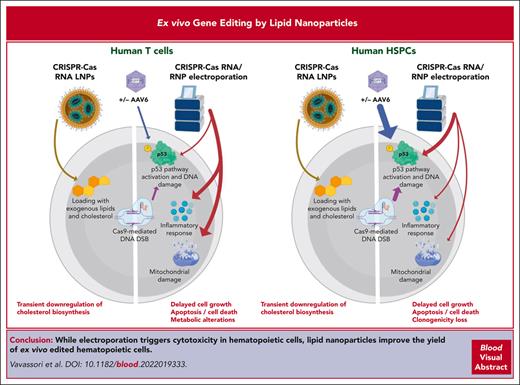
Key Points
Electroporation triggers significant cytotoxicity in gene-edited T cells and hematopoietic stem cells.
LNPs as a delivery method for editing reagents dampen cytotoxicity, increasing the yield of ex vivo–edited hematopoietic cells.
Abstract
Ex vivo gene editing in T cells and hematopoietic stem/progenitor cells (HSPCs) holds promise for treating diseases. Gene editing encompasses the delivery of a programmable editor RNA or ribonucleoprotein, often achieved ex vivo via electroporation, and when aiming for homology-driven correction of a DNA template, often provided by viral vectors together with a nuclease editor. Although HSPCs activate a robust p53-dependent DNA damage response upon nuclease-based editing, the responses triggered in T cells remain poorly characterized. Here, we performed comprehensive multiomics analyses and found that electroporation is the main culprit of cytotoxicity in T cells, causing death and cell cycle delay, perturbing metabolism, and inducing an inflammatory response. Nuclease RNA delivery using lipid nanoparticles (LNPs) nearly abolished cell death and ameliorated cell growth, improving tolerance to the procedure and yielding a higher number of edited cells compared with using electroporation. Transient transcriptomic changes upon LNP treatment were mostly caused by cellular loading with exogenous cholesterol, whose potentially detrimental impact could be overcome by limiting exposure. Notably, LNP-based HSPC editing dampened p53 pathway induction and supported higher clonogenic activity and similar or higher reconstitution by long-term repopulating HSPCs compared with electroporation, reaching comparable editing efficiencies. Overall, LNPs may allow efficient and harmless ex vivo gene editing in hematopoietic cells for the treatment of human diseases.
Introduction
Ex vivo gene editing of human T cells and hematopoietic stem/progenitor cells (HSPCs) holds therapeutic potential for the treatment of several diseases.1 Among several editing strategies, nuclease-based approaches have been most investigated. They encompass the transient delivery of nuclease messenger RNA (mRNA) or ribonucleoprotein (RNP) via electroporation to introduce a DNA double-stranded break (DSB) at a target site. Cellular repair of the DSB installs genetic edits through nonhomologous/microhomology-mediated end joining (NHEJ) or homology-driven recombination (HDR) using a DNA template often codelivered by means of viral vectors, such as adeno-associated viral vectors (AAVs) or integrase-defective lentiviral vectors.2 Although NHEJ allows disrupting gene coding frames or regulatory elements,3-6 HDR enables in situ correction of mutant alleles7-13 or targeted integration of the transgene cassette in safe harbors such as AAVS1.14
We reported that HSPCs activate a robust and cumulative p53-dependent DNA damage response (DDR) upon sensing the nuclease-induced DSBs and the viral genome carrying the HDR template, promoting cell cycle arrest and decreasing hematopoietic reconstitution potential and graft clonality in mice that underwent xenotransplantation. Transient expression of a dominant-negative p53 mutant (GSE56) dampens DDR and partially rescues its adverse effect.15-17 Whether gene editing can have a similar impact on T cells remains unknown.
Despite electroporation being commonly used, its toxicity is poorly investigated. Electroporation-induced physical permeabilization of cellular membranes and mitochondrial and DNA damage may cause cytotoxicity18,19 and contribute to the adverse impact of gene editing. Alternative delivery strategies have emerged based on lipid nanoparticles (LNPs), and some of them have been clinically validated to safely and effectively deliver mRNA in vivo for the prevention and treatment of various diseases.20-23 However, these platforms have yet to be investigated for ex vivo delivery of editing components into hematopoietic cells.24
Here, we interrogated the response to gene editing of human T cells using comprehensive multiomic analysis and found that electroporation is the main culprit of cytotoxicity. We then explored commercially available RNA LNPs and developed protocols for editing T cells and HSPCs at an efficiency comparable with that of electroporation but with significantly lower toxicity.
Methods
Gene editing
AAV6 production and single guide RNA (sgRNA) sequences were previously reported.11,15,25 RNPs were assembled by incubating at a 1:2 or 1:1.5 molar ratio of SpCas9 protein (Aldevron) with sgRNA (Synthego). CRISPR/Cas9 RNA was prepared by mixing CleanCap Cas9 mRNA (5 moU, Trilink) with sgRNA at a 2.49:1 mass ratio.
For electroporation, 5 × 105/106 T cells or 2 × 105/7.5 × 105 cord blood (CB)/mobilized peripheral blood (mPB) HSPCs for each condition were treated after 3 days of stimulation, as previously reported11,16 and detailed in supplemental Methods, available on the Blood website. A total of 3 μg GSE56 mRNA15 was electroporated along with RNP, as indicated. LNPs were formulated with the GenVoy-ILM T Cell Kit (Precision Nanosystems) and quantified as detailed in supplemental Methods. Empty LNPs were prepared by substituting RNA with water. LNPs were stored at 4°C for <1 week. After 3 days of stimulation, 5 × 105 T cells or 3 × 105 CB/mPB HSPCs for each condition were seeded at a concentration of 5 × 105 cells per mL. Recombinant human ApoE (0.1 μg/mL) was added to the medium, and cells were resuspended and incubated with different doses of CRISPR/Cas9 RNA LNPs or the same volume of empty LNPs. AAV6 transduction was performed 0 or 2 hours before the addition of LNPs at a dose of 5 × 104/2 × 104 vg per cell, unless otherwise specified. After 24 hours, cells were washed with Dulbecco's phosphate-buffered saline (DPBS) and reseeded.
Molecular analyses
Deep sequencing, digital droplet polymerase chain reaction (ddPCR), and T7 assay for HDR/NHEJ assessment were performed on purified DNA as reported previously.11,16,17 For gene expression, RNA was extracted, and 2 ng of complementary DNA were used for CDKN1A (Hs00355782_m1, Life Technologies) and APOBEC3H (Hs00419665_m1) ddPCR as previously reported.16 Primers and probes are listed in supplemental Table 2.
RNA-seq was performed as previously described using DESeq2.16 Proteomic analysis was performed on samples from 3 healthy donors (HDs), as described in supplemental Methods.
Additional information is reported in supplemental Methods.
Results
Impact of electroporation on human CD4+ T cells
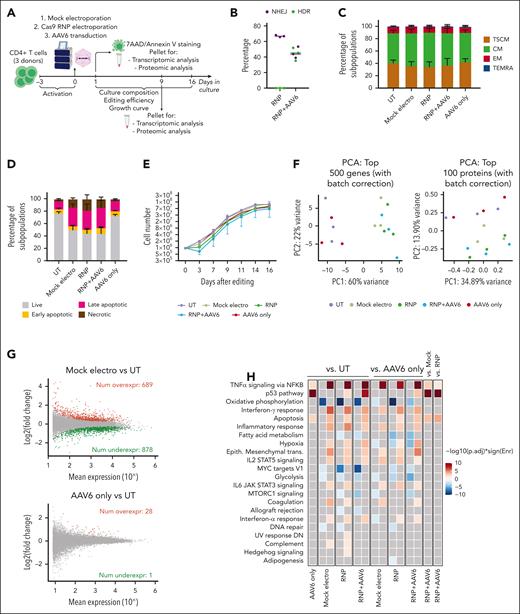
We performed a multiparametric analysis investigating the impact of each step of the gene editing procedure on human CD4+ T cells from 3 HDs, using previously optimized culture/editing protocols and reagents targeting CD40LG.11 We compared untreated cells (UTs), mock-electroporated cells (mock electro), cells electroporated with RNP without AAV (RNP-only) or with an AAV6 donor (RNP + AAV6), and AAV6-transduced cells (AAV6-only, no electroporation; Figure 1A). Editing efficiency was robust, reaching up to ∼75% NHEJ and ∼50% HDR, based on the treatment, preserving a substantial proportion of the clinically relevant T-stem memory and -central memory cells26 (Figure 1B-C). Strikingly, we found a major increase in apoptotic/necrotic cells after mock electroporation, reaching up to 50% of the cells, which slightly worsened only when introducing the editing components. Cell death in AAV6 only cells was nearly comparable with that in UTs (Figure 1D). Concordantly, electroporated cells showed an initial delay, up to halted cell growth upon addition of editing components, which recovered over time (Figure 1E). Unsupervised principal component analysis of short-term transcriptomics and proteomics separately clustered UTs and AAV6-only cells from all electroporated samples, regardless of other editing components (Figure 1F). We identified a large set of differentially expressed genes (DEGs) modulated by electroporation per se (Figure 1G, top), with the upregulation of genes belonging to apoptosis and inflammatory categories and downregulation of cell cycle and metabolism-related categories when comparing mock electro, RNP-only, or RNP + AAV6 samples with UTs (Figure 1H). Consistently, few changes were observed comparing RNP-only or RNP + AAV6 samples with mock electros, with positive enrichment of the p53 pathway, tumor necrosis factor α signaling, and apoptosis, suggesting cumulative induction by AAV6 (Figure 1H; supplemental Figure 3A). Accordingly, the same categories were positively enriched when comparing AAV6-only cells with UTs, although changes were limited to a few genes (Figure 1G, bottom). As a result, mock electro, RNP-only, and RNP + AAV6 samples showed similar patterns of p53/apoptotic/inflammatory gene upregulation, highlighting that the addition of editing components increased the response induced by electroporation cumulatively but limitedly, per se (Figure 1I, cluster 1). A more limited and different set of genes were upregulated via AAV6 treatment (Figure 1I, cluster 2), with 1 donor showing a markedly diverse pattern compared with the others. Cells from this donor also showed significant growth inhibition by AAV6 (supplemental Figure 3B), uncovering donor variability in AAV6 response. Transient inhibition of the p53 response by coelectroporating GSE56 mRNA15 did not improve cell viability and growth, despite maintaining similar editing efficiencies and phenotypes (supplemental Figure 3C-G).
Electroporation induces transient transcriptomic and proteomic changes that promote cell cycle arrest and trigger apoptosis in human CD4+ T cells. (A) Schematic representations of gene editing experimental procedure and multiparametric analysis performed in CD4+ T cells. (B) Percentage of HDR- and NHEJ-edited alleles after CD40LG editing with CRISPR/Cas9 with RNP only or RNP + AAV6 in CD4+ T cells from males (n = 3). Median. (C) Cell population composition of CD4+ T cells in the indicated conditions (n = 3), 16 days after treatment. Data are represented as mean ± standard error of the mean (SEM). CD4+ T-cell phenotypes were defined as follows: effector memory RA (TEMRA): CD45RA+CD62L–; effector memory (EM): CD45RA–CD62L-; central memory (CM): CD45RA–CD62L+; and T memory stem (TSCM): CD45RA+CD62L+. (D) Percentage of live, early/late apoptotic, and necrotic cells 24 hours after editing from panel C (n = 3). Data are represented as mean ± SEM. (E) Growth curve of CD4+ T cells from panel C (n = 3). Data are represented as median ± range. (F) Unsupervised principal component analyses (PCAs) of transcriptomic (left) and proteomic (right) data 12 hours after treatment of CD4+ T cells from panel C. (G) Plot of log intensity ratios vs the mean average signals (MA plot) showing significant DEGs in the mock electro vs UT (top) or AAV6 only vs UT (bottom) comparisons. (H) Heatmap showing enrichment results from all comparisons between conditions from panel C on the hallmark gene set (Molecular Signatures Database). Color intensity reflects statistical significance of the test (adjusted P values), whereas signs (positive or negative) correspond to the set of DEGs used in the statistical test (upregulated or downregulated, respectively). No significant enrichment was found when comparisons are missing. (I) Heatmaps showing normalized read counts for all genes belonging to the indicated categories in conditions from panel C. (J) Heatmap showing NES of proteomic gene set enrichment analysis on differentially expressed proteins (DEPs) pre-ranked based on the ratio of all comparisons between conditions from panel C vs the hallmark gene sets. Missing comparisons indicate that no significant differences were captured in the analysis. (K) Plots showing changes of expression for genes or proteins belonging to the core enrichment of oxidative phosphorylation (left) or fatty acid metabolism (right) categories from the RNP + AAV6 vs UT comparison. (L) Unsupervised PCA of transcriptomic (left) and proteomic (right) data 9 days after treatment of CD4+ T cells from panel C. AAV6 only, AAV6 transduced-only cells, without electroporation.
Electroporation induces transient transcriptomic and proteomic changes that promote cell cycle arrest and trigger apoptosis in human CD4+ T cells. (A) Schematic representations of gene editing experimental procedure and multiparametric analysis performed in CD4+ T cells. (B) Percentage of HDR- and NHEJ-edited alleles after CD40LG editing with CRISPR/Cas9 with RNP only or RNP + AAV6 in CD4+ T cells from males (n = 3). Median. (C) Cell population composition of CD4+ T cells in the indicated conditions (n = 3), 16 days after treatment. Data are represented as mean ± standard error of the mean (SEM). CD4+ T-cell phenotypes were defined as follows: effector memory RA (TEMRA): CD45RA+CD62L–; effector memory (EM): CD45RA–CD62L-; central memory (CM): CD45RA–CD62L+; and T memory stem (TSCM): CD45RA+CD62L+. (D) Percentage of live, early/late apoptotic, and necrotic cells 24 hours after editing from panel C (n = 3). Data are represented as mean ± SEM. (E) Growth curve of CD4+ T cells from panel C (n = 3). Data are represented as median ± range. (F) Unsupervised principal component analyses (PCAs) of transcriptomic (left) and proteomic (right) data 12 hours after treatment of CD4+ T cells from panel C. (G) Plot of log intensity ratios vs the mean average signals (MA plot) showing significant DEGs in the mock electro vs UT (top) or AAV6 only vs UT (bottom) comparisons. (H) Heatmap showing enrichment results from all comparisons between conditions from panel C on the hallmark gene set (Molecular Signatures Database). Color intensity reflects statistical significance of the test (adjusted P values), whereas signs (positive or negative) correspond to the set of DEGs used in the statistical test (upregulated or downregulated, respectively). No significant enrichment was found when comparisons are missing. (I) Heatmaps showing normalized read counts for all genes belonging to the indicated categories in conditions from panel C. (J) Heatmap showing NES of proteomic gene set enrichment analysis on differentially expressed proteins (DEPs) pre-ranked based on the ratio of all comparisons between conditions from panel C vs the hallmark gene sets. Missing comparisons indicate that no significant differences were captured in the analysis. (K) Plots showing changes of expression for genes or proteins belonging to the core enrichment of oxidative phosphorylation (left) or fatty acid metabolism (right) categories from the RNP + AAV6 vs UT comparison. (L) Unsupervised PCA of transcriptomic (left) and proteomic (right) data 9 days after treatment of CD4+ T cells from panel C. AAV6 only, AAV6 transduced-only cells, without electroporation.
Overrepresentation analysis (ORA) of proteomic data helped confirm perturbation of some pathways related to cell cycle and energy metabolism upon electroporation, regardless of the addition of editing components. Interestingly, the change in proteins belonging to the core enrichment of metabolic categories was often in the opposite direction compared with transcriptomic analysis performed at the same time, possibly reflecting temporal dissociation of response during state transition27 (Figure 1J-K; supplemental Figure 3H).
Importantly, unsupervised principal component analysis of transcriptomic and proteomic data showed no more subclustering of different treatments upon long-term culture (Figure 1L), indicating that edited cells surviving the procedure recovered the gene expression profile and proliferation of UTs (Figure 1E).
LNP-mediated CRISPR/Cas9 RNA delivery improves T-cell tolerance to gene editing
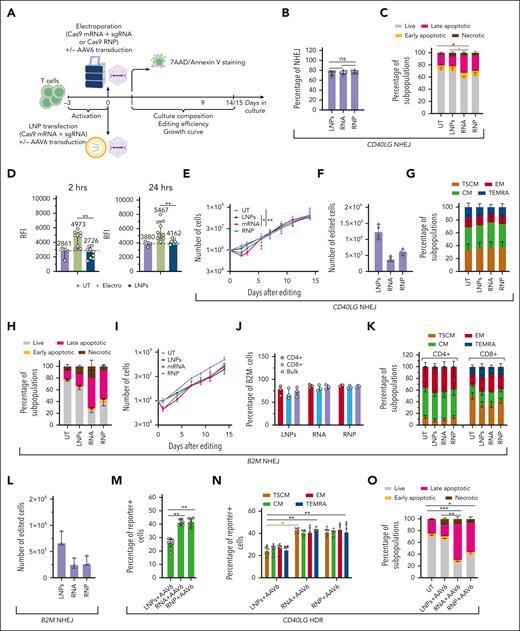
We then explored LNP-based delivery of the Cas9 mRNA and sgRNA targeting CD40LG using a commercial formulation for ex vivo T-cell transfection, showing a median size of ∼130 nm, a polydispersity index ≤ 0.3,28 and excellent transfection efficiency (supplemental Figure 4A-C). The presence of serum in the culture media nearly abolished editing efficiency (supplemental Figure 4D), likely because of low density lipoprotein (LDL)-induced downregulation and competition for binding to its receptor (LDLR)29 (supplemental Figure 4E). LDLR expression and editing efficiency were similar using different T-cell activation reagents (supplemental Figure 4D-E). We, then, optimized a good manufacturing practice–compliant serum-free culture protocol, yielding robust cell growth and increased LDLR expression (supplemental Figure 4E-F). LNP transfection was less efficient per RNA mass at lower doses but reached similar NHEJ efficiency at the highest doses, compared with electroporation of RNA/RNP (supplemental Figure 4G). Strikingly, LNP transfection did not increase the percentage of apoptotic/necrotic cells early after treatment, when compared with UTs (supplemental Figure 4H). However, we observed increasing growth arrest and cell death over time in cells treated with LNPs (supplemental Figure 4I). Washing out LNPs 24 hours after incubation prevented toxicity without affecting NHEJ efficiency (supplemental Figure 4G-I). Using this optimized protocol (Figure 2A), we performed a multiparametric side-by-side comparison of LNP- and RNA/RNP electroporation–mediated editing in CD4+ T cells from 6 HDs. Robust NHEJ editing (≤80%) was achieved with all treatments (Figure 2B), with a comparable distribution of indel sizes across the different delivery modalities at matched editing efficiencies (supplemental Figure 4J). The increased apoptosis/necrosis in electroporated samples was nearly absent in LNP-treated ones (Figure 2C). Measurement of the mitochondrial potential showed depolarization immediately after electroporation (supplemental Figure 4K), followed by sustained hyperpolarization (Figure 2D; supplemental Figure 4L), the earliest change associated with apoptosis.30 Conversely, LNP-treated cells did not show altered mitochondrial potential (Figure 2D; supplemental Figure 4L) but showed minimally decreased growth, even shortly after editing (Figure 2E). Consequently, the early yield of NHEJ-edited cells doubled with LNP treatment (Figure 2F), although electroporated samples recovered in the long term with faster kinetics than in earlier studies because of improved culture conditions (supplemental Figure 4F). None of the treatments induced significant phenotypic cell skewing (Figure 2G).
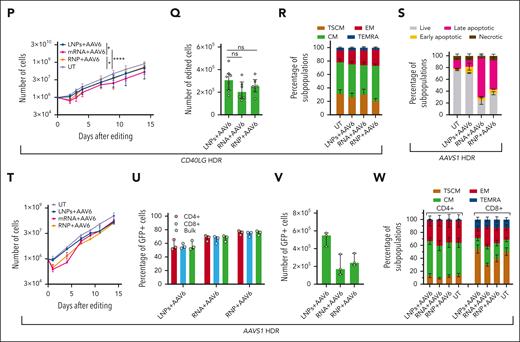
CRISPR/Cas9 RNA delivery using LNP improves T-cell tolerance to gene editing. (A) Schematic representations of gene editing experimental procedure and related analyses after CRISPR/Cas9 RNA delivery using LNPs or CRISPR/Cas9 RNP/RNA delivery via electroporation in T cells. (B) Percentage of NHEJ-edited CD40LG alleles after editing CD4+ T cells from males using 1.25 μg of LNPs, 25 pmol of RNP, or 1.25 μg of RNA (n = 6). Friedman test with Dunn multiple comparisons. Median ± interquartile range (IQR). (C) Percentage of live, early/late apoptotic, and necrotic cells 24 hours after treatments in B and in UT cells (n = 6). Friedman test with Dunn multiple comparisons performed at live cells. Mean ± SEM. (D) Relative fluorescence intensity (RFI) of Mitotracker Red (mitochondrial membrane potential) normalized on Mitotracker Green (mitochondrial mass) measured via flow cytometry 2 hours (left) or 24 hours (right) after indicated treatments from supplemental Figure 4L (n=3, 9, and 9). LNPs and electroporated conditions were pooled for this analysis; Wilcoxon test; median ± IQR. (E) Growth curve from panel C (n = 6). Friedman test with Dunn multiple comparisons performed on day 3; median ± IQR. (F) Number of edited cells 24 hours after editing in 4 experiments from panel C (n = 4); median ± IQR. (G) Cell population composition from panel C (n = 6), 14 days after treatment. Mean ± SEM. (H) Percentage of live, early/late apoptotic, and necrotic CD3+ T cells 24 hours after B2M editing using 1.5 μg of LNPs, 50 pmol of RNP, or 1.5 μg of RNA (n = 3). UTs were used as the control; mean ± SEM. (I) Growth curve of CD3+ T cells from panel H (n = 3); median ± range. (J) Percentage of B2M– cells (biallelic KO) measured via flow cytometry within CD4+, CD8+, and total CD3+ T-cell populations from panel H (n = 3); median ± range. (K) Cell population composition from panel H (n = 3), 14 days after treatment; mean ± SEM. (L) Number of B2M– cells from panel H 24 hours after editing (n = 3); median ± range. (M) Percentage of reporter-positive CD4+ T cells from male donors after CD40LG editing using 1.25 μg of LNPs, 25 pmol of RNP, or 1.25 μg of RNA, and the cognate HDR template provided by AAV6 (n = 8, 6, and 6). Kruskal-Wallis test followed by post hoc analysis with Dunn test; median ± IQR. (N) Percentage of reporter-positive cells within CD4+ T cells subpopulations from panel M (n=8, 6, and 6). Kruskal-Wallis test followed by post hoc analysis with Dunn test; median ± IQR. (O) Percentage of live, early/late apoptotic, and necrotic CD4+ T cells 24 hours after editing in experiments from panel M. UTs were used as controls (n = 8, 8, 6, and 6). Kruskal-Wallis test followed by post hoc analysis with Dunn test on live cells; mean ± SEM. (P) Growth curve of CD4+ T cells from panel O. Kruskal-Wallis test followed by post hoc analysis with Dunn test performed on day 3; median ± IQR. (Q) Number of reporter-positive cells from panel O, 24 hours after editing. Kruskal-Wallis test followed by post hoc analysis with Dunn test; median ± IQR. (R) Cell population composition from panel O (n = 3), 14 days after treatment; mean ± SEM. (S) Percentage of live, early/late apoptotic, and necrotic CD3+ T cells 24 hours after AAVS1 editing using 1.25 μg of LNPs, 25 pmol of RNP, or 1.25 μg RNA, and the cognate HDR template provided by AAV6 (n = 3). UTs were used as controls. Mean ± SEM. (T) Growth curve of CD3+ T cells from panel S (n = 3); median ± range. (U) Percentage of GFP+ CD3+ T cells within CD4+, CD8+, and total populations from panel S (n = 3). Median ± range. (V) Number of GFP+ cells from panel R 24 hours after editing. Median ± IQR. (W) Cell population composition from panel S (n = 3), 14 days after treatment. Mean ± SEM. ns, not significant.
CRISPR/Cas9 RNA delivery using LNP improves T-cell tolerance to gene editing. (A) Schematic representations of gene editing experimental procedure and related analyses after CRISPR/Cas9 RNA delivery using LNPs or CRISPR/Cas9 RNP/RNA delivery via electroporation in T cells. (B) Percentage of NHEJ-edited CD40LG alleles after editing CD4+ T cells from males using 1.25 μg of LNPs, 25 pmol of RNP, or 1.25 μg of RNA (n = 6). Friedman test with Dunn multiple comparisons. Median ± interquartile range (IQR). (C) Percentage of live, early/late apoptotic, and necrotic cells 24 hours after treatments in B and in UT cells (n = 6). Friedman test with Dunn multiple comparisons performed at live cells. Mean ± SEM. (D) Relative fluorescence intensity (RFI) of Mitotracker Red (mitochondrial membrane potential) normalized on Mitotracker Green (mitochondrial mass) measured via flow cytometry 2 hours (left) or 24 hours (right) after indicated treatments from supplemental Figure 4L (n=3, 9, and 9). LNPs and electroporated conditions were pooled for this analysis; Wilcoxon test; median ± IQR. (E) Growth curve from panel C (n = 6). Friedman test with Dunn multiple comparisons performed on day 3; median ± IQR. (F) Number of edited cells 24 hours after editing in 4 experiments from panel C (n = 4); median ± IQR. (G) Cell population composition from panel C (n = 6), 14 days after treatment. Mean ± SEM. (H) Percentage of live, early/late apoptotic, and necrotic CD3+ T cells 24 hours after B2M editing using 1.5 μg of LNPs, 50 pmol of RNP, or 1.5 μg of RNA (n = 3). UTs were used as the control; mean ± SEM. (I) Growth curve of CD3+ T cells from panel H (n = 3); median ± range. (J) Percentage of B2M– cells (biallelic KO) measured via flow cytometry within CD4+, CD8+, and total CD3+ T-cell populations from panel H (n = 3); median ± range. (K) Cell population composition from panel H (n = 3), 14 days after treatment; mean ± SEM. (L) Number of B2M– cells from panel H 24 hours after editing (n = 3); median ± range. (M) Percentage of reporter-positive CD4+ T cells from male donors after CD40LG editing using 1.25 μg of LNPs, 25 pmol of RNP, or 1.25 μg of RNA, and the cognate HDR template provided by AAV6 (n = 8, 6, and 6). Kruskal-Wallis test followed by post hoc analysis with Dunn test; median ± IQR. (N) Percentage of reporter-positive cells within CD4+ T cells subpopulations from panel M (n=8, 6, and 6). Kruskal-Wallis test followed by post hoc analysis with Dunn test; median ± IQR. (O) Percentage of live, early/late apoptotic, and necrotic CD4+ T cells 24 hours after editing in experiments from panel M. UTs were used as controls (n = 8, 8, 6, and 6). Kruskal-Wallis test followed by post hoc analysis with Dunn test on live cells; mean ± SEM. (P) Growth curve of CD4+ T cells from panel O. Kruskal-Wallis test followed by post hoc analysis with Dunn test performed on day 3; median ± IQR. (Q) Number of reporter-positive cells from panel O, 24 hours after editing. Kruskal-Wallis test followed by post hoc analysis with Dunn test; median ± IQR. (R) Cell population composition from panel O (n = 3), 14 days after treatment; mean ± SEM. (S) Percentage of live, early/late apoptotic, and necrotic CD3+ T cells 24 hours after AAVS1 editing using 1.25 μg of LNPs, 25 pmol of RNP, or 1.25 μg RNA, and the cognate HDR template provided by AAV6 (n = 3). UTs were used as controls. Mean ± SEM. (T) Growth curve of CD3+ T cells from panel S (n = 3); median ± range. (U) Percentage of GFP+ CD3+ T cells within CD4+, CD8+, and total populations from panel S (n = 3). Median ± range. (V) Number of GFP+ cells from panel R 24 hours after editing. Median ± IQR. (W) Cell population composition from panel S (n = 3), 14 days after treatment. Mean ± SEM. ns, not significant.
LNP transfection did not affect viability and only minimally decreased growth of CD3+ T cells edited at another locus (B2M exon 2), as opposed to electroporation, while preserving T-stem memory and -central memory cells and promoting efficient editing (75% biallelic knockout (KO), measured via flow cytometry) without fostering longer deletions (Figure 2H-K; supplemental Figure 4M). Because of the lower impact on cell growth, the early yield of edited cells was more than doubled compared with that with electroporation (Figure 2L).
Next, we investigated whether LNP delivery could be applied for HDR-based editing, targeting CD40LG with a therapeutic cassette containing a reporter.11 LNPs were less efficient than electroporation in promoting HDR at all doses tested (supplemental Figure 4N), likely because of competition for entering cells or interference with leaving endosomes. None of the tested permutations of LNP/AAV6 delivery improved HDR (supplemental Figure 4O-P). We performed a multiparametric comparison of LNP-based and RNA-RNP electroporation in CD4+ T cells from 8 HDs (Figure 2A) for CD40LG editing. Although the HDR efficiency was again one-third lower for LNP-treated cells (Figure 2M-N), there was a nearly absent increase in apoptotic/necrotic cells, mirrored by normal mitochondrial potential, as opposed to electroporated cells (Figure 2D,O; supplemental Figure 4L). Consistently, LNP-based editing only minimally delayed growth, even shortly after editing (Figure 2P). As a result, the yield of HDR-edited cells early after editing was similar or even increased for LNP-treated samples (Figure 2Q). None of the treatments induced significant phenotypic cell skewing (Figure 2R). Similar results were obtained when targeting a green fluorescent protein (GFP) expression cassette into AAVS1 using HDR15 in CD3+ cells (Figure 2S-W), confirming the advantages of LNPs.
LNP-mediated cholesterol overloading triggers an adverse response overcome after particle withdrawal
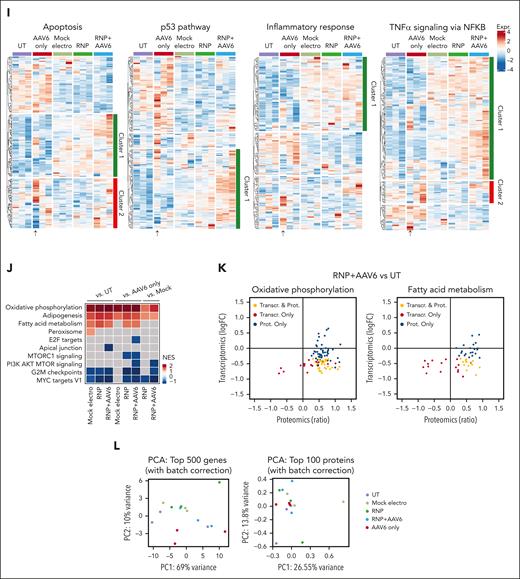
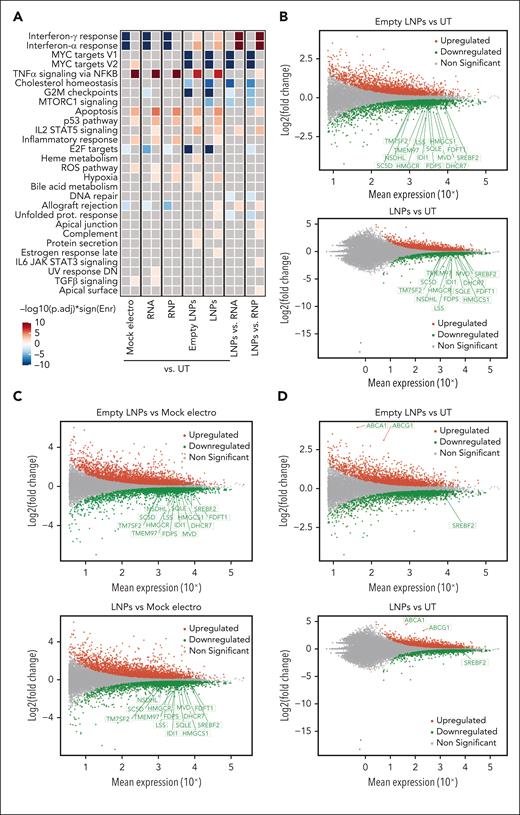
To characterize the impact of LNPs in T cells, we performed transcriptomic analysis 24 hours after CD40LG editing in CD4+ T cells. As controls, we used UTs, mock electros, or cells treated with LNPs formulated without RNA (empty LNPs; supplemental Figures 4A and 5A-E). As we improved culture conditions with respect to the previous analyses, we first focused on the mock electro–UT comparison. Again, ORA highlighted a significant upregulation of genes belonging to apoptosis, the p53 pathway, and inflammatory categories and downregulation of cell cycle target genes (Figure 3A). Interferon (IFN) responses were negatively enriched in electroporated cells, as opposed to previous findings. We reasoned that more robust activation by the new culture conditions may lead to robust IFN secretion.31 Consistent with the faster recovery of cell growth, downregulation of metabolism-related genes was no longer observed in electroporated samples, confirming the improvement in culture conditions.
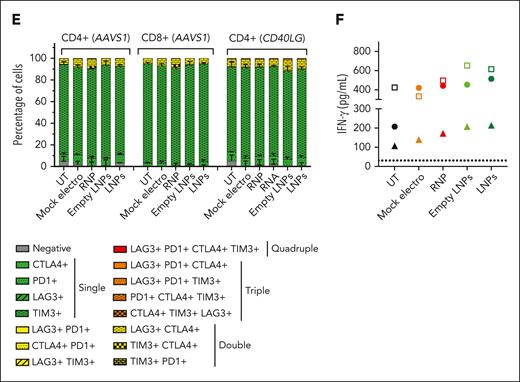
LNPs induce pervasive transcriptomic changes without affecting T-cell functionality. (A) Heatmap showing enrichment results from all comparisons between conditions between conditions from supplemental Figure 5A on the hallmark gene set (Molecular Signatures Database). Color intensity reflects statistical significance of the test (adjusted P values), whereas signs (positive or negative) correspond to the set of DEGs used in the statistical test (upregulated or downregulated, respectively). (B-D) MA plot showing significant DEGs in (B) empty LNPs vs UTs (top) or LNPs vs UTs (bottom); (C) empty LNPs vs mock electros (top) or LNPs vs mock electros (bottom); (D) empty LNPs vs UTs (top) or LNPs vs UTs (bottom) comparisons. Labels highlight DEGs from panels B-C belonging to the core enrichment of cholesterol homeostasis category, and (D) SREBF2, ABCA1, and ABCG1. (E) Percentage of T cells negative and single-, double-, triple-, and quadruple-positive for exhaustion markers 14 days after treatments (n = 3). Mean ± SEM. (F) IFN-γ secretion 3 days after anti-CD3 polyclonal stimulation, ie, 14 days after treatments (n = 3). Each symbol represents a different T-cell donor.
LNPs induce pervasive transcriptomic changes without affecting T-cell functionality. (A) Heatmap showing enrichment results from all comparisons between conditions between conditions from supplemental Figure 5A on the hallmark gene set (Molecular Signatures Database). Color intensity reflects statistical significance of the test (adjusted P values), whereas signs (positive or negative) correspond to the set of DEGs used in the statistical test (upregulated or downregulated, respectively). (B-D) MA plot showing significant DEGs in (B) empty LNPs vs UTs (top) or LNPs vs UTs (bottom); (C) empty LNPs vs mock electros (top) or LNPs vs mock electros (bottom); (D) empty LNPs vs UTs (top) or LNPs vs UTs (bottom) comparisons. Labels highlight DEGs from panels B-C belonging to the core enrichment of cholesterol homeostasis category, and (D) SREBF2, ABCA1, and ABCG1. (E) Percentage of T cells negative and single-, double-, triple-, and quadruple-positive for exhaustion markers 14 days after treatments (n = 3). Mean ± SEM. (F) IFN-γ secretion 3 days after anti-CD3 polyclonal stimulation, ie, 14 days after treatments (n = 3). Each symbol represents a different T-cell donor.
When comparing cells treated with empty or RNA-containing LNPs with UTs, ORA highlighted similar upregulation of p53, apoptosis, and inflammatory genes, comparable with that in electroporated samples. However, distinct sets of genes belonging to the p53 and apoptosis categories were subclustered based on their expression upon different treatments. Although electroporation is enriched for positive regulation of apoptosis, LNPs are enriched for genes implicated in its negative regulation, concordantly with the lower rate of cell death. Moreover, electroporation is enriched for DDR and cell cycle–related genes (supplemental Figure 5F-G). IFN pathways were positively enriched in empty LNPs/LNPs vs UTs as opposed to electroporated ones. Moreover, empty LNPs/LNP samples showed a downregulation of genes belonging to cholesterol homeostasis (eg, LDLR, SQLE, and SREBF2) when compared with UTs (Figure 3B) and electroporated samples (Figure 3C), likely in response to the cellular loading with exogenous lipids.32 In contrast to the preserved proliferation, empty LNPs/LNP samples showed downregulation of cell cycle–related genes compared with UT and electroporated samples (Figure 3A). It has been reported that cholesterol homeostasis in proliferating T cells is maintained through the opposite regulation of SREBF2 and LXR transcriptional programs. Although upon T-cell activation, SREBF2 transcribes genes involved in cholesterol biosynthesis and uptake,33 excess cholesterol activates LXR, which transcribes target genes such as ABCA1 and ABCG1, which are membrane transporters mediating cholesterol efflux. Enforced expression of ABCG1 engages a metabolic checkpoint, blocking proliferation.33,34 Empty LNPs/LNP samples showed the downregulation of SREBF2 and its target genes33 and upregulation of ABCA1 and ABCG1 compared with other conditions (Figure 3D), consistent with cholesterol loading from LNPs and possibly explaining the observed impact on cell cycle. Moreover, the induction of IFN response has been reported upon SREBF2 downregulation,35 possibly explaining upregulation of genes in these categories in LNP-treated vs electroporated cells. These adverse cellular responses may explain the irreversible growth arrest and toxicity observed in initial studies, when LNPs were left in the medium (supplemental Figure 4I). Because LNPs were washed out at the time of transcriptomic analysis, it is conceivable that such responses quickly resolved, as mirrored by the rapid engagement of LNP-treated cells in robust proliferation. In a long-term culture, LNP-edited cells did not show signs of exhaustion upon cell surface analysis, with negligible proportions of cells expressing multiple inhibitory receptors and responding to polyclonal restimulation and/or mixed leucocyte reaction with similar to higher IFN-γ production and proliferation levels compared with UT or electroporated cells (Figure 3E-F; supplemental Figure 5H-I), supporting preserved functionality as the other treatments.
LNP-mediated CRISPR/Cas9 delivery improves tolerance of human HSPCs to ex vivo gene disruption
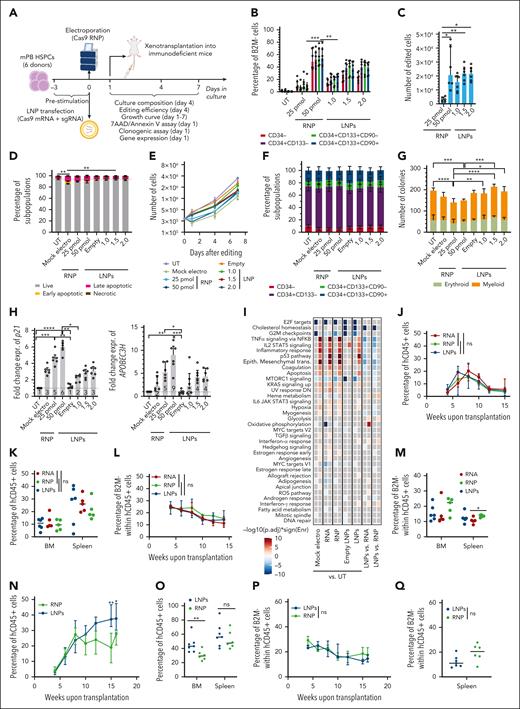
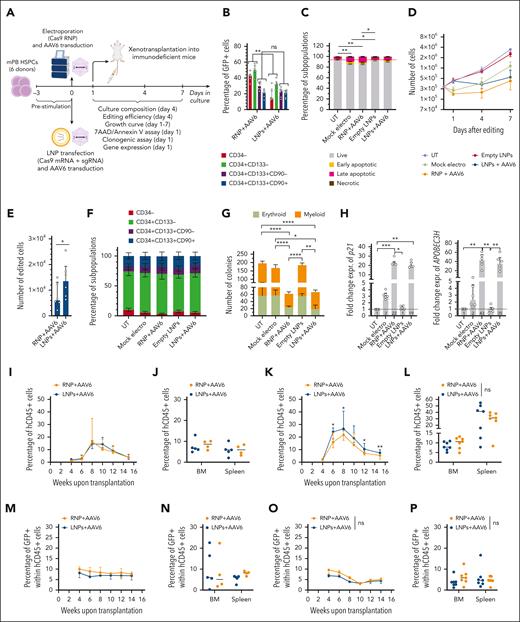
Because transfection efficiency of LNPs was also high in human HSPCs (supplemental Figure 6A-B), we investigated LNP-based gene editing. We compared LNPs and electroporation for B2M disruption in mPB HSPCs using previously optimized culture conditions15 that showed robust LDLR expression in all progenitor subsets (supplemental Figure 6C). LNP-treated cells showed higher B2M-KO efficiency compared with electroporation across all subpopulations at matched RNA doses, reaching ∼50% in the primitive compartment (CD34+CD133+CD90+). Shortening the LNP incubation period decreased B2M-KO (supplemental Figure 6D). Because electroporation of Cas9 RNP is the current state-of-the-art for HSPC editing, we compared LNP- and RNP-mediated editing in 6 mPB HSPC donors (Figure 4A). Despite RNA LNPs reaching lower editing efficiency than RNP at the highest doses, the yield of B2M-KO cells was comparable for both treatments (Figure 4B-C), given the improved viability and growth of LNP-treated cells (Figure 4D-E; supplemental Figure 6E). None of the treatments induced significant phenotypic cell skewing (Figure 4F). In agreement with previous reports,15 we observed a cumulative impact of electroporation and nuclease-induced DNA DSBs on HSPC clonogenic potential, which was not affected by LNPs (Figure 4G). Consistently, p21 and APOBEC3H induction 24 hours after editing was lower for LNP samples (Figure 4H; supplemental Figure 6F-G), reflecting an exclusive input on the p53 pathway from nuclease-induced DSBs rather than cumulative inputs from DSBs and electroporation. Similar results were obtained when treating HSPCs from the CB (supplemental Figure 6H-J) or when editing mPB HSPCs in the exon 1 of B2M using a more efficient sgRNA (supplemental Figure 6K-L). Of note, nuclease delivery using LNPs yielded a similar proportion of NHEJ-edited alleles and did not increase the proportion of on-target longer deletions at matched editing efficiencies compared with the other delivery modalities (supplemental Figure 6M-O).
CRISPR/Cas9 RNA delivery using LNP improves HSPC tolerance to gene editing. (A) Schematic representations of gene editing experimental procedure and related analyses after CRISPR/Cas9 RNA delivery using LNPs or CRISPR/Cas9 RNP/RNA delivery via electroporation in mPB HSPCs. (B) Percentage of B2M– HSPCs within subpopulations (n = 6). Cells were electroporated with 25 or 50 pmol of RNP or transfected with 1, 1.5, or 2 μg of LNPs. mPB HSPC phenotype (from committed to primitive progenitors) is defined as follows: CD34−; CD34+CD133−; CD34+CD133+; and CD34+CD133+CD90+. Friedman test with Dunn multiple comparisons performed on the latter subpopulation; median ± IQR. (C) Number of edited cells from panel B, 24 hours after editing (n = 6); Friedman test with Dunn multiple comparisons; median ± IQR. (D) Percentage of live, early/late apoptotic, and necrotic CD34+CD133+CD90+ HSPCs, 24 hours after B2M editing in experiments from panel B (n = 6). Mock electros and empty LNPs were added as controls (n = 6). Friedman test with Dunn multiple comparisons performed on live cells; mean ± SEM. (E) Growth curve of HSPCs from panel D (n = 6); median ± IQR. (F) Cell population composition from panel D, 4 days after treatment; mean ± SEM. (G) Number of colonies generated using mPB HPSCs from panel D (n = 6). Friedman test with Dunn multiple comparisons; median ± IQR. (H) Fold change expression of p21 (left) and APOBEC3H (right) relative to UT 24 hours after treatment from experiments in panel D (n = 6). Friedman test with Dunn multiple comparisons; median ± IQR. (I) Heatmap showing enrichment results from different comparisons between conditions from supplemental Figure 6P against the hallmark gene set (Molecular Signatures Database). Color intensity reflects statistical significance of the test (adjusted P values), whereas signs (positive or negative) correspond to the set of DEGs used in the statistical test (upregulated or downregulated, respectively). (J) Percentage of circulating human (h)CD45+ cells over time in mice that underwent transplant with mPB HSPCs edited with 50 pmol of RNP, 1.5 μg of mRNA, or 1.5 μg of LNPs (n = 5,5,6). Kruskal-Wallis test; median ± IQR. (K) Percentage of hCD45+ cells in the bone marrow (BM) and spleen of mice from panel J. Kruskal-Wallis test; median. (L) Percentage of B2M– cells over time, within the human graft in the PB of mice from panel J. Kruskal-Wallis test performed at the last time point; median ± IQR. (M) Percentage of B2M− cells within human graft in BM and spleen of mice from panel J. Kruskal-Wallis test; median. (N) Percentage of circulating human (h)CD45+ cells over time in mice that underwent transplantation with CB HSPCs edited with 50 pmol of RNP or 1.5 μg of LNPs (n = 6 and 7); Mann-Whitney test; median ± IQR. (O) Percentage of hCD45+ cells in BM and spleen of mice from panel N; Mann-Whitney test; median. (P) Percentage of B2M− cells over time, within the human graft in the PB of mice from panel N. Mann-Whitney test performed at the last time point; median ± IQR. (Q) Percentage of B2M– cells within human graft in spleen of mice from panel N; Mann-Whitney test; median.
CRISPR/Cas9 RNA delivery using LNP improves HSPC tolerance to gene editing. (A) Schematic representations of gene editing experimental procedure and related analyses after CRISPR/Cas9 RNA delivery using LNPs or CRISPR/Cas9 RNP/RNA delivery via electroporation in mPB HSPCs. (B) Percentage of B2M– HSPCs within subpopulations (n = 6). Cells were electroporated with 25 or 50 pmol of RNP or transfected with 1, 1.5, or 2 μg of LNPs. mPB HSPC phenotype (from committed to primitive progenitors) is defined as follows: CD34−; CD34+CD133−; CD34+CD133+; and CD34+CD133+CD90+. Friedman test with Dunn multiple comparisons performed on the latter subpopulation; median ± IQR. (C) Number of edited cells from panel B, 24 hours after editing (n = 6); Friedman test with Dunn multiple comparisons; median ± IQR. (D) Percentage of live, early/late apoptotic, and necrotic CD34+CD133+CD90+ HSPCs, 24 hours after B2M editing in experiments from panel B (n = 6). Mock electros and empty LNPs were added as controls (n = 6). Friedman test with Dunn multiple comparisons performed on live cells; mean ± SEM. (E) Growth curve of HSPCs from panel D (n = 6); median ± IQR. (F) Cell population composition from panel D, 4 days after treatment; mean ± SEM. (G) Number of colonies generated using mPB HPSCs from panel D (n = 6). Friedman test with Dunn multiple comparisons; median ± IQR. (H) Fold change expression of p21 (left) and APOBEC3H (right) relative to UT 24 hours after treatment from experiments in panel D (n = 6). Friedman test with Dunn multiple comparisons; median ± IQR. (I) Heatmap showing enrichment results from different comparisons between conditions from supplemental Figure 6P against the hallmark gene set (Molecular Signatures Database). Color intensity reflects statistical significance of the test (adjusted P values), whereas signs (positive or negative) correspond to the set of DEGs used in the statistical test (upregulated or downregulated, respectively). (J) Percentage of circulating human (h)CD45+ cells over time in mice that underwent transplant with mPB HSPCs edited with 50 pmol of RNP, 1.5 μg of mRNA, or 1.5 μg of LNPs (n = 5,5,6). Kruskal-Wallis test; median ± IQR. (K) Percentage of hCD45+ cells in the bone marrow (BM) and spleen of mice from panel J. Kruskal-Wallis test; median. (L) Percentage of B2M– cells over time, within the human graft in the PB of mice from panel J. Kruskal-Wallis test performed at the last time point; median ± IQR. (M) Percentage of B2M− cells within human graft in BM and spleen of mice from panel J. Kruskal-Wallis test; median. (N) Percentage of circulating human (h)CD45+ cells over time in mice that underwent transplantation with CB HSPCs edited with 50 pmol of RNP or 1.5 μg of LNPs (n = 6 and 7); Mann-Whitney test; median ± IQR. (O) Percentage of hCD45+ cells in BM and spleen of mice from panel N; Mann-Whitney test; median. (P) Percentage of B2M− cells over time, within the human graft in the PB of mice from panel N. Mann-Whitney test performed at the last time point; median ± IQR. (Q) Percentage of B2M– cells within human graft in spleen of mice from panel N; Mann-Whitney test; median.
We, then, performed whole transcriptomic analysis 24 hours after AAVS1 editing with LNPs or electroporation of RNA/RNP in mPB HSPCs. For controls, we used UTs, mock electros, or cells treated with empty LNPs. LNP transfection was as efficient as RNP electroporation in promoting NHEJ at AAVS1 and slightly more efficient than RNA electroporation at matching doses (supplemental Figure 6P-S). ORA highlighted significant upregulation of genes belonging to categories involved in DDR, apoptosis, and inflammation when comparing all electroporated samples with UTs, with increasing response in the RNP and RNA samples (Figure 4I; supplemental Figure 6T-U), in agreement with our previous studies of CB HSPCs.15 Among the downregulated DEGs, we observed a significant enrichment of the categories involved in cell cycle, oxidative phosphorylation, and fatty acid metabolism/adipogenesis. In contrast, empty LNPs did not activate DDR and inflammation, which were instead triggered by LNPs carrying RNA, thus by editing components and DSBs, when compared with UTs. The same comparison showed downregulation of cholesterol homeostasis and cell cycle–related genes, as already observed in T cells (Figure 4I; supplemental Figure 6V-W). Interestingly, distinct sets of p53-related genes subclustered based on the expression upon treatment, with electroporation enriching for DDR genes different from those specifically induced by nuclease exposure, and LNP vehicle enriching for a smaller subset involved in apoptosis regulation (supplemental Figure 6U-X).
To evaluate the impact and efficiency of editing on the fraction of repopulating HSPCs, immunodeficient mice received a transplant with the outgrowth of 7.5 × 105 mPB HSPCs seeded at the start of the culture (time 0 [t0]), edited at B2M with LNPs or electroporated with either RNA/RNP (supplemental Figure 7A-B) and found similar engraftment of human cells for all treatments, as monitored longitudinally in the PB and hematopoietic organs at the end of the experiment (Figure 4J-K; supplemental Figure 7C). Gene editing efficiencies, as revealed by B2M-KO, were also similar among groups, with a slight decrease from early to later times (Figure 4L-M). We, then, performed a similar experiment with CB HSPCs and transplanted the outgrowth of 5 × 104 t0-seeded cells edited either at B2M (supplemental Figure 7D-E) or AAVS1 using LNPs or via RNP electroporation. LNP-treated cells showed similar engraftment early after transplant but higher human graft size at later time points in the PB, spleen, and bone marrow in 1 of 2 experiments (Figure 4N-O; supplemental Figure 7F-H). B2M and AAVS1 editing efficiencies in human xenografts were confirmed via deep sequencing, with similar ranges reached by either RNA-delivery platform (Figure 4P-Q; supplemental Figure 7I-L).
LNP-mediated CRISPR/Cas delivery supports HDR editing of HSPCs
We investigated whether LNP delivery could be suitable for HDR-based editing, targeting AAVS115 with a GFP cassette in HSPCs. LNPs outperformed electroporation at matched low RNA doses and reached similar or better HDR at maximal doses across all subpopulations (supplemental Figure 7M-N). We compared LNP- and RNP-mediated editing in mPB HSPCs from 6 HDs (Figure 5A) and found similar HDR editing in CD90+ cells (Figure 5B). However, because electroporated HSPCs showed higher toxicity and delayed growth, the yield of CD90+GFP+ cells increased for LNP-treated samples (Figure 5C-E; supplemental Figure 7O). None of the treatments changed the subset composition compared with that of UTs (Figure 5F). In agreement with previous reports,15,16,36-38 we observed impaired clonogenic potential and robust p21 and APOBEC3H induction in cells edited with electroporation or LNPs, in which the dominant AAV6 effect masked the advantage provided by the latter platform (Figure 5G-H). Similar results were obtained when treating CB HSPCs (supplemental Figure 7P-R).
CRISPR/Cas9 RNA delivery using LNPs allows HDR-mediated editing in human HSPCs. (A) Schematic representations of gene editing experimental procedure, and related analyses, after CRISPR/Cas9 RNA delivery using LNPs or CRISPR/Cas9 RNP/RNA delivery via electroporation, in combination with the cognate AAV6, in mPB HSPCs. (B) Percentage of GFP+ cells within mPB HSPC subpopulations 96 hours after AAVS1 editing. Cells were edited with 25 pmol of RNP or 1 μg of LNPs and transduced with AAV6 immediately after electroporation or 2 hours before transfection, respectively. Wilcoxon test on individual subpopulations. Median ± IQR. (C) Percentage of live, early/late apoptotic, and necrotic CD34+CD133+CD90+ mPB HSPCs 24 hours after AAVS1 editing in experiments from panel B (n = 6). UTs, mock electros, and empty LNPs were added as controls (n = 6). Friedman test with Dunn multiple comparisons performed on live cells. Mean ± SEM. (D) Growth curve from panel C (n = 6). Median ± IQR. (E) Number of edited cells from panel C 24 hours after editing (n = 6). Wilcoxon test. Median ± IQR. (F) Cell population composition from panel C (n = 6), 4 days after treatment. Mean ± SEM. (G) Number of colonies generated by mPB HPSCs from panel C (n = 6). Median ± IQR. Friedman test with Dunn multiple comparisons. (H) Fold change expression of p21 and APOBEC3H relative to UTs 24 hours after treatment from experiments in panel C. Friedman test with Dunn multiple comparisons. Median ± IQR. (I) Percentage of circulating hCD45+ cells over time in mice that underwent transplantation with mPB HSPCs (from a pool of donors), edited with 25 pmol RNP or 1 μg of LNPs, in combination with AAV6 (n = 4 and 5). Median ± range. (J) Percentage of hCD45+ cells in the BM and spleen of mice from panel I. Median. (K) Percentage of circulating hCD45+ cells over time in mice that underwent transplantation with mPB HSPCs edited as in panel I but using a different pool of donors (n = 7). Mann-Whitney test. Median ± range. (L) Percentage of hCD45+ cells in BM and spleen of mice from panel K. Mann-Whitney test. Median. (M-P) Percentage of GFP+ cells over time within human graft in PB (M,O) or hematopoietic organs (N,P) of mice from panel I (M-N) and panel K (O-P). Mann-Whitney test. Median or median ± range.
CRISPR/Cas9 RNA delivery using LNPs allows HDR-mediated editing in human HSPCs. (A) Schematic representations of gene editing experimental procedure, and related analyses, after CRISPR/Cas9 RNA delivery using LNPs or CRISPR/Cas9 RNP/RNA delivery via electroporation, in combination with the cognate AAV6, in mPB HSPCs. (B) Percentage of GFP+ cells within mPB HSPC subpopulations 96 hours after AAVS1 editing. Cells were edited with 25 pmol of RNP or 1 μg of LNPs and transduced with AAV6 immediately after electroporation or 2 hours before transfection, respectively. Wilcoxon test on individual subpopulations. Median ± IQR. (C) Percentage of live, early/late apoptotic, and necrotic CD34+CD133+CD90+ mPB HSPCs 24 hours after AAVS1 editing in experiments from panel B (n = 6). UTs, mock electros, and empty LNPs were added as controls (n = 6). Friedman test with Dunn multiple comparisons performed on live cells. Mean ± SEM. (D) Growth curve from panel C (n = 6). Median ± IQR. (E) Number of edited cells from panel C 24 hours after editing (n = 6). Wilcoxon test. Median ± IQR. (F) Cell population composition from panel C (n = 6), 4 days after treatment. Mean ± SEM. (G) Number of colonies generated by mPB HPSCs from panel C (n = 6). Median ± IQR. Friedman test with Dunn multiple comparisons. (H) Fold change expression of p21 and APOBEC3H relative to UTs 24 hours after treatment from experiments in panel C. Friedman test with Dunn multiple comparisons. Median ± IQR. (I) Percentage of circulating hCD45+ cells over time in mice that underwent transplantation with mPB HSPCs (from a pool of donors), edited with 25 pmol RNP or 1 μg of LNPs, in combination with AAV6 (n = 4 and 5). Median ± range. (J) Percentage of hCD45+ cells in the BM and spleen of mice from panel I. Median. (K) Percentage of circulating hCD45+ cells over time in mice that underwent transplantation with mPB HSPCs edited as in panel I but using a different pool of donors (n = 7). Mann-Whitney test. Median ± range. (L) Percentage of hCD45+ cells in BM and spleen of mice from panel K. Mann-Whitney test. Median. (M-P) Percentage of GFP+ cells over time within human graft in PB (M,O) or hematopoietic organs (N,P) of mice from panel I (M-N) and panel K (O-P). Mann-Whitney test. Median or median ± range.
Finally, we transplanted 106 outgrowths of t0-seeded mPB HSPCs edited at AAVS1 via LNPs or electroporation (supplemental Figure 7S-T). PB, bone marrow, and spleen analyses revealed multilineage long-term engraftment for both treatments, with higher graft size for the LNP group in 1 of 2 experiments performed with different pools of HD (Figure 5I-L; supplemental Figure 7U-V). The percentage of GFP+ cells in organs was comparable between the conditions tested and showed a slight decrease at longer durations (Figure 5M-P).
Discussion
We report relevant acute toxicity of electroporation in human T cells. Although optimized culture conditions partially circumvented these detrimental effects, the usage of LNPs as ex vivo delivery vehicles for editing reagents substantially decreased toxicity and provided for higher and prompter yields of edited cells. Notably, LNP-based delivery also benefited ex vivo HSPC gene editing better preserving the clonogenic potential of edited cells. Overall, our findings provide mechanistic insights and scientific rationale for leveraging on LNPs to achieve efficient and harmless delivery of editing reagents and, more generally, RNA-based therapeutics, on hematopoietic cells.
Exposure of cells to electroporation was previously associated with cell injury and death, whose magnitude depends on cell type, voltage, number, and length of electric pulses. Mechanistically, it was variously ascribed to DNA damage and perturbation of the cytoplasm, lysosomal, and mitochondrial membranes, causing the influx or leakage, respectively, of proapoptotic molecules and decreased energy supply.19 Our findings confirm a rapid alteration of the mitochondrial potential after pulsing, followed by altered energy metabolism and activation of DDR. The impact of these responses on cell viability and the extent of recovery varied between T cells and HSPCs, with a higher proportion of apoptosis in the former, possibly owing to a higher dependence on mitochondria for activation and proliferation.39,40 In contrast, HSPCs showed less apoptosis but experienced a mostly reversible p53-dependent delay in growth, exacerbated in the fraction of clonogenic progenitors. Nuclease-induced DSB and AAV617 further aggravated these p53-dependent responses in HSPCs,15,16 however, being of limited impact in T cells, with donor variability for the response to AAV6. Accordingly, transient p53 inhibition was ineffective in alleviating T-cell cytotoxicity, as opposed to our reports in HSPCs.15
Although LNPs provided similar editing efficiencies as electroporation but with less toxicity, transcriptomic analysis revealed the sensing of LNPs by both T cells and HSPCs, which downregulated genes involved in cholesterol biosynthesis and regulation. Cholesterol metabolism has a broad impact on cellular activities, including cell cycle progression and interferon gene regulation,33-35 explaining the pervasive transcriptomic changes observed early after editing. Although LNPs circumvented electroporation-induced inflammatory and p53 responses in HSPCs, these pathways were activated in T cells, consistent with previous reports showing the induction of an inflammatory state upon intracellular cholesterol accumulation in the latter cell type.41,42 Despite cholesterol excess causing functional exhaustion of T cells43 and proliferation/differentiation of HSPCs,44-47 our experiments support preserved cell functionality in culture and in vivo, suggesting prompt resolution of LNP-induced responses. Of note, LNPs could not overcome detrimental responses induced by editing reagents, possibly underestimating their beneficial effect.
Our study highlights the added value and versatility of an LNP-based platform for efficient ex vivo hematopoietic cell engineering. An increase in cell recovery by reducing the acute toxicity of edited T cells may improve the efficacy of immunotherapies by shortening the manufacturing process48,49 and/or by manufacturing products that otherwise would not reach the established therapeutic dose owing to harvest constraints.50 Moreover, maintaining a broader T-cell receptor repertoire diversity by minimizing the loss of edited cells may be crucial for some immunotherapy applications.11,51-53 Similarly, reaching a higher number of edited HSPCs can be pivotal to obtaining highly polyclonal products and to providing safer therapeutic opportunities for patients yielding low/fragile HSPCs upon harvesting54-56 although we could not document a consistent improvement over electroporation in xenografts, likely because of donor variability.
Additional applications of LNPs may be the concurrent or asynchronous delivery of editing reagents and nucleic acids, endowing cells with specific functions.16,57 Multistep delivery may allow multiplex editing while minimizing the risk of translocations.58,59 Optimized LNP formulations may minimize the pervasive cholesterol-mediated response observed and the competition with viral vectors to improve HDR efficiency. Moreover, because Cas9 mRNA increases the burden of off-target60 and, possibly, undesired on-target events as compared with RNP, LNPs supporting delivery of Cas9-RNP could further improve the platform. However, we found comparable proportions of longer deletions at the target site for either RNA- or RNP-based delivery methods when achieving a similar extent of NHEJ editing. It is also conceivable that the advantages of LNPs extend to the delivery of base and prime editors, possibly upon fine-tuning of formulation to best accommodate larger RNA payloads. Overall, because LNP technology is currently flourishing and has already reached the clinical stage for in vivo applications, our analyses demonstrated the feasibility and advantage of its use for ex vivo gene editing in hematopoietic cells for the treatment of human diseases.
Acknowledgments
The authors thank all members of the laboratory of L.N. for discussion, the IRCCS San Raffaele Hospital Flow Cytometry and Center for Omics Sciences facilities, and A. Auricchio and M. Doria (Telethon Institute of Genetics and Medicine; TIGEM, Pozzuoli, Italy) for providing AAV6 vectors. The authors thank A. Tommassini (IEO, European Institute of Oncology IRCCS) for technical help with proteomic experiment; T. Plati for technical and logistical support in LNPs supply; and I. Cuccovillo, G. Desantis, K. Giannetti, C. Rossi, and T. Di Tomaso (SR-Tiget) for supply and purification of mPB HSPCs. The schematic illustrations were created with BioRender.com.
This work was supported by grants to L.N. from Telethon (SR-Tiget core grant) and the EU Horizon 2020 Program (UPGRADE); and to S.F. from European Hematology Association (2022 Junior Research Grant). T.B.’s group is funded by AIRC IG (21834) and EPIC-XS (project no. 823839), funded by the EU Horizon 2020 Program.
Authorship
Contribution: V.V. and S.F. conceived the study, performed research and interpreted data; S.B. and I.M. performed bioinformatic analyses; L.A. and C.G. provided technical support; C.A. contributed to good manufacturing practice culture protocol optimization in the process development laboratory, supervised by M.R.; A.V. helped with library preparation; A.A. designed and performed T-cell functional assays; M.S. and A.C. helped with proteomic experiment design and execution, supervised by T.B.; L.N. supervised research and coordinated the work; and V.V., S.F., and L.N. wrote the manuscript.
Conflict-of-interest disclosure: L.N., V.V., and S.F. are listed as inventors on patent applications of gene editing owned and managed by the San Raffaele Scientific Institute and the Telethon Foundation. L.N. is the founder and quota holder of GeneSpire, a startup company aiming to develop gene editing applications in hematopoietic cells. The remaining authors declare no competing financial interests.
Correspondence: Luigi Naldini, San Raffaele Telethon Institute for Gene Therapy, IRCCS San Raffaele Scientific Institute, Via Olgettina, 58, 20132 Milan, Italy; e-mail: naldini.luigi@hsr.it.
References
Author notes
∗V.V. and S.F. contributed equally to this study.
Sequencing data have been deposited in Gene Expression Omnibus (accession number GSE215250). Mass-spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE8 partner repository (accession number PXD037529). Scripts for RNA sequencing and proteomic data analysis are available at GitLab (http://www.bioinfotiget.it/gitlab/custom/vavassori_lnp2022).
Relevant data are included in the article; accession numbers and scripts are reported in the article and supplemental Methods. Reagents are available under material transfer agreement on request from the corresponding author, Luigi Naldini (naldini.luigi@hsr.it).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal