with that of imatinib.56,57 We aimed to compare the ex vivo TKI sensitivity of ALL cells from patients with different ABL-class tyrosine kinase fusion genes and to address the importance of intact SH2 and SH3 domains.
Material and methods
Primary ALL cells
Peripheral blood and/or bone marrow samples were collected from children with newly diagnosed or relapsed BCP-ALL or T-ALL harboring ABL-class or BCR::ABL1 fusions, using institutional review board-approved research protocols. We only included pediatric ALL samples because these are typically well-characterized, facilitating the identification of ABL-class fusions. In contrast, adult samples are less comprehensively characterized at diagnosis, making ABL-class fusions more likely to be overlooked, and the availability of such samples is limited. In addition, the prognosis and treatment approaches for ALL differ significantly between pediatric and adult patients. Before sample collection, written informed consent was obtained from the patients or their parents/guardians in accordance with the Declaration of Helsinki, as approved by the Medical Ethics Committee of the Erasmus Medical Center, the Biobank and Data Access Committee of the Princess Máxima Center for Pediatric Oncology, the Medical Ethical Committee of the University Medical Centers Hamburg Eppendorf, or the Children’s Oncology Group, as described.22,58,59 The study cohort consisted of pediatric patients enrolled in clinical trials conducted by the Dutch Childhood Oncology Group (ALL8, 9, 10, 11, and ALLTogether1), German Cooperative ALL group (COALL 06-97 and 07-03), and Children’s Oncology Group (COG P9906 and COG AALL0232). Patients with ABL-class alterations were identified either through break-apart fluorescence in situ hybridization or RNA sequencing performed by reference laboratories. Mononuclear cells were isolated from the collected samples using Lymphoprep density gradient centrifugation. Cytospins were prepared from leukemic cells and stained with May-Grünwald-Giemsa stain for the determination of leukemic blast percentage. RNA and DNA extraction from the samples was routinely performed using the TRIzol reagent. For all experiments, the leukemic blast percentage in the samples was at least 80%.
Drug sensitivity assays
The survival of primary and primary or serially PDX-expanded ALL samples exposed to 4 different concentrations of TKIs in an ex vivo leukemic niche was measured using a flow cytometry-based viability readout. Moreover, survival after TKI exposure of transduced Ba/F3 cells with lentiviral constructs of ABL-class fusions was measured in a metabolic assay using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT). For both methods, after 3 days of exposure to imatinib, dasatinib, or bosutinib, survival was measured and corrected for the untreated control. The Z-score of the area under the dose-response curve (AUC) or half-maximal inhibitory concentration (IC50) was used as a measure of TKI sensitivity. The sensitivity of ABL-class ALL samples was compared with that of a reference cohort of BCR::ABL1-positive ALL samples. For more detailed methods, please refer to the supplemental Material and Methods, available on the Blood website.
Mutation and deletion analysis
Mutations in leukemia-associated genes were detected by whole-exome sequencing. To study copy number alterations, multiplex ligation-dependent probe amplification (MLPA) or single nucleotide polymorphism (SNP) array results were requested from the diagnostic departments of the involved study groups. Further details on the sample and data processing can be found in the supplemental Material and Methods.
Results
Patient cohort
We conducted our study using cryopreserved primary ALL cells, primary xenografted ALL cells, or serially xenografted ALL cells obtained at the time of diagnosis or relapse from 20 pediatric ABL-class ALL samples, comprising 16 BCP-ALL and 3 T-ALL cases before any TKI treatment and 1 BCP-ALL sample obtained 33 days after the diagnosis of a patient treated with dasatinib for 2 weeks (supplemental Table 1). PDGFRB fusion was the most common (50%, 3 different partners), followed by ABL1 (35%, 4 different partners), CSF1R (10%, 1 partner), and ABL2 (5%, 1 partner), consistent with the incidence reported in our previous clinical study.2 As a reference cohort, we included 32 BCR::ABL1-positive pediatric BCP-ALL (n = 31) and T-ALL (n = 1) samples.
Effect of ABL-class tyrosine kinase gene on ex vivo TKI sensitivity
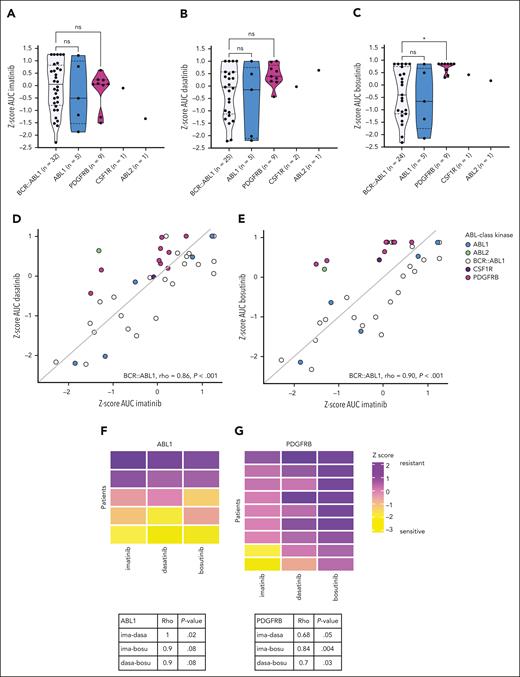
To study the efficacy of imatinib, dasatinib, and bosutinib in ALL samples with different ABL-class tyrosine kinase fusion genes, we determined the ex vivo sensitivity (n = 16) of primary and primary xenografted ALL samples and compared these data with those of the BCR::ABL1-positive reference cohort. Similar to the BCR::ABL1-positive ALL samples, the ABL1- and PDGFRB-fused ALL samples exhibited considerable variability in sensitivity to imatinib, ranging from high sensitivity (Z-score imatinib <0.59) to complete lack of response (Z-score imatinib >0.47) (Figure 1A). For ABL1-fused ALL samples, sensitivity toward the second-generation TKIs dasatinib and bosutinib was in the same range as the BCR::ABL1-positive ALL samples (Figure 1A-C). However, none of the PDGFRB-fused ALL samples exhibited high sensitivity to dasatinib, and none were sensitive to bosutinib (P = .01; Figure 1A-C). Owing to the limited number of samples with CSF1R (n = 1) or ABL2 (n = 1) fusions, no conclusions can be drawn regarding TKI sensitivity.
Ex vivo TKI sensitivity of ABL-class ALL samples. (A-C) Z-score of the area under the dose-response curve (AUC) for imatinib, dasatinib, and bosutinib sensitivity, respectively, of primary and primary xenografted ALL samples with ABL-class fusions compared with primary and primary xenograft ALL samples of pediatric BCR::ABL1-positive ALL. (D-E) Correlation between sensitivity to imatinib (ima) and dasatinib (dasa) or bosutinib (bosu). The Spearman correlation coefficient (Rho) is calculated for the reference cohort of BCR::ABL1-positive ALL samples. The x = y line is displayed in gray. (F-G) Correlation between imatinib and dasatinib or bosutinib in ABL1- or PDGFRB-fused ALL samples, respectively. The Spearman correlation coefficient (Rho) and P value are calculated and are displayed in the tables below.
Ex vivo TKI sensitivity of ABL-class ALL samples. (A-C) Z-score of the area under the dose-response curve (AUC) for imatinib, dasatinib, and bosutinib sensitivity, respectively, of primary and primary xenografted ALL samples with ABL-class fusions compared with primary and primary xenograft ALL samples of pediatric BCR::ABL1-positive ALL. (D-E) Correlation between sensitivity to imatinib (ima) and dasatinib (dasa) or bosutinib (bosu). The Spearman correlation coefficient (Rho) is calculated for the reference cohort of BCR::ABL1-positive ALL samples. The x = y line is displayed in gray. (F-G) Correlation between imatinib and dasatinib or bosutinib in ABL1- or PDGFRB-fused ALL samples, respectively. The Spearman correlation coefficient (Rho) and P value are calculated and are displayed in the tables below.
To study whether ALL samples sensitive to imatinib also responded to dasatinib or bosutinib, referred to as cross-TKI sensitivity, we compared the ex vivo response to imatinib with the ex vivo response to dasatinib and bosutinib in ABL1- and PDGFRB-fused ALL samples. Our reference cohort with BCR::ABL1-positive ALL samples showed significant cross-TKI sensitivity, as represented by a high correlation between ex vivo sensitivity to imatinib and dasatinib (Rho = 0.86; P < .001; Figure 1D) and imatinib and bosutinib (Rho = 0.90; P < .001; Figure 1E). Similar to BCR::ABL1-positive ALL samples, ABL1-fused ALL samples exhibited high cross-TKI sensitivity; ex vivo imatinib sensitivity was correlated with dasatinib (Rho = 1; P = .02; Figure 1D,F) and bosutinib (Rho = 0.9; P = .08; Figures 1E and 2F). In contrast, PDGFRB-fused ALL samples exhibited lower cross-TKI sensitivity, as represented by a lower correlation between imatinib and dasatinib (Rho = 0.68; P = .05; Figure 1D,G) and bosutinib (Rho = 0.7; P = .03; Figure 1E,G). These differences were observed at clinically relevant concentrations (supplemental Figure 1A-C). These data suggest that the specific ABL-class tyrosine kinase gene involved in the fusion plays an important role in sensitivity to dasatinib and bosutinib and may also have clinical implications in TKI selection.
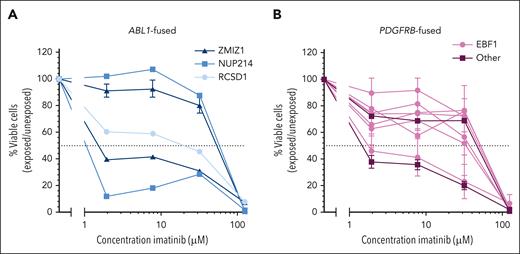
Ex vivo TKI sensitivity per ABL-class tyrosine kinase gene colored by the 5′ fusion partner. (A) Dose-response curves for imatinib treatment of ZMIZ1::ABL1- (dark blue), NUP214::ABL1- (medium blue), or RCSD1::ABL1-fused (light blue) primary or primary xenografted ALL samples. (B) Dose-response curves for imatinib in primary or primary xenografted ALL samples with EBF1::PDGFRB fusion (pink) or other PDGFRB fusions (purple), JAKMIP2, or CCDC88C). Leukemic cell survival relative to untreated controls, mean ± standard error of the mean (SEM) of 1 to 3 biological replicates.
Ex vivo TKI sensitivity per ABL-class tyrosine kinase gene colored by the 5′ fusion partner. (A) Dose-response curves for imatinib treatment of ZMIZ1::ABL1- (dark blue), NUP214::ABL1- (medium blue), or RCSD1::ABL1-fused (light blue) primary or primary xenografted ALL samples. (B) Dose-response curves for imatinib in primary or primary xenografted ALL samples with EBF1::PDGFRB fusion (pink) or other PDGFRB fusions (purple), JAKMIP2, or CCDC88C). Leukemic cell survival relative to untreated controls, mean ± standard error of the mean (SEM) of 1 to 3 biological replicates.
Effect of the fusion partner on ex vivo TKI sensitivity
To explore the potential factors influencing imatinib sensitivity in ALL samples with the same ABL-class tyrosine kinase gene, we investigated whether these differences could be linked to the partner gene. The partner gene can for example affect the expression level of the fusion gene or include certain functional domains. Among the ABL1-fused ALL samples, ZMIZ1 and NUP214 were recurrent fusion partners, and RCSD1 was identified as a fusion partner in 1 ALL sample. We found no consistent difference in imatinib sensitivity between the various fusion partners of ABL1-fused ALL samples (Figure 2A). For PDGFRB-fused ALL samples, the fusion partner was either EBF1 or another, which included CCDC88C and JAKMIP2, and no relationship between the fusion partner and ex vivo imatinib sensitivity was observed (Figure 2B). This suggests that, in contrast to the type of ABL-class tyrosine kinase gene, the 5′ fusion partner does not play a role in sensitivity to imatinib.
Sample characteristics and ex vivo TKI sensitivity
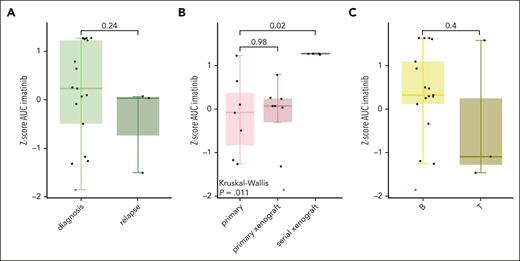
The use of samples with different origins and disease statuses has made it challenging to compare TKI sensitivity between studies. To investigate the effect of disease status, we examined samples obtained at diagnosis and relapse. We found that relapse samples without prior TKI exposure exhibited sensitivity similar to that of diagnostic samples (Figure 3, Figure 4A). Moreover, the leukemic cells of patient 2 were obtained after 2 weeks of dasatinib treatment and were highly sensitive to all 3 TKIs (Figure 3). To investigate the effect of sample origin, we evaluated the responsiveness to ex vivo imatinib treatment between primary leukemic cells, primary xenografted samples, and serially xenografted samples. Interestingly, primary leukemic cells and primary xenografted samples exhibited similar sensitivities, as evidenced by the highly overlapping drug response curves of patient 9 (supplemental Figure 2). However, serially xenografted samples consistently showed resistance to the 3 tested TKIs (Figures 3 and 4B). Lastly, no relationship between immunophenotype (BCP-ALL vs T-ALL) and TKI sensitivity was observed (Figures 3 and 4C).
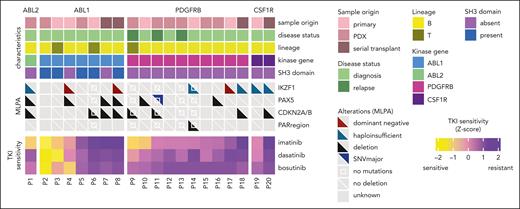
Heat map of patient characteristics and secondary lesions. The primary, primary xenografted, and serial xenografted ALL samples are displayed by the ABL-class tyrosine kinase gene from the most sensitive (left) to the least sensitive (right) based on the area under the dose-response curve (AUC) for imatinib measured using an ex vivo imatinib sensitivity assay. Deletions and other aberrations in leukemia-associated genes, PAX5, IKZF1, CDKNA2A/B, and pseudoautosomal (PAR) regions, based on MLPA assays or whole-exome sequencing are shown. IKZF1 deletions are subdivided into deletions resulting in haploinsufficiency (in dark green, affecting the start codon in exon 2) and deletions that affect the DNA-binding domain, exerting a dominant-negative effect on the unaffected allele (in dark red, deletions of exons 4-7). A white square in the middle denotes no mutations but whole-exome sequencing data available, a diagonal white line denotes no deletions but MLPA data available, and gray squares denote no data available.
Heat map of patient characteristics and secondary lesions. The primary, primary xenografted, and serial xenografted ALL samples are displayed by the ABL-class tyrosine kinase gene from the most sensitive (left) to the least sensitive (right) based on the area under the dose-response curve (AUC) for imatinib measured using an ex vivo imatinib sensitivity assay. Deletions and other aberrations in leukemia-associated genes, PAX5, IKZF1, CDKNA2A/B, and pseudoautosomal (PAR) regions, based on MLPA assays or whole-exome sequencing are shown. IKZF1 deletions are subdivided into deletions resulting in haploinsufficiency (in dark green, affecting the start codon in exon 2) and deletions that affect the DNA-binding domain, exerting a dominant-negative effect on the unaffected allele (in dark red, deletions of exons 4-7). A white square in the middle denotes no mutations but whole-exome sequencing data available, a diagonal white line denotes no deletions but MLPA data available, and gray squares denote no data available.
Comparison of sample characteristics with sensitivity to imatinib. (A) Box plots of diagnostic and relapse samples and Z-score of the area under the dose-response curve (AUC). (B) Box plots of primary ALL samples, primary xenograft ALL samples, and serial PDX-transplanted ALL samples and the Z-score of the AUC for imatinib. The mean AUC of imatinib was compared using the Kruskal-Wallis test with Dunn post hoc test. (C) Box plots of ALL cells from B-lineage (indicated by “B”) or T-lineage (indicated by “T”) and the Z-score of the AUC for imatinib. The box plots show the median Z-score ± 1.5× interquartile range in the Tukey style. The red dots represent patient 2, from whom ALL cells were obtained from day 33 after 2 weeks of TKI treatment.
Comparison of sample characteristics with sensitivity to imatinib. (A) Box plots of diagnostic and relapse samples and Z-score of the area under the dose-response curve (AUC). (B) Box plots of primary ALL samples, primary xenograft ALL samples, and serial PDX-transplanted ALL samples and the Z-score of the AUC for imatinib. The mean AUC of imatinib was compared using the Kruskal-Wallis test with Dunn post hoc test. (C) Box plots of ALL cells from B-lineage (indicated by “B”) or T-lineage (indicated by “T”) and the Z-score of the AUC for imatinib. The box plots show the median Z-score ± 1.5× interquartile range in the Tukey style. The red dots represent patient 2, from whom ALL cells were obtained from day 33 after 2 weeks of TKI treatment.
Secondary lesions and ex vivo TKI sensitivity
We studied the role of secondary lesions in B-cell differentiation factors in ex vivo TKI sensitivity of ABL-class ALL. This revealed common deletions in IKZF1 (44%), CDKN2A/B (44%), and PAX5 (35%), whereas deletions in the pseudoautosomal region (PAR) (7%) were less frequent (Figure 3). IKZF1 deletions were absent in the T-ALL cases (n = 3). No association was observed between IKZF1, PAX5, CDKN2A/B deletions, or ex vivo response to imatinib (Figure 3). One sample obtained from a patient with relapsed PDGFRB-fused ALL had an inactivating mutation in the tumor suppressor gene TP53, and the cells were intermediately imatinib-sensitive (AUC 80-120 AU). Our data indicate that neither deletions in B-cell differentiation factors nor the identified TP53 mutations were associated with ex vivo TKI resistance. This suggests that other factors may affect the response to TKIs and contribute to the variability within each ABL-class tyrosine kinase group.
A model system for ABL-class TKI sensitivity
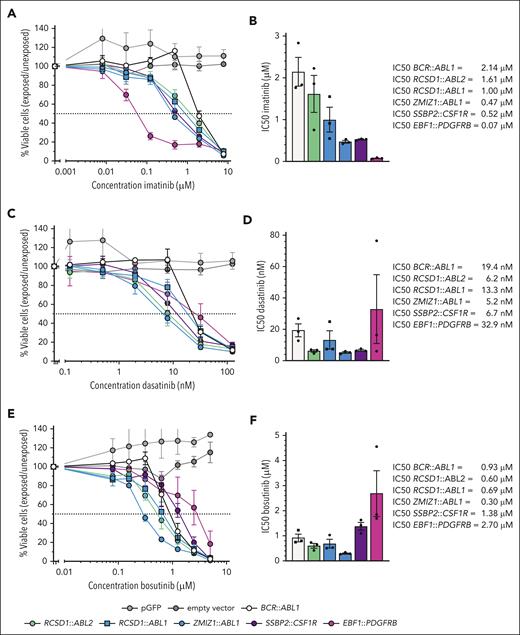
To study the role of the type of ABL-class tyrosine kinase gene on imatinib, dasatinib, and bosutinib response in a homogeneous genetic background, we generated transduced Ba/F3 cell lines with 1 or 2 representative fusions for each ABL-class tyrosine kinase gene. Cell lines became mouse interleukin-3-independent upon transduction with each of these constructs, like previously reported.5 In correspondence with BCR::ABL1-positive and ABL1-fused ALL samples, the transduced Ba/F3 cell lines expressing RCSD1::ABL1, ZMIZ1::ABL1, or RCSD1::ABL2 fusions exhibited sensitivity to all 3 TKIs (Figure 5A-F). The Ba/F3 cell line expressing EBF1::PDGFRB exhibited superior imatinib sensitivity compared with the Ba/F3 model expressing BCR::ABL1, defined by a >10-fold lower half-maximal inhibitory concentration (Figure 5A-B [IC50]). In contrast, the Ba/F3 cell line expressing EBF1::PDGFRB was less sensitive to dasatinib and bosutinib than the other Ba/F3 cell lines expressing an ABL-class fusion (Figure 5C-F). The Ba/F3 cell line expressing SSBP2::CSF1R was sensitive to imatinib and dasatinib, but less sensitive to bosutinib (Figure 5D-E). The cell line data confirmed that the difference in sensitivity to different TKIs depends on the type of ABL-class tyrosine kinase gene.
TKI sensitivity of transduced Ba/F3 cell lines expressing ABL-class fusions. (A) Dose-response curves of transduced Ba/F3 cell lines with various ABL-class fusions for imatinib. (B) Bar graph of half-maximal inhibitory concentration (IC50) values of imatinib obtained from dose-response curves. (C) Dose-response curves of transduced Ba/F3 cell lines with various ABL-class fusions of dasatinib. (D) Bar graph of IC50 values of dasatinib obtained from dose-response curves. (E) Dose-response curves of transduced Ba/F3 cell lines with various ABL-class fusions for bosutinib. (F) Bar graph of IC50 values of bosutinib obtained from dose-response curves. For the dose-response curves, the values are normalized against untreated controls for each cell line and represent the mean ± SEM (n = 3). Bar graphs represent the mean ± SEM.
TKI sensitivity of transduced Ba/F3 cell lines expressing ABL-class fusions. (A) Dose-response curves of transduced Ba/F3 cell lines with various ABL-class fusions for imatinib. (B) Bar graph of half-maximal inhibitory concentration (IC50) values of imatinib obtained from dose-response curves. (C) Dose-response curves of transduced Ba/F3 cell lines with various ABL-class fusions of dasatinib. (D) Bar graph of IC50 values of dasatinib obtained from dose-response curves. (E) Dose-response curves of transduced Ba/F3 cell lines with various ABL-class fusions for bosutinib. (F) Bar graph of IC50 values of bosutinib obtained from dose-response curves. For the dose-response curves, the values are normalized against untreated controls for each cell line and represent the mean ± SEM (n = 3). Bar graphs represent the mean ± SEM.
Effect of SH3 domain on TKI sensitivity
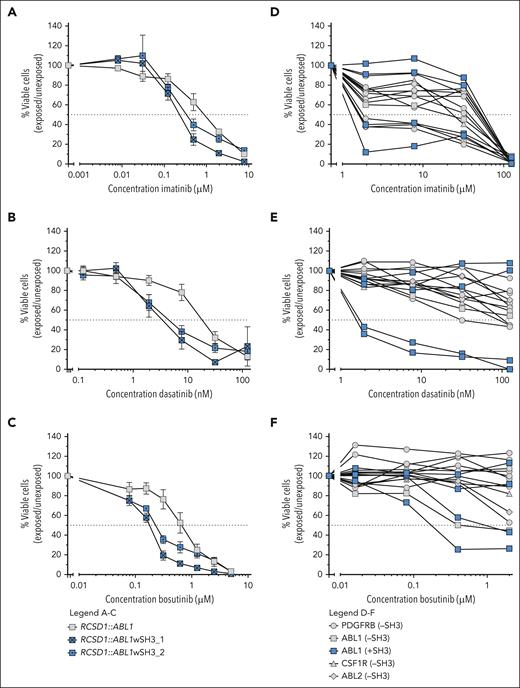
To investigate the impact of intact SH3 and SH2 domains on TKI sensitivity, we introduced these domains into the SH3-lacking RCSD1::ABL1 construct. We generated 2 distinct constructs comprising the ABL1 segment, including the SH3 and SH2 domains. One includes ABL1 from amino acid 27 of exon 2 onwards, denoted RCSD1::ABL1wSH3_1, and the other includes ABL1 from amino acid 62 of exon 2 onwards, designated RCSD1::ABL1wSH3_2 (supplemental Figure 3). The Ba/F3 cell lines expressing the RCSD1::ABL1 constructs with intact SH3 and SH2 domain were slightly more sensitive to imatinib, dasatinib, and bosutinib compared with Ba/F3 cells expressing the SH3-lacking original RCSD1::ABL1 construct (Figure 6A-C). In contrast to the Ba/F3 results, no impact of the presence or absence of intact SH3 and SH2 domains was observed on TKI sensitivity of ABL-class ALL cells derived from patients (Figure 6D-E). The sensitivity of the SH3-containing ABL-class ALL samples was variable and comparable to that of SH3-lacking ABL-class ALL samples. This suggests that mechanisms other than the effect of the SH3 and SH2 domains affect the ex vivo response to TKI in ABL-class ALL.
TKI sensitivity of cells with ABL-class fusions with and without (intact) SH3 and SH2 domains. (A-C) Dose-response curves for imatinib, dasatinib, and bosutinib, respectively, of Ba/F3 models expressing SH3 domain-lacking RCSD1::ABL1 (in gray) or modified constructs including an intact SH3 and SH2 domain (RCSD1::ABL1wSH3_1/2, in blue). Values are normalized against untreated controls for each cell line and represent mean ± SEM (n = 3). (D-F) Dose-response curves for imatinib, dasatinib, and bosutinib, respectively, of primary or primary xenografted ABL-class ALL samples either with SH3 domain (in blue) or without SH3 domain (in gray). Leukemic cell survival relative to untreated controls represents the mean of 1 to 3 biological replicates.
TKI sensitivity of cells with ABL-class fusions with and without (intact) SH3 and SH2 domains. (A-C) Dose-response curves for imatinib, dasatinib, and bosutinib, respectively, of Ba/F3 models expressing SH3 domain-lacking RCSD1::ABL1 (in gray) or modified constructs including an intact SH3 and SH2 domain (RCSD1::ABL1wSH3_1/2, in blue). Values are normalized against untreated controls for each cell line and represent mean ± SEM (n = 3). (D-F) Dose-response curves for imatinib, dasatinib, and bosutinib, respectively, of primary or primary xenografted ABL-class ALL samples either with SH3 domain (in blue) or without SH3 domain (in gray). Leukemic cell survival relative to untreated controls represents the mean of 1 to 3 biological replicates.
TKI sensitivity of ABL-class fusions relative to BCR::ABL1 controls
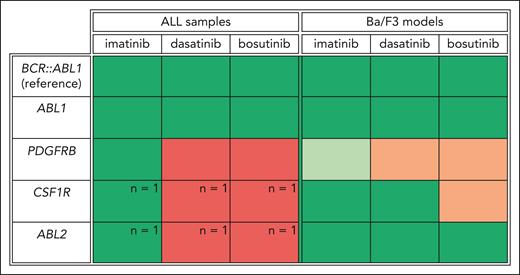
To gain a comprehensive overview of TKI sensitivity based on the involved tyrosine kinase gene, we calculated the TKI sensitivity of ABL-class ALL samples and cell models relative to BCR::ABL1-positive samples per TKIs. This comparison showed that the transduced Ba/F3 model recapitulated the sensitivity observed in ABL-class ALL samples (Figure 7). The Ba/F3 model confirmed that bosutinib exhibited a lower potency against CSF1R-fused cells, whereas both dasatinib and bosutinib demonstrated reduced potency against PDGFRB-fused cells. Similar to BCR::ABL1-positive samples, ABL1- and ABL2-fused samples exhibited cross-TKI sensitivity (Figure 7). For PDGFRB-fused samples, the sensitivity to imatinib in the ALL samples and the Ba/F3 models was similar to or superior to the corresponding BCR::ABL1-positive references, respectively. In contrast, dasatinib and bosutinib sensitivity in the PDGFRB-fused samples was lower than that in the corresponding BCR::ABL1-positive references. This observation suggests that imatinib treatment may be the best TKI choice for PDGFRB-fused ALL and that current or planned clinical trials of dasatinib-based therapies could be less effective for patients with ALL with PDGFRB-fusion.
Comprehensive overview of TKI sensitivity based on the involved tyrosine kinase in ALL samples and transduced Ba/F3 models relative to BCR::ABL1-positive cells. For primary or primary xenografted ALL samples, the percentage of ALL samples by tyrosine kinase gene (ABL1, PDGFRB, CSF1R, or ABL2) with an area under the dose-response curve (AUC) above the median AUC of BCR::ABL1-positive ALL samples was calculated for each TKI (imatinib, dasatinib, or bosutinib). Colors were assigned based on the calculated percentages; green means ≤75% of the samples are above the median AUC of BCR::ABL1-positive ALL samples. Red indicates that >75% of the samples are above the median AUC of the BCR::ABL1-positive ALL samples. For CSF1R and ABL2, only 1 ALL sample was included in this study. This is denoted in the figure. For the transduced Ba/F3 models, green indicates that the half-maximal inhibitory concentration (IC50) is below the IC50 of the transduced Ba/F3 model expressing the BCR::ABL1 fusion, orange indicates an IC50 above that of the BCR::ABL1 Ba/F3 model, and light green indicates a >10-fold lower IC50 than that of the BCR::ABL1 Ba/F3 model.
Comprehensive overview of TKI sensitivity based on the involved tyrosine kinase in ALL samples and transduced Ba/F3 models relative to BCR::ABL1-positive cells. For primary or primary xenografted ALL samples, the percentage of ALL samples by tyrosine kinase gene (ABL1, PDGFRB, CSF1R, or ABL2) with an area under the dose-response curve (AUC) above the median AUC of BCR::ABL1-positive ALL samples was calculated for each TKI (imatinib, dasatinib, or bosutinib). Colors were assigned based on the calculated percentages; green means ≤75% of the samples are above the median AUC of BCR::ABL1-positive ALL samples. Red indicates that >75% of the samples are above the median AUC of the BCR::ABL1-positive ALL samples. For CSF1R and ABL2, only 1 ALL sample was included in this study. This is denoted in the figure. For the transduced Ba/F3 models, green indicates that the half-maximal inhibitory concentration (IC50) is below the IC50 of the transduced Ba/F3 model expressing the BCR::ABL1 fusion, orange indicates an IC50 above that of the BCR::ABL1 Ba/F3 model, and light green indicates a >10-fold lower IC50 than that of the BCR::ABL1 Ba/F3 model.
Discussion
We investigated the ex vivo response to 3 different TKIs in relation to the tyrosine kinase gene affected in 23 ABL-class ALL samples. A previous study related the outcome of patients with ABL-class ALL in the pre-TKI era to the involved tyrosine kinase gene. In that study, patients with CSF1R-fused ALL demonstrated the best event-free survival, whereas patients with ABL2-fused ALL exhibited the poorest outcomes with a chemotherapy regimen. Patients with ABL1- and PDGFRB-fused ALL fell in between.2 This suggests that although patients with ABL-class ALL are considered as 1 group, there are differences, at least in the chemotherapy response related to the involved tyrosine kinase gene.
We assessed sensitivity to the TKIs imatinib, dasatinib, and bosutinib in primary ALL cells and PDX model-origin cells. We importantly detected variability in ex vivo TKI sensitivity related to the type of ABL-class tyrosine kinase genes involved. ABL1-fused ALL samples exhibited strong cross-TKI sensitivity, whereas PDGFRB-fused ALL samples did not exhibit cross-TKI sensitivity. PDGFRB-fused ALL samples and Ba/F3 models exhibited lower sensitivity to dasatinib and bosutinib than ABL1-fused ALL samples. In addition, in our PDGFRB-fused ALL samples, the ex vivo sensitivity to imatinib was similar to that of BCR::ABL1-positive ALL samples, whereas PDGFRB-fused ALL samples were less sensitive to dasatinib and bosutinib at clinically relevant concentrations. Reduced efficacy of bosutinib has been reported previously in Ba/F3 models expressing PDGFRB or CSF1R fusions.4 This reduced efficacy may be explained by the difference in the conformations of PDGFRB and CSF1R associated with the involved protein domains. Notably, bosutinib was designed to target ABL1 specifically and is thus unlikely to have meaningful activity against PDGFRB or CSF1R.16 In contrast to our data, previous studies have suggested sensitivity to dasatinib in Ba/F3 models expressing PDGFRB-fusions.4,60,61 Moreover, various case reports of patients with PDGFRB-fused ALL indicated a response to dasatinib.5,35,40 However, multiple studies using kinase selectivity assays have shown a lower affinity of dasatinib for wild-type PDGFRB than for BCR::ABL1 or wild-type ABL1.62,63 These data indicated that a higher dose of dasatinib is required for PDGFRB-fused ALL to obtain the same level of inhibition as in ABL1-fused ALL. In contrast, our Ba/F3 model expressing EBF1::PDGFRB was extremely sensitive to imatinib compared with other ABL-class fusions. This finding suggests that imatinib may be the best choice for PDGFRB-fused ALL. Our data suggest that although ABL-class fusions are considered as 1 group, the choice of TKI may be guided by the involved tyrosine kinase gene.
In addition to the influence of the type of tyrosine kinase gene involved in ABL-class fusions, other mechanisms seem to play a role in TKI sensitivity, because not all patient samples with the same ABL-class tyrosine kinase gene responded to ex vivo TKI exposure. For instance, some studies have shown the prognostic significance of IKZF1 deletions in pediatric patients with BCR::ABL1-positive ALL with inferior clinical outcomes after imatinib-based treatment.64 These pediatric patients especially showed a poor outcome when having the haploinsufficient form of the IKZF1 deletion.64 Moreover, the IKZF1plus profile, which is characterized by deletions of IKZF1 and PAX5, CDKN2A/B, and/or PAR, has been associated with a poor risk of pediatric BCP-ALL in general.65 In addition, in BCR::ABL1-positive ALL cases, the IKZF1plus profile was previously associated with ex vivo imatinib-resistance.66 For patients with ABL-class ALL, a high frequency of IKZF1 deletions was reported in the pre-TKI era, although neither IKZF1 deletions nor the IKZF1plus profile have been associated with the outcome.2 We also reported a high frequency of IKZF1 deletions (44%) that were not associated with ex vivo imatinib sensitivity in ABL-class ALL samples. Due to the limited cohort size, we were not able to compare the effect on TKI response of deletions causing haploinsufficiency with the effect of dominant-negative isoforms. These data suggest that, in contrast to BCR::ABL1-positive ALL,67IKZF1 deletions do not affect ex vivo TKI sensitivity in ABL-class ALL.
Although, in BCR::ABL1-positive ALL, the SH3 and SH2 domains of ABL1 are generally retained, some ABL1- and ABL2-fused ABL-class ALL cases lack these protein-regulating domains. We hypothesized that these domains might influence TKI sensitivity by their effect on the conformation of the fusion protein. A previous study showed that Ba/F3 cell lines expressing atypical BCR::ABL1 transcripts lacking the SH3 domain exhibited reduced sensitivity to TKIs imatinib, dasatinib, and ponatinib, but not nilotinib, compared with cell lines expressing typical BCR::ABL1 transcripts.68 We showed in a controlled model system that the introduction of the SH3 domain sensitized Ba/F3 cells expressing RCSD1::ABL1 to imatinib, dasatinib, and bosutinib. However, the TKI sensitivity of the SH3 domain-lacking RCSD1::ABL1 was in the same range as that of other Ba/F3 models expressing ABL1 and ABL2 fusions. In our ALL samples without the SH3 domain, TKI sensitivity was not different from that of ALL samples with an intact SH3 domain. Taken together, our data suggest that there may be a subtle effect of the SH3 domain on TKI sensitivity in controlled Ba/F3 model systems but that the lack of an SH3 domain in patient ALL cells does not correlate with imatinib-resistance. This indicates that other mechanisms underlie the variable ex vivo response to TKIs for patients with ABL-class ALL.
In addition to the underlying secondary lesions and involved domains, we tested whether the sample origin or disease status influenced ex vivo TKI response. We showed that there was no difference in ex vivo TKI sensitivity between ABL-class BCP-ALL and T-ALL cells or between samples taken at diagnosis and those taken at relapse. However, our serial transplanted PDX models exhibited high ex vivo TKI resistance compared with the primary material and primary xenografted samples. This observation suggests that the serial transplantation process may lead to the selection of TKI-resistant clones irrespective of their previous exposure to TKIs. Previously, primary xenografted models have been described as representative of the original leukemia.69,70 However, serial transplanted xenografts have been shown to result in decreased clonal complexity owing to clone-dependent properties that confer a proliferative advantage.70 This suggests that even in the absence of TKIs, cells can have a proliferation benefit, resulting in the selection of TKI-resistant clones by serial transplantation. Consequently, primary xenografts better preserve the clonal diversity of primary ALL, making them a more favorable model system for studying ex vivo TKI sensitivity.
The clinical trials treating patients with ABL-class ALL combine treatment with a TKI with a chemotherapy backbone.51 However, there is no established ex vivo setup that enables exposure to various chemotherapeutic agents in combination with TKI, especially when dealing with a limited number of patient cells that exhibit poor survival ex vivo. The combination of chemotherapy with TKI may induce apoptosis in cells that were not or only minimally affected by TKI. Further studies using PDX models and clinical trials are required to elucidate the effect of involved tyrosine kinase in an in vivo or clinical setting.
In conclusion, although patients with ABL-class ALL have been treated clinically as a uniform entity with TKI-based therapies, our new preclinical findings suggest that the type of involved tyrosine kinase is the main determinant of the ex vivo TKI response. Similar to the approaches used for TKI selection for patients with CML and specific ABL1 kinase domain mutations causing TKI resistance,71 we posit that patients with ABL-class ALL with different tyrosine kinases could have differential responses to specific TKIs, a hypothesis that merits careful exploration in clinical trials.
Acknowledgments
The authors thank Fleur Laponder and Laura van der Griend at the Princess Máxima Center for assistance with cloning experiments, Cesca van de Ven at the Princess Máxima Center for generating and characterizing BCR::ABL1 and ABL-class ALL patient–derived primary xenograft models, Alex Hoogkamer for next-generation sequencing analysis of primary ALL samples and the primary xenograft model specimens, Marilyn Li at the Children’s Hospital of Philadelphia for next-generation sequencing analysis of serial transplanted ALL PDX model specimens, and Kathryn Roberts and Charles Mullighan at St. Jude Children’s Research Hospital for provision of ABL-class constructs.
S.K.T. is a scholar of the Leukemia & Lymphoma Society and holds the Joshua Kahan Endowed Chair in Pediatric Leukemia Research at the Children’s Hospital of Philadelphia. The work described in this article was funded by the Dutch Cancer Society (grant KWF-11117; M.L.d.B.), Kika Foundation (program funding, M.L.d.B.), National Institutes of Health, National Cancer Institute awards U01 CA232486 and CA243072 (S.K.T.), Department of Defense Translational Team Science award CA180683P1 (S.K.T.), and the V Foundation for Childhood Cancer Research (S.K.T.).
Authorship
Contribution: I.v.O., J.M.B., and M.L.d.B. conceived the project; I.v.O., S.K.T., C.E.J.R., and A.B. designed and performed the experiments and analyzed the results; S.K.T., H.A.d.G.-K., and G.E. contributed to the samples and clinical data; I.v.O., J.M.B., and M.L.d.B. drafted the manuscript; S.K.T. edited the manuscript; and all authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Monique L. den Boer, Princess Máxima Center, Heidelberglaan 25, 3584 CS Utrecht, The Netherlands; email: m.l.denboer@prinsesmaximacentrum.nl.
References
Author notes
All data generated or analyzed during this study are included in this published article and its supplemental Information Files.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal