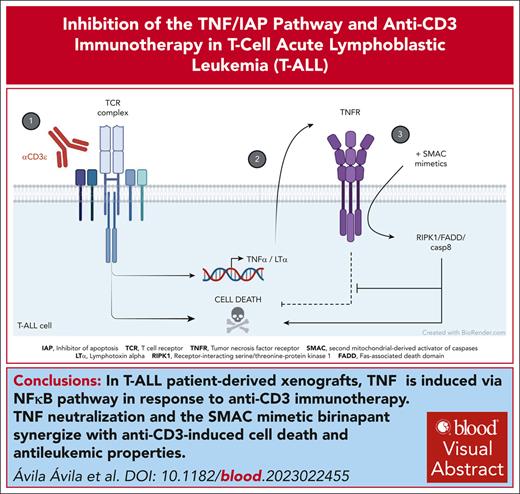
TNF via NF-κB pathway is induced in response to anti-CD3 immunotherapy in patient-derived T-ALL xenografts.
TNF neutralization and the SMAC mimetic birinapant synergize with anti-CD3–induced cell death and antileukemic properties.
Visual Abstract
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematological malignancy. Current treatments, based on intensive chemotherapy regimens provide overall survival rates of ∼85% in children and <50% in adults, calling the search of new therapeutic options. We previously reported that targeting the T-cell receptor (TCR) in T-ALL with anti-CD3 (αCD3) monoclonal antibodies (mAbs) enforces a molecular program akin to thymic negative selection, a major developmental checkpoint in normal T-cell development; induces leukemic cell death; and impairs leukemia progression to ultimately improve host survival. However, αCD3 monotherapy resulted in relapse. To find out actionable targets able to re-enforce leukemic cells’ vulnerability to αCD3 mAbs, including the clinically relevant teplizumab, we identified the molecular program induced by αCD3 mAbs in patient-derived xenografts derived from T-ALL cases. Using large-scale transcriptomic analysis, we found prominent expression of tumor necrosis factor α (TNFα), lymphotoxin α (LTα), and multiple components of the “TNFα via NF-κB signaling” pathway in anti-CD3–treated T-ALL. We show in vivo that etanercept, a sink for TNFα/LTα, enhances αCD3 antileukemic properties, indicating that TNF/TNF receptor (TNFR) survival pathways interferes with TCR-induced leukemic cell death. However, suppression of TNF-mediated survival and switch to TNFR-mediated cell death through inhibition of cellular inhibitor of apoptosis protein-1/2 (cIAP1/2) with the second mitochondrial-derived activator of caspases (SMAC) mimetic birinapant synergizes with αCD3 to impair leukemia expansion in a receptor-interacting serine/threonine-protein kinase 1–dependent manner and improve mice survival. Thus, our results advocate the use of either TNFα/LTα inhibitors, or birinapant/other SMAC mimetics to improve anti-CD3 immunotherapy in T-ALL.
Introduction
T-cell lineage acute lymphoblastic leukemia (T-ALL) derive from the transformation of thymic T-cell progenitors and is an aggressive disease characterized by the proliferation and accumulation of T-cell precursors abnormally arrested in differentiation in the blood, bone marrow, and lymphoid organs. Current first-line chemotherapy regimens provide overall survival rates of ∼85% to 90% in children, and ∼50% in adults. Refractory and relapsed T-ALL cases have a dismal prognosis that is improved by use of nelarabine either as single agent or combined with chemotherapy.1 Immunotherapy, including chimeric antigen receptor (CAR) T-cell therapies, which have proven their efficacy in B-cell leukemias, lags behind in T-ALL because targetable surface antigens are shared between leukemic cells and normal T cells, rendering specific targeting of T-ALL difficult. For example, CAR T-cell approaches in T-ALL are limited by CAR T-cell fratricide activity, depletion of host T cells, and risk of graft-versus-host disease when performed in an allogeneic setting, that need to be corrected by multiple editing of the CAR T-cell genome that may turn out clinically problematic. It is thus essential to identify new, alternative therapeutic tools to offer a wider variety of options to treat patients with T-ALL.
Our previous genetic studies using a mouse T-cell receptor (TCR)-positive T-ALL model driven by an activated Janus kinase allele show that signaling from a negative-selecting mouse TCR, but not from a positively selecting TCR, severely interferes with T-ALL development2 and progression.3 We further extended the antileukemic properties of TCR activation to patient-derived T-ALL xenografts by showing that engagement of leukemic cells’ TCRs by either the well-characterized murine monoclonal antibody (mAb) to human CD3ε, OKT33 or teplizumab,4 a humanized and non–Fcγ-receptor binding version of OKT3, both induced rapid T-ALL cell death and a strong delay in leukemia progression. Furthermore, OKT3 and teplizumab synergized with chemotherapy to improve leukemic mice survival, although most cases eventually relapse.4 Anti-CD3 mAbs thus represent a potential therapeutic option for patients with T-ALL, but it is essential to understand the molecular basis of their antileukemic properties to identify potential new vulnerabilities to reinforce their therapeutic potential. Here, we report that the molecular signature induced in vivo in T-ALL cells in response to OKT3 and teplizumab identify the tumor necrosis factor (TNF) via NF-κB signaling pathway as the top deregulated pathway in gene set enrichment analysis (GSEA) canonical pathways. Importantly, we demonstrate that both anti-CD3 mAbs induce TNF-mediated survival of patient-derived T-ALL xenografts in vivo, a program that can be switched toward cell death and synergy in antileukemic properties upon administration of birinapant, a second mitochondrial-derived activator of caspases (SMAC) mimetic that leads to cellular inhibitor of apoptosis protein-1 or 2 (cIAP1/2) ubiquitin ligases degradation. We also demonstrate both in vitro and in vivo that the anti-CD3/birinapant synergy relies upon receptor-interacting serine/threonine-protein kinase 1 (RIPK1) kinase activity, a key player of cell death pathways downstream of TNF receptor 1 (TNFR1). These results advocate that the combination of anti-CD3 mAbs with inhibitors of IAP1/2 proteins will offer new therapeutic options in T-ALL.
Methods
Patients
T-ALL diagnostic samples were included in the GRAALL-2005 study or in the FRALLE protocol and informed consent was obtained from all patients at trial entry (ClinicalTrials.gov identifier: NCT00327678). Studies were conducted in accordance with the Declaration of Helsinki and approved by local and multicenter research ethical committees.
Mice
NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (abbreviated NSG) female mice (2 months old; Charles River Laboratories) were maintained under specific pathogen-free conditions. Experiments were carried out in accordance with the European Union and French National Committee recommendations, under agreement APAFIS no. 32902-20210908816114788 version 1.
Mice were tail vein injected (106 cells per mouse) with leukemic cells (defined as FSChi; hCD45+; hCD7+) obtained from the bone marrow of NSG mice xenografted with T-ALL diagnostic cases. Leukemia expansion in blood was followed-up, as previously described.4 For long-term survival experiments, treatments were started when tumor burden reached ∼1% in the blood, that is, >10% in bone marrow. For short-term experiments, treatments were started at higher tumor burden (∼10% in the blood, that is, >50% leukemic cells in bone marrow). Control mice were always treated with the respective solvents or control antibodies.
In vivo antibodies and inhibitors treatments
Antibodies are listed in supplemental Methods, available on the Blood website. Mice were tail-vein intravenously injected with the quantity of antibody indicated in the figure legends. Etanercept (10 mg/kg) was injected intraperitoneally 6 hours before anti-CD3 treatment. Birinapant (MedChem Tronica, 20 mg/kg) was administrated intraperitoneally every 3 to 4 days, 2 hours after anti-CD3 injection. End point for survival experiment was defined as previously described.4
Transcriptomic analysis
Affymetrix
Total RNA was isolated from flow cytometry–sorted UPNT776 leukemic cells (hCD45+/hCD7+) 6 hours after treatment with either 40 μg immunoglobulin G2a (IgG2a; n = 3), OKT3 (n = 3), or teplizumab (n = 3), using the RNeasy kit (Qiagen). Complementary RNA synthesis and hybridization of Affymetrix GeneChip Human Gene 2.0 ST array were according to the manufacturer’s instructions, as described (http://www-microarrays.u-strasbg.fr). Raw data were processed as described in supplemental Methods.
RNA sequencing (RNAseq)
Total RNA was isolated from flow cytometry–sorted UPNT420 leukemic cells (hCD45+/hCD7+) 6 hours after treatment of leukemic mice with either 4 μg IgG2a (n = 6) or OKT3 (n = 6) using the RNeasy mini kit (Qiagen). Total RNA (1 μg) was used for the construction of sequencing libraries. RNA libraries were prepared using the Illumina Stranded mRNA Prep Ligation kit following manufacturer's protocols. Sequencing was carried out on a NovaSeq 6000 instrument from Illumina (S1-PE100, paired-end reads). Detailed description of analysis is reported in supplemental Methods. Counts were normalized using the trimmed mean of M values normalization from EdgeR (version 3.32.1). Differential gene expression was assessed with the Limma voom framework (version 3.46.0). Genes with an absolute fold change of ≥1.5 and an adjusted P value < .05 were labeled significant.
Functional analyses
GSEA (http://software.broadinstitute.org/gsea) was run using signal-to-noise for the ranking gene metric and 1000 permutations. The negative selection signature used as gene set reference has been described by Baldwin and Hogquist5 and in unpublished information by T.A. Baldwin. Enrichment analysis from differentially expressed genes has been performed with the Kyoto Encyclopedia of Genes and Genomes (KEGG) and illustrated by using the KEGG mapper tool.
Nanostring
Primary xenografts obtained from 8 independent T-ALL cases (see supplemental Table 5 for description) were engrafted into NSG mice. When tumor burden reached 10% in the blood, mice were either treated with 40 μg IgG2a (n = 3) or OKT3 (n = 3) for 12 hours. For the kinetic experiments, UPNT776-engrafted mice were treated with either 40 μg IgG2a (n = 3) or 40 μg OKT3 (n = 3, for each time point), and mice were euthanized at different time points after treatment. Total RNA was isolated from flow cytometry–sorted leukemic cells (hCD45+/hCD7+) using the RNeasy kit (Qiagen), and RNA from each group of 3 mice pooled for further analysis. Nanostring custom panel of 97 genes notably encompassing TNFR superfamily receptors, ligands, and effectors, and comprising 5 housekeeping genes (see supplemental Methods for exhaustive list of genes) was constructed and hybridized as recommended by the manufacturer’s instructions.
Western blot
Cells were washed in cold phosphate-buffered saline and lysed in RadioImmunoPrecipitation Assay (RIPA) buffer supplemented with protease and phosphatase inhibitors (Halt Protease and Phosphatase Inhibitor Cocktail, Thermo Scientific). Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Bio-Rad). Antibodies are listed in supplemental Methods.
Statistics
Leukemic load statistical analysis was performed using Prism (version 5.0; GraphPad Software). The data are expressed as mean ± standard deviation of n = 3 determinations. In short-term in vivo and in vitro assays, we used the analysis of variance and the Tukeys multiple comparison test or the Student t test. Kaplan-Meier survival curves were compared using the log-rank test. Differences were considered statistically significant at ∗P < .05, ∗∗P < .01, or ∗∗∗P < .001.
Results
We previously found that anti-CD3 antibodies are strongly antileukemic in TCR+ T-ALL mouse models and patient-derived T-ALL xenografts in immune-deficient NSG mice. Although combination of anti-CD3 mAbs with chemotherapy can result in eradication of leukemic cells in patient-derived T-ALL xenografts, anti-CD3 monotherapy ultimately resulted in relapse in all cases tested,4 calling for search of actionable vulnerabilities able to re-enforce anti-CD3 antileukemic potential.
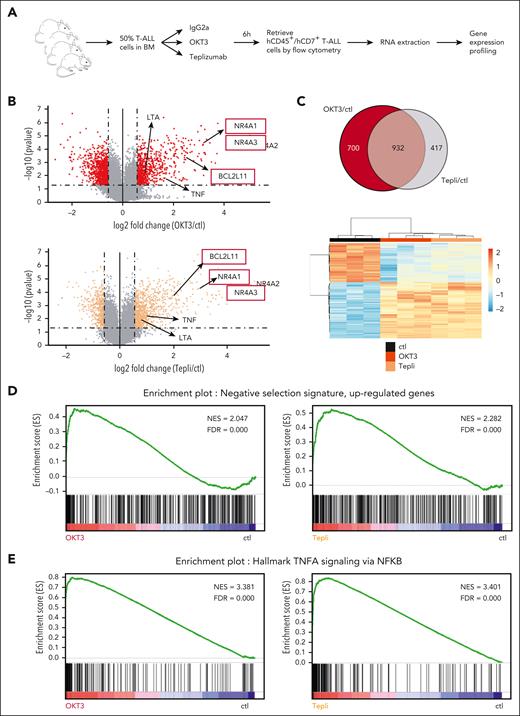
To this end, we identified the in vivo molecular response of T-ALL cells to anti-CD3 mAbs. For this, groups of NSG mice xenografted with patient-derived T-ALL xenograft UPNT776 were treated at high tumor burden with either anti-CD3 mAbs OKT3 or the clinically relevant teplizumab, or with the respective control antibodies. Mice were followed-up over time for leukemia burden, induction of caspase activity, and cell death (see supplemental Figure 1A for a scheme of the experiment). In line with our published results,4 anti-CD3 administration induced a fast decrease in leukemia burden, which becomes significant at 12 hours (supplemental Figure 1B) associated with significant increase of apoptosis at 8 hours, as illustrated both by increased annexin V staining (supplemental Figure 1C) and increased activity of caspase 3 and 8 (supplemental Figure 1D-E). To identify the molecular programs associated with cell death induction, T-ALL UPNT776-xenografted leukemic mice were treated for 6 hours with either 40 μg of either control antibody or anti-CD3 mAbs OKT3 or teplizumab, after which mice were euthanized, and leukemic cells flow cytometry sorted and analyzed for gene expression using Affymetrix GeneChip Human Gene 2.0 ST array (see Figure 1A for a scheme of the experiment). As shown in Figure 1B, both OKT3 and teplizumab induced a clear molecular signature with 1632 and 1349 differentially expressed genes, respectively (fold change of >1.5, P < .05; see supplemental Tables 1 and 2 for an exhaustive list of differentially expressed genes). Of these, 932 genes were found commonly deregulated, as shown in the Venn diagram and heat map of Figure 1C, the latter illustrating how the common OKT3/teplizumab signature clusters away from the transcriptomic signature of control cells. In line with our previous results,3 GSEA highlighted a very strong and significant enrichment for TCR-induced genes related to apoptotic clonal deletion/negative selection of normal thymic progenitors5 in both OKT3- and teplizumab-treated leukemic cells (Figure 1D), consistent with their induction toward apoptosis. Of note, NR4A1, NR4A3, and BCL2L11, the only genes so far demonstrated important for thymic negative selection,6 were found strongly upregulated in response to OKT3 and teplizumab (Figure 1B). A similar anti-CD3–induced gene expression signature was also found by RNAseq in another independent T-ALL case, UPNT 420 (supplemental Figure 2A-B), that also highlighted enrichment for the clonal/negative selection in anti-CD3–stimulated leukemic cells (supplemental Figure 2C). Because induction of apoptosis could involve BCL2L11/BIM upregulation, we thus investigated whether the B-cell lymphoma 2 (BCL2)/Bcl-X inhibitor navitoclax administered either alone or in combination with anti-CD3 could affect T-ALL expansion. Data in supplemental Figure 3B indeed show that ABT263 synergizes with anti-CD3 to impair T-ALL expansion and to prolong mice survival. Interestingly, when investigating the anti-CD3–induced gene expression signature among 50 GSEA hallmarks to identify which other pathways could be significantly enriched, the “TNF⍺ signaling via NF-κB” hallmark consistently ranked first (supplemental Tables 3 and 4). This enrichment was found profound and significant in both UPNT776 leukemic cells collected from OKT3- or teplizumab-treated mice (Figure 1E) and UPNT420 leukemic cells collected from OKT3-treated mice (supplemental Figure 2D). When analysis was performed using Human MSigDB collection C2-CP (canonical pathways that comprises 3090 gene sets from Biocarta, Kyoto Encylopedia of Genes and Genomes, PID subset of CP, Reactome, and WikiPathways subset of CP), “TNF⍺ signaling pathway” signature was also significantly enriched in OKT3-treated (normalized enrichment score = 2.57, false discovery rate = 0) or teplizumab-treated (normalized enrichment score = 2.55, false discovery rate = 0) cells. As illustrated in the schematic representation of TNF/TNFR signaling (supplemental Figure 4A-B), genes encoding ligands, adapters, signaling proteins, transcription factors, and regulators of TNF/TNFR pathway were found induced in T-ALL cells from anti-CD3–treated mice.
Anti-CD3 mAbs OKT3 and teplizumab share a commonly induced transcriptomic signature in T-ALL. (A) Schematic representation of the experiment. NSG mice infused with 106 leukemic cells from T-ALL case UPNT776, were monitored for leukemia expansion over time. Once leukemic, mice (n = 3 per group) were treated with 40 μg of either control mIgG2a, or OKT3 or teplizumab. Six hours later, mice were euthanized, hCD45+/hCD7+ leukemic cells sorted from the bone marrow by flow cytometry, and RNA extracted and analyzed using Affymetrix GeneChip Human Gene 2.0 ST array. (B) Volcano plot of probe sets differentially expressed between control and either OKT3- (top panel) or teplizumab (bottom panel) -treated leukemic cells described in panel A. Fold change (x-axis) is plotted against statistical significance (y-axis) for each probe set. Genes with a fold change of ≥1.5 and P < .05 are highlighted as red (OKT3) or orange (teplizumab) dots, respectively. (C) Top panel: Venn diagram of differentially expressed genes between control, OKT3-treated (red), and teplizumab-treated (grey) leukemic cells described in panel A. Commonly deregulated genes are highlighted in greyish red; lower panel: heat map representation of the 932 genes commonly deregulated by OKT3 and teplizumab as compared with the transcriptomic signature of control cells. (D) GSEA of the negative selection transcriptional signature of mouse thymocytes undergoing negative selection as published by Baldwin and Hogquist5 with the transcriptomic signature induced in vivo in T-ALL cells in response to OKT3 (left) or teplizumab (right), showing a strong and significant enrichment of this thymic signature in anti-CD3–treated samples. (E) GSEA of the “TNF⍺ signaling via NF-κB” signature with that induced in response to OKT3 (left) or teplizumab (right) in T-ALL cells, showing a strong and significant enrichment of this signature in anti-CD3–treated samples. ctl, control; FDR, false discovery rate; NES, normalized enrichment score; Tepli, teplizumab.
Anti-CD3 mAbs OKT3 and teplizumab share a commonly induced transcriptomic signature in T-ALL. (A) Schematic representation of the experiment. NSG mice infused with 106 leukemic cells from T-ALL case UPNT776, were monitored for leukemia expansion over time. Once leukemic, mice (n = 3 per group) were treated with 40 μg of either control mIgG2a, or OKT3 or teplizumab. Six hours later, mice were euthanized, hCD45+/hCD7+ leukemic cells sorted from the bone marrow by flow cytometry, and RNA extracted and analyzed using Affymetrix GeneChip Human Gene 2.0 ST array. (B) Volcano plot of probe sets differentially expressed between control and either OKT3- (top panel) or teplizumab (bottom panel) -treated leukemic cells described in panel A. Fold change (x-axis) is plotted against statistical significance (y-axis) for each probe set. Genes with a fold change of ≥1.5 and P < .05 are highlighted as red (OKT3) or orange (teplizumab) dots, respectively. (C) Top panel: Venn diagram of differentially expressed genes between control, OKT3-treated (red), and teplizumab-treated (grey) leukemic cells described in panel A. Commonly deregulated genes are highlighted in greyish red; lower panel: heat map representation of the 932 genes commonly deregulated by OKT3 and teplizumab as compared with the transcriptomic signature of control cells. (D) GSEA of the negative selection transcriptional signature of mouse thymocytes undergoing negative selection as published by Baldwin and Hogquist5 with the transcriptomic signature induced in vivo in T-ALL cells in response to OKT3 (left) or teplizumab (right), showing a strong and significant enrichment of this thymic signature in anti-CD3–treated samples. (E) GSEA of the “TNF⍺ signaling via NF-κB” signature with that induced in response to OKT3 (left) or teplizumab (right) in T-ALL cells, showing a strong and significant enrichment of this signature in anti-CD3–treated samples. ctl, control; FDR, false discovery rate; NES, normalized enrichment score; Tepli, teplizumab.
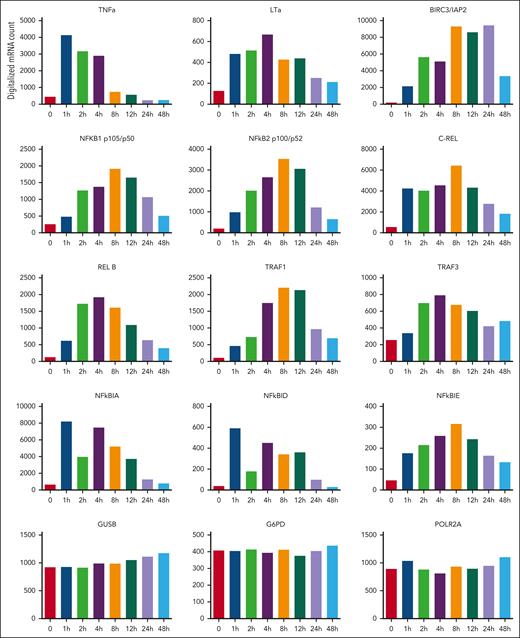
Using multiplex gene expression technology (ncounter Nanostring), we confirmed anti-CD3–mediated upregulation of these genes in a panel of 8 independent patient-derived T-ALL xenografts, most of them being known NF-κB targets (supplemental Figure 5). We also analyzed the induction kinetics of the TNF⍺, lymphotoxin ⍺ (LT⍺) ligands, and of the adapters/effectors TNFR-associated factor 1 (TRAF1), TRAF3, BIRC3/c-IAP2, NF-κBIA, NF-κBID, NF-κBIE, NF-κB1, NF-κB2, c-REL, and REL-B (Figure 2). Of note, TNF⍺ was rapidly and transiently induced whereas LT⍺ expression was more sustained (Figure 2). Taken together, these results show that critical players in TNF⍺/LT⍺ signaling are upregulated in T-ALL cells in response to anti-CD3 mAbs, suggesting a possible impact of TNF/TNFR signaling on the antileukemic potential of these antibodies.
TNF signaling components are induced in vivo in response to anti-CD3 in T-ALL. RNA obtained from leukemic cells (T-ALL case UPNT776) of engrafted mice that were either left untreated, or treated for the indicated time periods (hours) with anti-CD3 OKT3 were analyzed using a Nanostring custom panel of 97 genes comprising 5 housekeeping genes. Histograms show digitalized messenger RNA expression for TNF⍺, LT⍺, BIRC3, NF-κB1, NF-κB2, c-REL, RELB, TRAF1, TRAF3, NF-κBIA, NF-κBID, and NF-κBIE after normalization with Nsolver software. Glucuronidase beta (GUSB), glucose-6-phosphate dehydrogenase (G6PD), and RNA polymerase 2 subunit A (POLR2A) were used as control housekeeping genes.
TNF signaling components are induced in vivo in response to anti-CD3 in T-ALL. RNA obtained from leukemic cells (T-ALL case UPNT776) of engrafted mice that were either left untreated, or treated for the indicated time periods (hours) with anti-CD3 OKT3 were analyzed using a Nanostring custom panel of 97 genes comprising 5 housekeeping genes. Histograms show digitalized messenger RNA expression for TNF⍺, LT⍺, BIRC3, NF-κB1, NF-κB2, c-REL, RELB, TRAF1, TRAF3, NF-κBIA, NF-κBID, and NF-κBIE after normalization with Nsolver software. Glucuronidase beta (GUSB), glucose-6-phosphate dehydrogenase (G6PD), and RNA polymerase 2 subunit A (POLR2A) were used as control housekeeping genes.
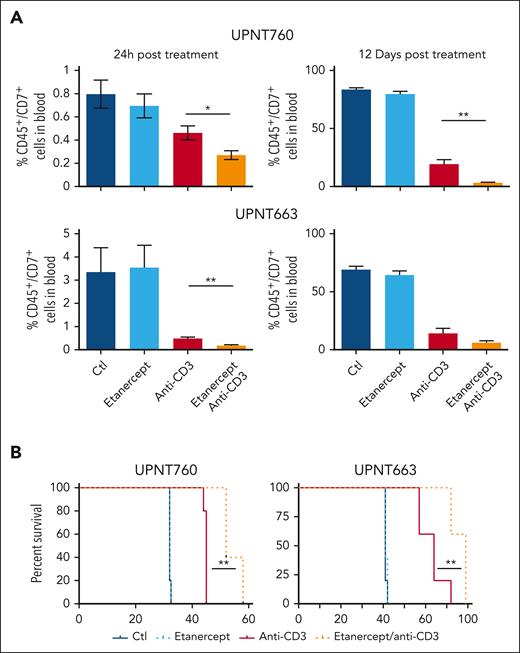
To investigate the role of TNF⍺/LT⍺ in the antileukemic properties of anti-CD3 mAbs, we thought to interfere with this pathway using etanercept, a recombinant, dimeric fusion protein consisting of 2 molecules of the extracellular ligand-binding portion of human TNFR2 linked to the Fc portion of human IgG1, that binds TNF and LTα, thus acting as a soluble decoy receptor and sink for TNF⍺/LT⍺.7 This allows investigating the consequences of TNF⍺/LT⍺ neutralization on the response of T-ALL cells to anti-CD3 mAbs in vivo. Thus, groups of leukemic mice xenografted with independent T-ALL cases were pretreated with etanercept and, 6 hour later, treated or not with OKT3. Leukemic load was then compared over time in all groups. As shown in Figure 3, etanercept did not significantly affect tumor load whereas, as expected, OKT3 treatment resulted in leukemic load inhibition over time. Importantly, etanercept administration further enhanced the antileukemic properties of OKT3 as shown by decreased tumor burden in etanercept/OKT3 treated mice as compared with leukemic mice treated with OKT3 alone (Figure 3A; supplemental Figure 6). Expectedly, etanercept/OKT3 combinatorial treatment resulted in improved leukemic mice survival as compared with mice treated with OKT3 as single agent (Figure 3B). These data indicate that TNF⍺/LT⍺, which are induced in T-ALL cells in response to OKT3, interferes with the implementation of anti-CD3–induced cell death in T-ALL cells, and that TNF⍺/LT⍺ inhibition improves anti-CD3 antileukemic potential in vivo.
Neutralization of TNF⍺/LT⍺ by etanercept administration improves the antileukemic properties of anti-CD3. (A) Leukemia tumor burden (hCD45+/hCD7+) at the indicated time point (24 hours, 12 days) after treatment of leukemic mice with either 10 mg/kg etanercept, or 4 μg of OKT3 in presence or absence of 10 mg/kg etanercept. Experiments were performed using independent patient-derived xenografts (PDXs) derived from UPNT760 and UPNT663 diagnostic T-ALL cases, as indicated. Dark blue bar: control mice treated with control IgG2a and carrier solvent; light blue bar: etanercept-treated mice; red bar: anti-CD3–treated mice; orange bar: anti-CD3/etanercept cotreated mice. (B) Kaplan-Meier survival curves of NSG mice transplanted with T-ALL UPNT-760 and UPNT-663 as indicated and treated as in panel A. ∗P < 0.05; ∗∗P < 0.01.
Neutralization of TNF⍺/LT⍺ by etanercept administration improves the antileukemic properties of anti-CD3. (A) Leukemia tumor burden (hCD45+/hCD7+) at the indicated time point (24 hours, 12 days) after treatment of leukemic mice with either 10 mg/kg etanercept, or 4 μg of OKT3 in presence or absence of 10 mg/kg etanercept. Experiments were performed using independent patient-derived xenografts (PDXs) derived from UPNT760 and UPNT663 diagnostic T-ALL cases, as indicated. Dark blue bar: control mice treated with control IgG2a and carrier solvent; light blue bar: etanercept-treated mice; red bar: anti-CD3–treated mice; orange bar: anti-CD3/etanercept cotreated mice. (B) Kaplan-Meier survival curves of NSG mice transplanted with T-ALL UPNT-760 and UPNT-663 as indicated and treated as in panel A. ∗P < 0.05; ∗∗P < 0.01.
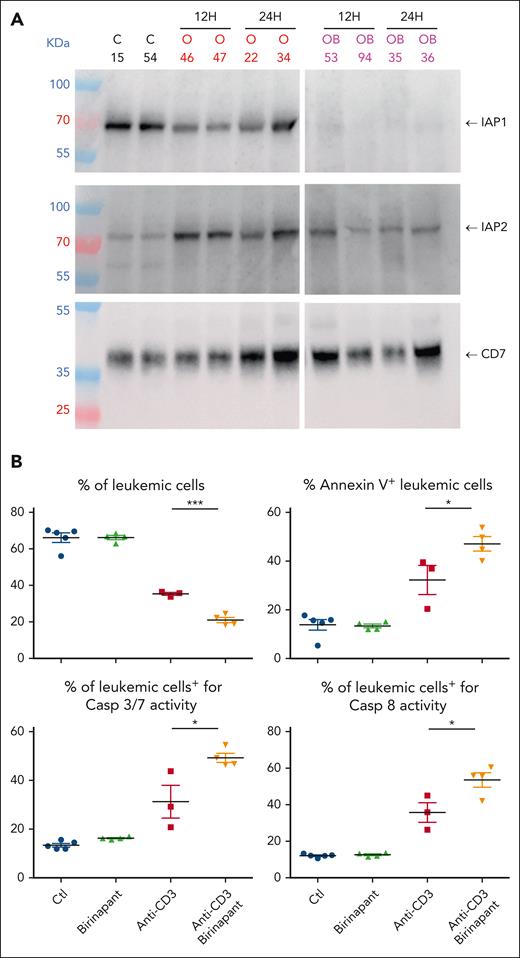
Engagement of TNFR1 by TNF⍺/LT⍺ triggers the formation of different signaling complexes, each resulting in distinct cellular responses. Complex I is assembled at the plasma membrane through recruitment via the tumor necrosis factor receptor type1-associated death domain protein adapter to the TNFR1 death domain followed by other components, including TRAF2, RIPK1, the ubiquitin E3 ligases cIAP1/2, and linear ubiquitin chain assembly complex. Multiple types of ubiquitination events in the TNFR1 complex ultimately lead to IκB kinase and NF-κB activation to promote survival. Disruption of this complex, for example by SMAC-mediated inactivation of the ubiquitin E3 ligases cIAP1/2, results in the formation of several cytoplasmic complexes that mediate RIPK1-dependent and independent cell death (as previously reviewed8). Given that TNFR1 and cIAP1 are expressed in patient-derived T-ALL xenografts (Figure 4A; supplemental Figure 7A-B) and BIRC3/cIAP2 is induced upon anti-CD3 treatment (Figures 2 and 4A; supplemental Figures 2A, 5, and 7), we next investigated whether inhibition of cIAP1/2 could indeed synergize with anti-CD3 mAbs to induce T-ALL cell apoptosis in vivo. Groups of leukemic mice xenografted with either T-ALL case UPTN760 (Figure 4B) or UPNT420 (supplemental Figure 8A) were treated for 6 hours with either OKT3, or the SMAC mimetic birinapant, or the combination of OKT3 and birinapant, or left untreated as control. Birinapant binds to cIAP1 and cIAP2 and induces their auto/transubiquitination followed by their proteasomal degradation.9 As expected, birinapant administration suppressed cIAP1 expression in T-ALL cells (Figure 4A, top panel) and blunted the OKT3-induced upregulation of cIAP2 (Figure 4A, middle panel). As shown in Figure 4B, although administration of birinapant as single agent had little effect on leukemia burden, induction of caspase 3/8 activity, and apoptosis of leukemic cells, the coadministration of OKT3 + birinapant induced higher levels of caspase activity and apoptosis of T-ALL cells than the level induced after OKT3-only administration, that resulted in further inhibition of leukemic load (Figure 4B; supplemental Figure 8A). We conclude from these results that anti-CD3–induced cell death and antileukemic properties can be re-enforced by impairing cIAP1/2 expression using SMAC mimetics.
The SMAC mimetic birinapant synergizes with anti-CD3 to induce leukemic cell apoptosis. (A) Western blot analysis of IAP1 (top panel) and IAP2 (middle panel) expression in leukemic cells of UPNT760 T-ALL xenografted mice at 12 or 24 hours after administration of either 4 μg OKT3 (O, red writing) or OKT3/birinapant (20 mg/kg) (OB, purple writing). Control mice were treated with IgG2a/carrier solvent. Results from 2 independent mice are shown. Anti-hCD7 expression is used as loading control (lower panel). (B) NSG mice were injected with 106 UPNT760 T-ALL cells. At high leukemia burden, mice were treated with either control IgG2a/solvent (blue dots), IgG2a/birinapant (20 mg/kg, green triangles), 4 μg OKT3/solvent (red squares), or combination of OKT3 and birinapant (orange triangles) for 6 hours, after which time leukemia burden (hCD45/hCD7+ cells), annexin V/propidium iodide (PI) staining, and caspase 3/7 and caspase 8 activities were analyzed by flow cytometry. Ctl, control. ∗ P < 0.05; ∗∗∗ P < 0.001.
The SMAC mimetic birinapant synergizes with anti-CD3 to induce leukemic cell apoptosis. (A) Western blot analysis of IAP1 (top panel) and IAP2 (middle panel) expression in leukemic cells of UPNT760 T-ALL xenografted mice at 12 or 24 hours after administration of either 4 μg OKT3 (O, red writing) or OKT3/birinapant (20 mg/kg) (OB, purple writing). Control mice were treated with IgG2a/carrier solvent. Results from 2 independent mice are shown. Anti-hCD7 expression is used as loading control (lower panel). (B) NSG mice were injected with 106 UPNT760 T-ALL cells. At high leukemia burden, mice were treated with either control IgG2a/solvent (blue dots), IgG2a/birinapant (20 mg/kg, green triangles), 4 μg OKT3/solvent (red squares), or combination of OKT3 and birinapant (orange triangles) for 6 hours, after which time leukemia burden (hCD45/hCD7+ cells), annexin V/propidium iodide (PI) staining, and caspase 3/7 and caspase 8 activities were analyzed by flow cytometry. Ctl, control. ∗ P < 0.05; ∗∗∗ P < 0.001.
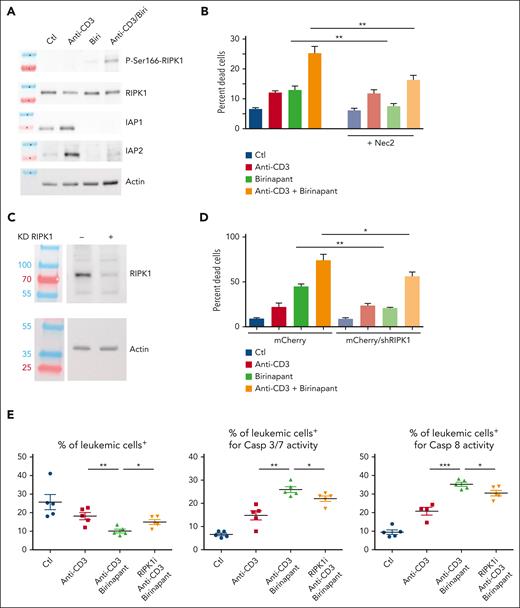
TNFR1-mediated cell death relies, at least in part, upon the protein kinase activity of RIPK1. The only substrate known so far for RIPK1 is RIPK1 itself on Ser166 in its activation loop.10 We thus investigated whether increased apoptosis induced in response to anti-CD3/birinapant is associated with increased RIPK1 activity. For this, the phosphorylation status of Ser166 of RIPK1 was assessed in CEM T-ALL cells (Figure 5A) and a patient-derived T-ALL xenograft (supplemental Figure 8B) treated or not with either anti-CD3, or birinapant, or the combination of both compounds. Although anti-CD3 treatment failed to induce RIPK1 autophosphorylation, its combination with birinapant treatment enhanced RIPK1 Ser166 phosphorylation (Figure 5A; supplemental Figure 8B). In line with our in vivo data, treatment with birinapant enhanced anti-CD3–induced cell death of CEM cells in vitro (Figure 5B). Pharmacological inhibition of RIPK1 activity by necrostatin 2 (Figure 5B) or knock down of RIPK1 expression by small-hairpin RNA–mediated silencing (Figure 5C-D) both reduced anti-CD3/birinapant-induced cell death. Importantly, pharmacological inhibition of RIPK1 activity also impaired the in vivo synergy of the anti-CD3 + birinapant combination in patient-derived T-ALL xenograft as shown by decreased inhibition of leukemia burden and decreased caspase 3/7 and caspase 8 activities upon RIPK1 inhibition in vivo (Figure 5E). We conclude from these results that T-ALL cell death induction by anti-CD3/birinapant treatment relies on RIPK1 kinase activity.
Anti-CD3/birinapant-induced cell death is dependent on RIPK1. (A) Western blot analysis of IAP1 and IAP2 expression and RIPK1 phosphorylation in CEM T-ALL cells stimulated for 4 hours with anti-CD3, birinapant (2.5 nM), or the anti-CD3/birinapant combination. RIPK1 and actin expression are used as normalizers. (B) CEM T-ALL cells were either left untreated (blue), or treated for 24 hours with either anti-CD3 (red), birinapant (2.5 nM, green), or the anti-CD3 + birinapant combination (orange), in either the absence (bright columns) or presence (faded columns) of the RIPK1 inhibitor necrostatin 2. Cell death (PI+ cells) was measured by flow cytometry, 24 hours after start of the treatment. The average of 3 independent experiments is shown. (C) Western blot analysis of RIPK1 expression in control (−) vs CEM T-ALL cells stably expressing a RIPK1 short-hairpin RNA (+). Actin is used as a loading control. (D) Percentage of cell death as measured by PI positivity by flow cytometry in nontreated cells (blue), or 24 hours after treatment with either anti-CD3 (red) or birinapant (100 nM; green) or the combination of anti-CD3 + birinapant (orange). The average of 3 independent experiments is shown. (E) NSG mice were injected with 106 UPNT420 T-ALL cells. At high leukemia burden, mice were treated with either control IgG2a/carrier solvent (blue dots), or 4 μg OKT3/carrier solvent (red squares), or the combination of OKT3/birinapant/carrier solvent (green triangles), or the same combination together with the RIPK1 inhibitor (50 mg/kg, orange triangles) for 12 hours, after which time leukemia burden (hCD45+/hCD7+ cells), caspase 3/7 and caspase 8 activities were analyzed by flow cytometry. Ctl, control. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
Anti-CD3/birinapant-induced cell death is dependent on RIPK1. (A) Western blot analysis of IAP1 and IAP2 expression and RIPK1 phosphorylation in CEM T-ALL cells stimulated for 4 hours with anti-CD3, birinapant (2.5 nM), or the anti-CD3/birinapant combination. RIPK1 and actin expression are used as normalizers. (B) CEM T-ALL cells were either left untreated (blue), or treated for 24 hours with either anti-CD3 (red), birinapant (2.5 nM, green), or the anti-CD3 + birinapant combination (orange), in either the absence (bright columns) or presence (faded columns) of the RIPK1 inhibitor necrostatin 2. Cell death (PI+ cells) was measured by flow cytometry, 24 hours after start of the treatment. The average of 3 independent experiments is shown. (C) Western blot analysis of RIPK1 expression in control (−) vs CEM T-ALL cells stably expressing a RIPK1 short-hairpin RNA (+). Actin is used as a loading control. (D) Percentage of cell death as measured by PI positivity by flow cytometry in nontreated cells (blue), or 24 hours after treatment with either anti-CD3 (red) or birinapant (100 nM; green) or the combination of anti-CD3 + birinapant (orange). The average of 3 independent experiments is shown. (E) NSG mice were injected with 106 UPNT420 T-ALL cells. At high leukemia burden, mice were treated with either control IgG2a/carrier solvent (blue dots), or 4 μg OKT3/carrier solvent (red squares), or the combination of OKT3/birinapant/carrier solvent (green triangles), or the same combination together with the RIPK1 inhibitor (50 mg/kg, orange triangles) for 12 hours, after which time leukemia burden (hCD45+/hCD7+ cells), caspase 3/7 and caspase 8 activities were analyzed by flow cytometry. Ctl, control. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
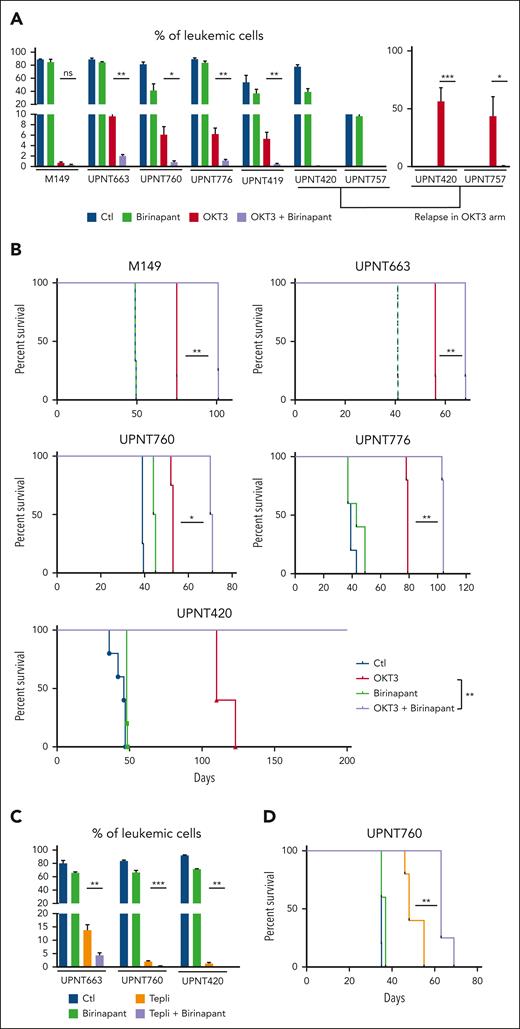
Because we found increased leukemic cell apoptosis in response to anti-CD3/birinapant treatment, we investigated whether this could translate into improved anti-CD3 therapeutic effect. For this, groups of leukemic mice xenografted with either a TCRγδ+ T-ALL or 6 independent T-ALL TCR⍺β+ cases (supplemental Table 5) were treated with OKT3, or the SMAC mimetic birinapant, or the combination of OKT3 and birinapant, or left untreated as control. As shown in Figure 6A and consistent with our previous results (Figure 4B), birinapant alone hardly affected tumor expansion, whereas, as expected, OKT3 treatment resulted in profound inhibition of leukemic load in all of the 7 T-ALL analyzed. Combining OKT3 with birinapant clearly further reduced leukemic load in all cases (Figure 6A, compare purple with red bars). Treatment of patient-derived T-ALL xenografts derived from T-ALL UPNT420 and UPNT757 with either OKT3 or the OKT3 + birinapant combination resulted in leukemia eradication at the time control, nontreated mice were terminally ill and had to be euthanized (Figure 6A). Relapse was observed to occur from ∼10 weeks in the OKT3-treated arm (Figure 6A, right panel) whereas mice from the OKT3 + birinapant–treated group remained leukemia free (Figure 6A). Expectedly, this synergy translated into a strongly improved survival of leukemic mice cotreated with OKT3 and birinapant (Figure 6B, purple line) as compared with OKT3-treated mice (red line). In case of T-ALL UPTN420, OKT3/birinapant combination even resulted in the cure of mice (Figure 6B). Importantly, synergy between anti-CD3 and birinapant was also observed when using the clinically relevant anti-CD3 teplizumab instead of OKT3 (Figure 6C-D). These results demonstrate that the immunotherapeutic properties of anti-CD3 antibodies can be improved by SMAC mimetics such as birinapant. Of note, anti-CD3/birinapant cooperation was found more efficient as compared with etanercept/anti-CD3 combination (supplemental Figure 9), suggesting that, although interfering with the prosurvival TNF/NF-κB pathway re-enforces anti-CD3 antileukemic effects, rerouting TNF signaling to RIPK1-dependent cell death using birinapant further improves anti-CD3 immunotherapeutic properties.
Birinapant synergizes with anti-CD3 to inhibit leukemia expansion. (A) NSG mice infused with 106 leukemic cells from the PDXs derived from the indicated T-ALL cases were monitored for leukemia expansion over time. Once leukemic, mice (n = 4 per group) were treated 6 times with either 4 μg of control IgG2a/carrier solvent (blue), or OKT3/carrier solvent (red), or IgG2a/birinapant (20 mg/kg, green), or OKT3 + birinapant (purple), every 3 to 4 days. In the left part of the panel, leukemic load is measured at the end of treatment. For UPNT420- and 757-derived T-ALL xenografts, mice treated with OKT3/carrier solvent were found to be in remission for 70 (UPNT420) and 110 (UPNT757) days, respectively, after beginning of the treatment but relapsed thereafter. The right part of the panel shows leukemia load at day 102 days (UPNT420) and day 171 (UPNT757), respectively, after beginning of treatment. Note that at these time points, OKT3 + birinapant–treated mice are leukemia free. (B) Kaplan-Meier survival curves of NSG mice transplanted with T-ALL M149, UPNT663, UPNT-760, UPNT776, or UPNT-420 of the cohorts described in panel A. (C) The same experimental protocol was used for PDXs derived from T-ALL UPNT663, UPNT760, and UPNT420 using the clinically relevant anti-CD3 teplizumab. (D) Kaplan-Meier survival curves of NSG mice transplanted with T-ALL UPNT-760 and treated as in panel C. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
Birinapant synergizes with anti-CD3 to inhibit leukemia expansion. (A) NSG mice infused with 106 leukemic cells from the PDXs derived from the indicated T-ALL cases were monitored for leukemia expansion over time. Once leukemic, mice (n = 4 per group) were treated 6 times with either 4 μg of control IgG2a/carrier solvent (blue), or OKT3/carrier solvent (red), or IgG2a/birinapant (20 mg/kg, green), or OKT3 + birinapant (purple), every 3 to 4 days. In the left part of the panel, leukemic load is measured at the end of treatment. For UPNT420- and 757-derived T-ALL xenografts, mice treated with OKT3/carrier solvent were found to be in remission for 70 (UPNT420) and 110 (UPNT757) days, respectively, after beginning of the treatment but relapsed thereafter. The right part of the panel shows leukemia load at day 102 days (UPNT420) and day 171 (UPNT757), respectively, after beginning of treatment. Note that at these time points, OKT3 + birinapant–treated mice are leukemia free. (B) Kaplan-Meier survival curves of NSG mice transplanted with T-ALL M149, UPNT663, UPNT-760, UPNT776, or UPNT-420 of the cohorts described in panel A. (C) The same experimental protocol was used for PDXs derived from T-ALL UPNT663, UPNT760, and UPNT420 using the clinically relevant anti-CD3 teplizumab. (D) Kaplan-Meier survival curves of NSG mice transplanted with T-ALL UPNT-760 and treated as in panel C. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
Discussion
To improve the proapoptotic and antileukemic activity of anti-CD3 mAbs in T-ALL, we identified the molecular programs induced in vivo by these antibodies, the aim being to unravel pharmacologically actionable targets able to further enhance leukemic cells’ vulnerability to anti-CD3 immunotherapy. RNAseq and gene chip array expression profiling in patient-derived xenografts generated from 2 distinct T-ALL cases extended our previous in vitro observation that both OKT3 and teplizumab induce the expression of actors of physiological clonal deletion, central to negative selection of developing thymocytes, notably NR4A1/Nur776,11,12 and BCL2L11/BIM.6,13,14 BCL2L11/BIM repression is a known survival signal in T-ALL15 whereas constitutive expression of NR4A1 is antagonistic to patient-derived T-ALL xenograft expansion in vivo (B. Zaniboni, C. Tran Quang, and J. Ghysdael, unpublished data, June 2017).
Interestingly, transcriptomic profiling analyses also show prominent activation in anti-CD3–treated T-ALL of expression of TNF⍺, LT⍺, and multiple components of the “TNF⍺ via NF-κB signaling” pathway.
Binding of TNF⍺/LT⍺ to TNFR1 triggers the formation of a signaling complex, which recruits, among others, RIPK1, TRAF2, and cIAP1 and cIAP2, leading to cIAP-dependent RIPK1 ubiquitination. Ubiquitinated RIPK1 serves as a platform for the ultimate recruitment of the scaffold protein NEMO and the IκB kinase, leading to NF-κB activation.16,17 However, in the absence of cIAP proteins, RIPK1 cannot be ubiquitinated, becomes active as a protein kinase, and allows the formation of RIPK1/Fas-associated death domain protein (FADD)/caspase 8 and RIPK1/RIPK3 cytoplasmic complexes that result in cell death.10,16,17 Therefore, IAP antagonist binding to cIAP1/2 and their subsequent proteolytic degradation both prevents cIAP-mediated ubiquitination leading to NF-κB activation, and acts like a switch to transform prosurvival TNF signaling into RIPK1-dependent cell death (as previously reviewed17).
In patient-derived T-ALL xenografts, we found the net activity of TNF⍺/LT⍺ induction in response to anti-CD3 is to interfere with anti-CD3–induced cell death, associated with upregulation of NF-κB target genes. Indeed, neutralization of TNF⍺/LT⍺ by etanercept synergized with anti-CD3 mAb to further impair leukemia progression and extend survival of mice with patient-derived T-ALL xenografts. Activation of NF-κB has previously been reported to support leukemia progression in mouse models of T-ALL18 and acute myeloid leukemia.19 Of note, increased TNF⍺ serum levels appear to correlate with decreased patient survival in acute leukemias.20
A number of small molecules acting as mimics of the mitochondrial cell death effector SMAC/Diablo (SMAC mimetics) have been developed that bind to the E3-ubiquitin ligases cIAP1 and cIAP2, activate their ubiquitin ligase activity, resulting in their proteolytic degradation. Unlike its cell killing activity as a single agent in a subset of B-cell ALL cases21-23 and in solid tumors,9 we found that the SMAC mimetic birinapant administered alone has no or only mild antileukemic activity in 7 of 7 patient-derived T-ALL xenografts tested, suggesting that cIAP1/2 have a limited role in cell death regulation in T-ALL. This is in line with the fact that cIAP1/2, in contrast to the X-linked inhibitor of apoptosis protein, are poor direct inhibitors of caspases24 but are critical to TNF/TNFR extrinsic cell death signaling.8,16 This contrasts with the high dependance of T-ALL on central actors of the mitochondrial/intrinsic cell death pathway linked to high levels of BCL2 (immature T-ALL) or Bcl-X (mature T-ALL) expression/activity and the antileukemic effects provided by BH3 mimetics targeting these antiapoptotic effectors in vitro and in patient-derived T-ALL xenografts.25,26 Accordingly, available clinical data indicate promising activity of BH3 mimetics combined with chemotherapy in the treatment of resistant/relapse T-ALL cases (as reviewed in27).
Interestingly, although weakly antileukemic on its own, we found that administration of birinapant strongly synergizes with anti-CD3 treatment, inducing rapid leukemic cell death in patient-derived T-ALL xenografts in vivo and T-ALL cell lines in vitro through a mechanism associated with, and reliant upon, the activation of RIPK1 protein kinase activity. This synergy resulted in increased antileukemic potential and in near doubling in host survival time in the anti-CD3 + birinapant arm as compared with anti-CD3 treatment alone in the 5 of 5 patient-derived T-ALL xenografts investigated. The antileukemic synergy of anti-CD3 mAbs and birinapant was observed with both anti-CD3 OKT3 and teplizumab.
Unlike OKT3 (muromonab), the first mouse mAb generated to control acute transplant rejection, which is associated with sometimes severe side effects linked to its immunogenicity and mitogenic potential, teplizumab is a IgG1-humanized version of OKT3 that carries 2 mutations (L234A;L235A) in its Fc domain and is thereby inefficient for binding FcR+ myeloid cells to induce complement-dependent cytotoxicity/antibody dependent cellular cytotoxicity and is poorly mitogenic for normal T cells (reviewed elsewhere28). Clinical trials using teplizumab in various autoimmune conditions show efficacy and low level adverse events29,30 and has been recently approved by the US Food and Drug Administration to delay stage III type 1 diabetes in adults and pediatric patients. A number of SMAC mimetics have been developed that promote cell death, notably of cancer cells. SMAC mimetics are presently evaluated in phase 1/2 clinical trials, mostly with patients with solid tumor cancer. These compounds show efficient on-target activity, are well tolerated, but appear to display limited antitumor activity as a single agent except in a subset of B-cell ALL cases.21-23 However, anticancer activity of SMAC mimetics can be improved when combined with several chemotherapies, in particular those leading to TNF⍺ production by tumor cells (reviewed elsewhere31,32).
Our observations in patient-derived T-ALL xenografts that birinapant-induced RIPK1 activation strongly enhances the antileukemic potential of anti-CD3 mAbs advocates the use of birinapant, or other SMAC mimetics, to improve anti-CD3 immunotherapy, notably as a salvage therapy and bridge to allogeneic hematopoietic stem cell transplantation, the only potentially curative option in refractory and relapsed T-ALL, to date.
Acknowledgments
The authors thank C. Lasgi (Institut Curie cytometry platform), E. Belloir, C. Alberti, and P. Dubreuil (Institut Curie animal facility) for expert technical assistance; V. Skory and C. Thibaud from the Plateforme Biopuces et Séquençage de l’IGBMC, Strasbourg, France; P. de la Grange from Genosplice for Affymetrix micro-array performance and help in analyses; D. Gentien, A. Rapinat, and E. Henry (Institut Curie Sequencing platform) for performing RNAseq and nanostring analyses; A. Hurgon (IC Central Pharmacy) for etanercept, and T. A. Baldwin and K. A. Hogquist for sharing unpublished data on the transcriptome associated with the negative selection signature of thymocytes.5
This work was supported by funds from Institut Curie, Centre National de la Recherche Scientifique, INSERM, Institut National du Cancer (INCa grants 2018 PRTK18-071 and PLBIO-04-ICR1), Ligue contre le Cancer (Programme de Recherche Enfants, Adolescents et Cancer et Ligue de l’Essonne), and the European Union (EU) ITN2020 ARCH consortium (grant agreement 813091). A.Á.Á. was supported by predoctoral fellowship from EU ITN2020-ARCH and la Fondation ARC pour la Recherche sur le Cancer; B.Z. was supported by predoctoral fellowships from the University Paris-Diderot and Ligue Nationale Contre le Cancer.
Authorship
Contribution: A.Á.Á., and C.T.Q. designed the study, carried out the experiments, performed data analysis and interpretation, organized the data, and wrote the manuscript; K.N., M.L.O., B.Z., and S.D. participated to the experimental work and data analysis; E.L. and V.A. provided primary patient material and associated immunophenotypes and molecular data, and J.G. designed the study, carried out data analysis and interpretation, organized the data, and wrote the manuscript.
Conflict-of-interest disclosure: C.T.Q. and J.G. have research agreements with Servier and Autolus Ltd. The remaining authors declare no competing financial interests.
Correspondence: Christine Tran Quang, Centre National de la Recherche Scientifique UMR3348, Institut Curie, Centre Universitaire, Bat 110, 91405 Orsay, France; email: christine.tran-quang@curie.fr; and Jacques Ghysdael, Centre National de la Recherche Scientifique UMR3348, Institut Curie, Centre Universitaire, Bat 110, 91405 Orsay, France; email: jacques.ghydael@curie.fr.
References
Author notes
J.G. and C.T.Q. are joint last authors.
All microarray data have been submitted to the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE240261. RNA sequencing (RNAseq) data are available under Gene Expression Omnibus series accession number GSE240296.
Data are available on request from the corresponding authors, Christine Tran Quang (christine.tran-quang@curie.fr) and Jacques Ghysdael (jacques.ghydael@curie.fr)
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal