In this issue of Blood, Durand et al describe a functional score that identifies cells with loss of TP53 and demonstrate a role for TP53 function in the response to BH3 mimetics in multiple myeloma (MM).1
MM is a disease that is defined in part by structural variations that occur during the germinal center reaction, consistent with dysfunctional class switch recombination. Approximately 50% of patients with MM harbor translocations that result in the juxtaposition of oncogenes to the strong enhancers of the IGH locus.2 However, these truncal events are soon followed by additional alterations that help drive disease progression and can influence disease outcome. These secondary events include copy number variations, non-IGH translocations, and single-nucleotide variations (SNVs). Independently, few of these have a significant impact on outcome, with the notable exceptions of gains/amplifications of chromosome 1q (1q+ or amp) and loss of chromosome 17p (del17p).3 The latter is where the tumor suppressor TP53 is found and is the only gene where SNVs have been shown to be a predictor of outcome,4 likely due to the fact that most TP53 SNVs occur in patients who harbor del17p and reflect biallelic loss of function.5,6 Therefore, identifying patients with biallelic loss of TP53 to be able to consider alternative treatment options, given their poor response to standard therapies, is necessary.
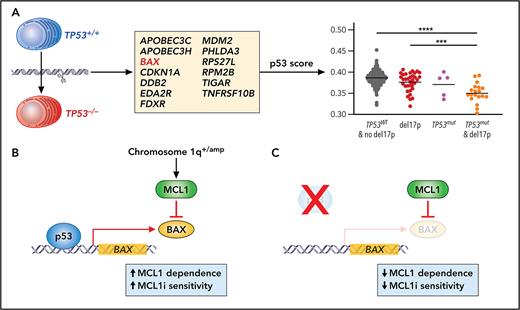
To address these issues, Durand et al aimed to identify a gene expression signature that could be used to functionally identify biallelic loss of TP53. Previous work on RAS/RAF mutations in MM has demonstrated that a functional output, in that case gene expression associated with mitogen-activated protein kinase pathway activation, can be more accurate in predicting outcome than just determining if there are mutations in NRAS, KRAS, or BRAF.7 For the current studies, a TP53 gene signature was derived by comparing expression of cells where TP53 expression was inhibited by gene editing in 2 human myeloma cell lines with wild-type TP53. The authors found downregulation of 16 genes (including TP53). To derive a functional readout of p53 activity, they focused on 13 genes that have been previously shown to be early targets in the pathway (see figure panel A). More important, when similar editing was performed in cells that lacked a functional p53 due to mutations and deletions, no change in the expression of these genes was observed. In addition, MDM2 inhibition with nutlin3a only increased the p53 score in cell lines that had wild-type TP53, and a low score also accurately predicted biallelic loss of TP53 in cell lines from other tumor types.
(A) Using gene editing and transcriptional profiling in human MM cell lines, Durand et al defined a 13 gene signature to generate a measure of p53 activity. This p53 score could discriminate patient samples that had biallelic loss of TP53. (B-C) The authors then focused on the function of 1 gene in the p53 score, BAX, and demonstrated that BAX expression resulted in increased dependence of cells on MCL1 and MCL1 inhibitor (MCL1i) sensitivity. Loss of TP53 and subsequent lower BAX expression render cells less dependent on MCL1; therefore, they are less sensitive to MCL1i. Professional illustration by Patrick Lane, ScEYEnce Studios.
(A) Using gene editing and transcriptional profiling in human MM cell lines, Durand et al defined a 13 gene signature to generate a measure of p53 activity. This p53 score could discriminate patient samples that had biallelic loss of TP53. (B-C) The authors then focused on the function of 1 gene in the p53 score, BAX, and demonstrated that BAX expression resulted in increased dependence of cells on MCL1 and MCL1 inhibitor (MCL1i) sensitivity. Loss of TP53 and subsequent lower BAX expression render cells less dependent on MCL1; therefore, they are less sensitive to MCL1i. Professional illustration by Patrick Lane, ScEYEnce Studios.
Once validated in cell lines, the authors then determined if their p53 score had prognostic value for determining outcomes in patients with MM. They applied their score to 2 clinical studies with newly diagnosed patients (CASSIOPEA and MMRF-CoMMpass) and in both cases found that the cohort of patients with the lowest p53 score were more likely to have biallelic loss of TP53 and experienced significantly worse outcomes. More important, in both studies, a significant proportion of patients with del17p and no second hit were in all groups defined by the p53 score, despite likely being defined as high risk using current guidelines.3 In addition, there was also a small proportion of patients with both TP53 SNVs and del17p who were classified in the higher p53 score cohorts, consistent with findings demonstrating that not all TP53 SNVs result in complete loss of p53 function.8
The authors then turn their attention to the potential role of the genes in the p53 score in disease biology and how this may be targeted in MM. They focused on BAX because of its central role in apoptosis and potential impact in the response to a wide variety of therapies. To determine the consequence of decreased BAX expression in cells that lack TP53, they used specific inhibitors of anti-apoptotic BCL2 proteins (aka, BH3 mimetics) as well as BH3 profiling and found that loss of TP53 selectively effects MCL1 priming, resulting in decreased sensitivity to the MCL1 inhibitor S63845 (see figure panels B-C). At first glance this is somewhat surprising as BAX can also bind to BCL2 and BCL-XL and could be activated by BH3-only proteins, such as BIM, that could be released from any of these prosurvival proteins. However, all the cell lines used in this study have t(4;14) and are already resistant to venetoclax due to a lack of BCL2 priming; therefore, it is impossible to change their resistance status. In addition, t(4;14) MM is associated with 1q+ and 1q amp and MCL1 is found at 1q21.4,9 Although correlative data presented here in a larger set of cell lines support the hypothesis that loss of p53 function is more likely to influence MCL1 dependence, future testing in cells with other backgrounds, such as t(14;16) and t(11;14), is warranted to determine the effects of p53 and BAX in BCL2- or BCL-XL–dependent cells.
Finally, taking into consideration the heterogeneity of MM cells in patients,10 the authors investigated if the p53 score was altered by BH3 mimetic treatment. Using single-cell RNA sequencing on 24 samples, they were able to determine that following treatment, the p53 score was lower in approximately one-third of the clusters, and this was more common in clusters that did not have del17p. This suggests that treatment with BH3 mimetics could select for loss of p53 function, having implications for how and when to use them in the MM therapy.
There are pros and cons to both the genomic and functional approaches for determining the consequences of TP53 loss. The functional approach can identify samples that are functionally equivalent to biallelic loss of TP53 without having the genetic lesions and determine if the mutations actually result in a functional loss. However, unlike the genomic approach, detection of a small subclonal fraction of cells is difficult unless one incorporates single-cell analysis, which may not be practical for routine use. Thus, the p53 score will likely be best used in concert with current approaches to assess the prognostic impact and aid most accurately in treatment decisions.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal