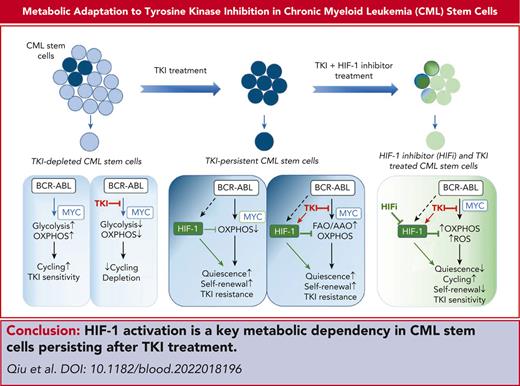
Key Points
TKI therapy alters CML stem and progenitor cell metabolism through both selection and metabolic reprogramming of specific subpopulations.
HIF-1 activation plays a critical role in CML stem cell adaptation to TKI treatment by restricting OXPHOS and maintaining stem cell dormancy.
Abstract
Tyrosine kinase inhibitors (TKIs) are very effective in treating chronic myelogenous leukemia (CML), but primitive, quiescent leukemia stem cells persist as a barrier to the cure. We performed a comprehensive evaluation of metabolic adaptation to TKI treatment and its role in CML hematopoietic stem and progenitor cell persistence. Using a CML mouse model, we found that glycolysis, glutaminolysis, the tricarboxylic acid cycle, and oxidative phosphorylation (OXPHOS) were initially inhibited by TKI treatment in CML-committed progenitors but were restored with continued treatment, reflecting both selection and metabolic reprogramming of specific subpopulations. TKI treatment selectively enriched primitive CML stem cells with reduced metabolic gene expression. Persistent CML stem cells also showed metabolic adaptation to TKI treatment through altered substrate use and mitochondrial respiration maintenance. Evaluation of transcription factors underlying these changes helped detect increased HIF-1 protein levels and activity in TKI-treated stem cells. Treatment with an HIF-1 inhibitor in combination with TKI treatment depleted murine and human CML stem cells. HIF-1 inhibition increased mitochondrial activity and reactive oxygen species (ROS) levels, reduced quiescence, increased cycling, and reduced the self-renewal and regenerating potential of dormant CML stem cells. We, therefore, identified the HIF-1–mediated inhibition of OXPHOS and ROS and maintenance of CML stem cell dormancy and repopulating potential as a key mechanism of CML stem cell adaptation to TKI treatment. Our results identify a key metabolic dependency in CML stem cells persisting after TKI treatment that can be targeted to enhance their elimination.
Introduction
Chronic myelogenous leukemia (CML) results from hematopoietic stem cell (HSC) transformation caused by the BCR-ABL gene. BCR-ABL tyrosine kinase inhibitors (TKIs) are effective in inducing deep remissions in CML, preventing disease progression, and enhancing survival. However, TKIs fail to eliminate primitive, quiescent CML stem cells responsible for disease propagation.1 Most patients with CML require continued TKI treatment to prevent recurrence, with risks of toxicity, teratogenicity, financial burden, and nonadherence. Improved understanding of mechanisms of stem cell persistence is required to enhance treatment-free remission and cure.
CML stem cell persistence may be related to cell-extrinsic mechanisms involving bone marrow (BM) niches that regulate dormancy and self-renewal,2-4 cell-intrinsic alterations in signaling pathways,5-8 transcriptional networks,9 and epigenetics.10 However, there is limited knowledge about the effects of TKI treatment on CML stem cell metabolism and the role of metabolic reprogramming in their persistence after TKI treatment. Normal self-renewing HSCs limit mitochondrial oxidative phosphorylation (OXPHOS) to maintain quiescence, relying on anaerobic glycolysis to support adenosine triphosphate (ATP) production. OXPHOS-generated reactive oxygen species (ROSs) are triggers for HSC differentiation.11-13 Cancer cells undergo metabolic reprogramming to meet energy and biosynthetic requirements, commonly demonstrating increased fermentation of glucose to lactate, even in the presence of oxygen, known as the Warburg effect.14 Cancer stem cells variably rely on either OXPHOS or glycolysis for growth and maintenance, depending on the specific cancer type. Acute myeloid leukemia (AML) stem cells use amino acid metabolism to maintain OXPHOS and survival.15 BCL-2 inhibition reduces OXPHOS and depletes quiescent AML stem cells,16,17 but upregulation of fatty acid oxidation (FAO) can lead to resistance.18 Primitive CML cells also show upregulated OXPHOS and may be depleted by a mitochondrial protein translation inhibitor.19,20 However, whether increased OXPHOS extends to the most primitive quiescent CML stem cell population, and the mechanisms underlying increased OXPHOS are unclear.
The metabolic response of cancer stem cells to treatment and the role of metabolic adaptations in stem cell persistence have received limited study. Here, we performed an integrated analysis of metabolic changes induced by TKI treatment in CML stem and progenitor cells. Our results provide new insights into mechanisms of adaptation to TKI treatment, and suggest potential approaches to deplete persistent, quiescent CML stem cells.
Methods
Mice
C57BL/6J mice and NOD.Cg-Rag1tm1Mom -Il2rgtm1Wjl Tg(CMV-IL-3, CSF2, KITLG) (NRGS) mice were obtained from Jackson Laboratory (Bar Harbor, ME) and C57BL/6.SJL mice from Charles River Laboratories (Frederick, MD). SCL-tTA-BCR-ABL mice were maintained on doxycycline-containing food. To induce CML, doxycycline administration was withdrawn, and leukemia development was confirmed after 4 or 8 weeks. Experiments were performed using 8- or 10-week-old mice of both sexes. Mouse care was in accordance with federal guidelines and protocols approved by the University of Alabama at Birmingham (UAB) Institutional Animal Care and Use Committee.
Human samples
BM and peripheral blood (PB) were obtained from patients with CML who received consultation at the UAB. Sample acquisition was approved by the UAB institutional review board in accordance with assurances filed with the Department of Health and Human Services and requirements of the Declaration of Helsinki. Informed consent was obtained from patients.
In vivo drug treatment
BM cells from CML mice (CD45.1/CD45.2 or CD45.2) were transplanted into irradiated recipient mice (CD45.1 or CD45.2) to generate leukemia. Mice were treated with nilotinib or vehicle for 2 days to 2 weeks; or with vehicle, nilotinib, echinomycin, or combination, for 2 weeks. BM cells from patients with CML were transplanted into irradiated NRGS mice. Mice that underwent engraftment were treated with vehicle, nilotinib, echinomycin, or combination for 2 weeks. See supplemental Methods, available on Blood website.
Extracellular flux analysis (mito stress test)
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using the Seahorse XF96 analyzer as previously described.20 See supplemental Methods.
Metabolite profiling
13C labeling and analysis
c-Kit–enriched cells were cultured with [U-13C6]glucose or [U-13C5]glutamine (CLM-1822) for 24 hours, after which intracellular metabolites were extracted, and analyzed using liquid chromatography–mass spectrometry. Correction for 13C natural abundance was done as previously described.23 See supplemental Methods.
Scenith
The single-cell energetic metabolism by profiling translation inhibition (SCENITH) assay protocol was performed as previously described.24 BM cells were treated with 2-deoxyglucose, oligomycin, or both, in sequence, in the presence of puromycin, labeled with cell surface marker antibodies, followed by intracellular staining with antipuromycin antibodies, and analyzed via flow cytometry (supplemental Methods).
TMRM labeling
BM cells were labeled with antibodies of surface markers and tetramethylrhodamine methyl ester (TMRM), and analyzed via flow cytometry (supplemental Methods).
scRNA data analysis
Sequencing files were processed using the 10x Genomics Cell Ranger pipeline (version 2.0.1). Scanpy (version 1.8) was used for quality control, normalization, dimensionality reduction (uniform manifold approximation and projection), clustering, and data integration.25 Cell clustering was based on Louvain algorithm after computing a shared nearest neighbor graph. Differentially expressed genes were analyzed using Wilcoxon rank-sum test. Enrichment analysis was performed using gene set enrichment analysis. Transcription factor modules were analyzed via pySCENIC version 0.11.2 (supplemental Methods).
Statistical analyses
Results are displayed as the mean ± standard error of mean. Significance values were calculated using Prism version 5.0 software (Prism; GraphPad, La Jolla, CA) using unpaired, nonparametric t tests (the Mann-Whitney test), or a one- or two-way analysis of variance, as appropriate. For RNA sequencing experiments, the P values were adjusted using the Benjamini-Hochberg method.
Results
TKI treatment leads to dynamic alterations in glycolysis and OXPHOS in CML progenitors
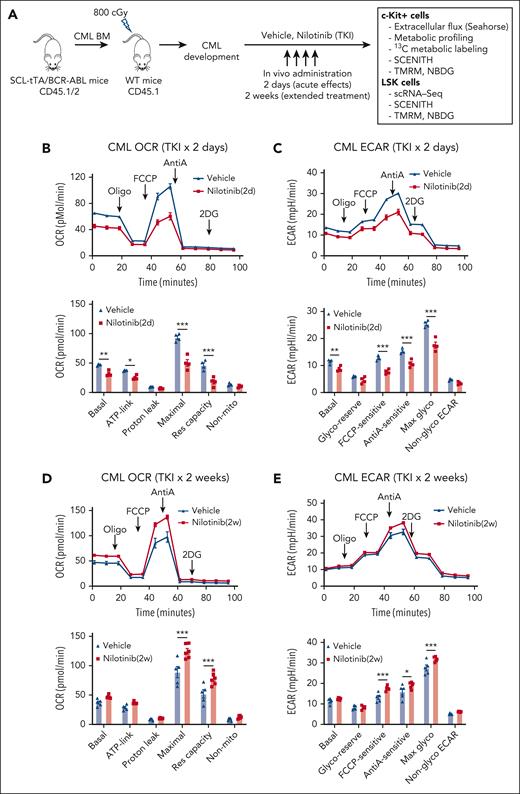
We studied changes in energy metabolism in CML stem and progenitor cells after TKI treatment, using an inducible, transgenic SCL-tTA/BCR-ABL (BA) mouse model. Leukemic BM cells (CD45.1/CD45.2 dual positive) were transplanted into CD45.1 recipient mice to obtain a cohort of mice with similar onsets of leukemia 6 or 8 weeks after transplantation. Mice were treated with nilotinib (TKI) or vehicle for 2 days or 2 weeks. Donor leukemia cell chimerism (>80%) was maintained after TKI treatment. Bioenergetics and metabolomics assays were performed using BM c-Kit+ cells to obtain sufficient cells for these analyses. Because c-Kit+ populations largely consist of progenitor cells, we also studied the effects of TKI treatment on metabolic gene expression and metabolic dependencies in BM Lin−Sca-1+c-Kit+ (LSK) populations enriched for stem cells (Figure 1A).
TKI treatment leads to dynamic alterations in glycolysis and OXPHOS in CML progenitors. (A) Overall experimental strategy. BM cells (2 × 106) from BCR-ABL transgenic mice (CD45.1/2) in which leukemia had been induced by tetracycline withdrawal were transplanted into CD45.1 recipient mice. Once mice had developed leukemia 6 or 8 weeks after transplantation, they were treated with nilotinib (TKI) or vehicle for 2 days or 2 weeks. BM c-Kit+ cells and LSK cells were selected and studied as shown. (B-C) Extracellular flux analysis of OCR (B) and ECAR (C) in CML BM c-Kit+ cells after 2 days of nilotinib treatment (n = 4 mice each). (D-E) Extracellular flux analysis of OCR (D) and ECAR (E) in CML BM c-kit+ cells after 2 weeks of nilotinib treatment (n = 5-6 mice each). Significance values: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. Results represent the mean ± standard error of mean (SEM) of multiple replicates.
TKI treatment leads to dynamic alterations in glycolysis and OXPHOS in CML progenitors. (A) Overall experimental strategy. BM cells (2 × 106) from BCR-ABL transgenic mice (CD45.1/2) in which leukemia had been induced by tetracycline withdrawal were transplanted into CD45.1 recipient mice. Once mice had developed leukemia 6 or 8 weeks after transplantation, they were treated with nilotinib (TKI) or vehicle for 2 days or 2 weeks. BM c-Kit+ cells and LSK cells were selected and studied as shown. (B-C) Extracellular flux analysis of OCR (B) and ECAR (C) in CML BM c-Kit+ cells after 2 days of nilotinib treatment (n = 4 mice each). (D-E) Extracellular flux analysis of OCR (D) and ECAR (E) in CML BM c-kit+ cells after 2 weeks of nilotinib treatment (n = 5-6 mice each). Significance values: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. Results represent the mean ± standard error of mean (SEM) of multiple replicates.
Bioenergetics analysis was conducted using the Agilent Seahorse XF96 system. CML c-Kit+ cells demonstrate significantly increased OCR and ECAR compared with those of healthy cells.19,20 In CML c-Kit+ cells, after 2 days of nilotinib treatment, a significantly reduction in basal, ATP-linked, and maximal OCRs and reserve capacity was observed (Figure 1B) as well as a significant reduction in basal, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP)-sensitive, anti-A–sensitive, and maximal ECAR (Figure 1C). In contrast, a modest increase of maximal OCR and reserve capacity (Figure 1D; supplemental Figure 1A), and FCCP-sensitive, anti-A–sensitive, and maximal ECAR (Figure 1E; supplemental Figure 1B) was observed after 2 weeks of nilotinib treatment. We conclude that TKI treatment leads to initial inhibition followed by restoration of OXPHOS and glycolysis in CML c-Kit cells. In contrast, c-Kit+ cells from healthy mice showed a modest increase in maximal OCR and ECAR after both 2 days and 2 weeks of treatment (supplemental Figure 1C-F).
TKI treatment leads to dynamic alterations in energy metabolism in CML progenitors
To understand the basis for altered OCR and ECAR, we analyzed metabolite levels in c-Kit+ cells freshly isolated from TKI- and vehicle-treated mice, using high-resolution mass spectrometry.22 Cells from mice treated with vehicle, nilotinib for 2 days, or nilotinib for 2 weeks clustered separately (supplemental Figure 2A). We observed significant reduction in glycolytic (Figure 2A; supplemental Figure 2B) and tricarboxylic acid (TCA) cycle intermediates (Figure 2B; supplemental Figure 2C) after 2 days of TKI treatment, with reduced ATP/adenosine diphosphate and guanosine triphosphate/guanosine diphosphate ratios (Figure 2C). However, glycolytic (Figure 2D; supplemental Figure 2D) and TCA cycle intermediate ratios (Figure 2E; supplemental Figure 2E) as well as ATP/adenosine diphosphate and guanosine triphosphate/guanosine diphosphate ratios (Figure 2F; supplemental Figure 2F) were similar to those in vehicle-treated controls after 1 and 2 weeks of TKI treatment.
TKI treatment leads to dynamic alterations in energy metabolism in CML progenitors. Mice with CML were treated with nilotinib or vehicle for 2 days or 2 weeks, and BM c-Kit+ cells were selected and metabolomic profiling was performed. (A) The relative abundance of glycolytic intermediates after 2 days of nilotinib treatment is shown (n = 3-4). (B) The relative abundance of TCA cycle intermediates after 2 days of nilotinib treatment is shown (n = 3-4). (C) The ratio of ATP/ADP and GTP/GDP after 2 days of nilotinib treatment is shown (n = 3-4). (D) The relative abundance of glycolytic intermediates after 2 weeks of nilotinib treatment is shown (n =3). (E) The relative abundance of TCA cycle intermediates after 2 weeks of nilotinib treatment is shown (n = 3). (F) The ratio of ATP/ADP and GTP/GDP after 2 weeks of nilotinib treatment is shown (n = 3). (G) BM c-Kit+ cells were labeled with [U-13C6]glucose in vitro (n = 3). The percent labeling fraction of glycolytic end products (pyruvate, lactate, and alanine) and citric acid cycle intermediates (citrate/isocitrate, α-ketoglutarate, and malate) after 2 days and after 2 weeks of vehicle (Veh) or nilotinib (NIL) treatment is shown. The labeling fractions are corrected for 13C natural abundance. Significance values: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. Results represent mean ± SEM of multiple replicates. 3PG/2PG, 3-phosphoglyceric acid/2-phosphoglyceric acid; ADP, adenosine diphosphate; a-KG, α-ketoglutarate; F-1,6-BP, fructose 1,6-bisphosphate; G6P/F6P, glucose 6-phosphate/fructose 6-phosphate; GDP, guanosine diphosphate; GSH, glutathione; GTP, guanosine triphosphate; ns, not significant; PEP, phosphoenolpyruvic acid.
TKI treatment leads to dynamic alterations in energy metabolism in CML progenitors. Mice with CML were treated with nilotinib or vehicle for 2 days or 2 weeks, and BM c-Kit+ cells were selected and metabolomic profiling was performed. (A) The relative abundance of glycolytic intermediates after 2 days of nilotinib treatment is shown (n = 3-4). (B) The relative abundance of TCA cycle intermediates after 2 days of nilotinib treatment is shown (n = 3-4). (C) The ratio of ATP/ADP and GTP/GDP after 2 days of nilotinib treatment is shown (n = 3-4). (D) The relative abundance of glycolytic intermediates after 2 weeks of nilotinib treatment is shown (n =3). (E) The relative abundance of TCA cycle intermediates after 2 weeks of nilotinib treatment is shown (n = 3). (F) The ratio of ATP/ADP and GTP/GDP after 2 weeks of nilotinib treatment is shown (n = 3). (G) BM c-Kit+ cells were labeled with [U-13C6]glucose in vitro (n = 3). The percent labeling fraction of glycolytic end products (pyruvate, lactate, and alanine) and citric acid cycle intermediates (citrate/isocitrate, α-ketoglutarate, and malate) after 2 days and after 2 weeks of vehicle (Veh) or nilotinib (NIL) treatment is shown. The labeling fractions are corrected for 13C natural abundance. Significance values: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. Results represent mean ± SEM of multiple replicates. 3PG/2PG, 3-phosphoglyceric acid/2-phosphoglyceric acid; ADP, adenosine diphosphate; a-KG, α-ketoglutarate; F-1,6-BP, fructose 1,6-bisphosphate; G6P/F6P, glucose 6-phosphate/fructose 6-phosphate; GDP, guanosine diphosphate; GSH, glutathione; GTP, guanosine triphosphate; ns, not significant; PEP, phosphoenolpyruvic acid.
We analyzed the substrate and pathway contributions to metabolite levels using steady-state metabolic labeling with [U-13C6]glucose for 24 hours of c-Kit cells from vehicle- or TKI-treated CML mice and measured the labeled and unlabeled metabolites via high-resolution mass spectrometry. TKI treatment for 2 days resulted in the reduced labeling of the glycolytic end product, pyruvate, and alanine (Figure 2G). Together with the reduction in levels of glycolytic intermediates, these observations indicate reduced glycolytic flux. We also observed reduced labeling of TCA cycle intermediates. In contrast, TKI treatment for 2 weeks led to the increased labeling of lactate, alanine, and TCA cycle intermediates compared with that in controls. Increased 13C labeling of serine, glycine, and glutathione (supplemental Figure 3A-B) was also observed, indicating an increased use of glucose carbons for amino acid synthesis. [U-13C5]glutamine labeling of α-ketoglutarate, malate, and citrate had also reduced after 2 days of TKI treatment, indicating the reduced contribution of glutamine oxidation to TCA cycle intermediates (supplemental Figure 3C). However, labeling was increased or normalized after 2 weeks of TKI treatment, along with increased glutathione labeling (supplemental Figure 3D-E).
We conclude that CML progenitor cells show initial reduction in glycolytic flux, glutamine oxidation, and TCA cycle activity after TKI treatment but show restored glycolytic and glutaminolytic activity, and increased glucose-mediated lactate generation, TCA cycle activity, and amino acid synthesis, with continued TKI exposure.
TKI treatment leads to metabolic reprogramming and selection of CML progenitor subpopulations
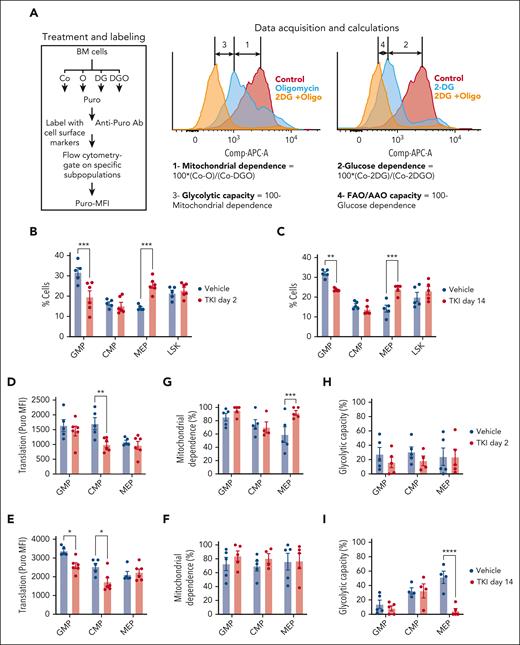
We used the SCENITH assay to evaluate dynamic changes in metabolism in c-Kit+ subpopulations treated with TKI.24,26,27 In SCENITH, we use flow cytometry to assess metabolic activities in multiple populations in parallel. Puromycin incorporation is used to measure protein synthesis, which tightly correlates with ATP levels, reflecting the global metabolic activity (Figure 3A). Metabolic parameters are calculated based on changes in the puromycin mean fluorescence intensity after addition of metabolic inhibitors. Mitochondrial and glucose dependency are calculated as a proportion of protein synthesis dependent on OXPHOS and glucose oxidation, respectively. Glycolytic capacity and FAO and/or amino acid oxidation capacity are calculated as a proportion of protein synthesis sustained after inhibition of OXPHOS and glucose oxidation, respectively. We adapted this assay to study metabolic alterations in hematopoietic stem and progenitor cell subpopulations, confirming that mitochondrial dependence was lowest in healthy long-term HSCs (LTHSCs), intermediate in multipotent progenitors (MPPs), and highest in granulocytic-macrophage progenitors (GMPs) (supplemental Figure 4A).
TKI treatment leads to metabolic reprogramming and selection of CML progenitor subpopulations. (A) Overview of the SCENITH assay performed using the BM of CML mice treated with vehicle or nilotinib (TKI) for 2 days and 2 weeks. The SCENITH assay uses flow cytometry to measure changes in the level of translation in response to inhibitors as a measurement for cellular metabolism. BM cells were divided and separately treated with 2-deoxyglucose (DG), oligomycin (O), and DG + O, together with controls (Cos) incubated without inhibitors, and labeled with puromycin (Puro). After cell surface labeling and intracellular labeling for Puro, different subpopulations were analyzed via flow cytometry for Puro-MFI response to the various inhibitors. Calculations of metabolic dependencies and capacities based on Puro-MFI are shown. (B-C) GMP, common myeloid progenitor (CMP), MEP, and LSK cell frequency within CML c-Kit+ cells (n = 5-6), as assessed via flow cytometry, after treatment with TKI for 2 days (B) or 2 weeks (C). (D-E) Protein synthesis (Puro-MFI) within committed progenitors (GMPs, CMPs, and MEPs) after 2 days (D) and 14 days (E) of TKI treatment. (F-I) Mitochondrial dependence and glycolytic capacity within GMPs, CMPs, and MEPs after 2 days (F,H) and 14 days (G,I) of TKI treatment. Significance values: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001. Results represent mean ± SEM of multiple replicates. AAO, amino acid oxidation; MFI, mean fluorescence intensity.
TKI treatment leads to metabolic reprogramming and selection of CML progenitor subpopulations. (A) Overview of the SCENITH assay performed using the BM of CML mice treated with vehicle or nilotinib (TKI) for 2 days and 2 weeks. The SCENITH assay uses flow cytometry to measure changes in the level of translation in response to inhibitors as a measurement for cellular metabolism. BM cells were divided and separately treated with 2-deoxyglucose (DG), oligomycin (O), and DG + O, together with controls (Cos) incubated without inhibitors, and labeled with puromycin (Puro). After cell surface labeling and intracellular labeling for Puro, different subpopulations were analyzed via flow cytometry for Puro-MFI response to the various inhibitors. Calculations of metabolic dependencies and capacities based on Puro-MFI are shown. (B-C) GMP, common myeloid progenitor (CMP), MEP, and LSK cell frequency within CML c-Kit+ cells (n = 5-6), as assessed via flow cytometry, after treatment with TKI for 2 days (B) or 2 weeks (C). (D-E) Protein synthesis (Puro-MFI) within committed progenitors (GMPs, CMPs, and MEPs) after 2 days (D) and 14 days (E) of TKI treatment. (F-I) Mitochondrial dependence and glycolytic capacity within GMPs, CMPs, and MEPs after 2 days (F,H) and 14 days (G,I) of TKI treatment. Significance values: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001. Results represent mean ± SEM of multiple replicates. AAO, amino acid oxidation; MFI, mean fluorescence intensity.
The number and frequency of GMPs within c-Kit+ cells was reduced, and megakaryocytic-erythroid progenitors (MEPs) was increased, after TKI treatment (Figure 3B-C; supplemental Figure 4B-D). Focusing on committed progenitors, protein synthesis was reduced in GMPs and common myeloid progenitors, but not MEPs, after 2 weeks of TKI treatment (Figure 3D-E). Mitochondrial dependence in MEPs was unchanged after 2 days but was increased after 2 weeks of TKI treatment, with a reduction in glycolytic capacity (Figure 3F-I). Glucose dependency was not altered (supplemental Figure 4E-H). We conclude that TKI treatment leads to both selection of c-Kit+ subpopulations and metabolic reprogramming within the selected populations.
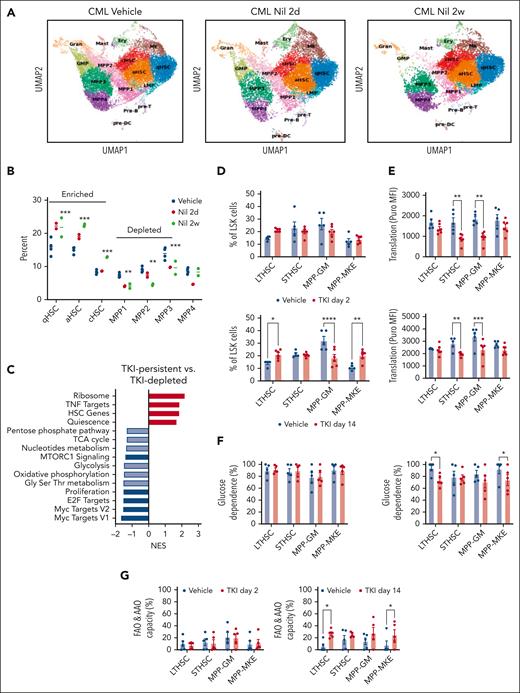
Primitive, quiescent CML stem cell subpopulations with downregulated metabolic gene signatures are enriched after TKI treatment
We performed single-cell RNA (scRNA) sequencing of BM LSK cell populations to determine the effects of TKI on expression of metabolism-related genes in primitive stem cells. Clustering and cell type identification showed a continuum of quiescent HSCs (qHSCs), activated HSCs, cycling HSCs (cHSCs), MPPs, lymphoid-biased MPPs, GMPs, and megakaryocytic progenitors within LSK cells (Figure 4A). Treatment with TKI for 2 weeks increased the frequency and number of qHSCs, activated HSCs, and cHSCs (TKI-persistent), and reduced frequency and number of MPPs (TKI-depleted), (Figure 4A-B; supplemental Figure 5A-B). TKI-persistent clusters from untreated mice were characterized by reduced OXPHOS, glycolysis, and nucleotide metabolism gene signatures; reduced MYC, E2F, and proliferation signatures; and enriched HSC and quiescence signatures (Figure 4C; supplemental Figure 5C). TKI-sensitive and TKI-persistent subpopulations both showed reduced expression of OXPHOS-, MYC-, and E2F-related signatures after 2 days of TKI treatment (supplemental Figure 5D). Further reduction in glycolysis, TCA cycle, and amino acid signatures was observed in TKI-depleted, but not TKI-persistent subpopulations, after 2 weeks of TKI treatment. TKI-persistent subpopulations showed partial restoration of OXPHOS, MYC, and E2F signatures after 2 weeks of treatment (supplemental Figure 5D-F).
Enrichment of primitive CML stem cell subpopulations after TKI treatment. LSK cells were isolated from the BM of CML mice treated with vehicle or nilotinib (Nil) for 2 days or 2 weeks, and scRNA sequencing was performed using the 10x Genomics platform. Cluster identification was based on gene profiles. Clusters are color coded. (A) Uniform manifold approximation and projection (uMAP) display of scRNA sequencing from CML LSK cells treated with vehicle (n = 42 238 single cells, 4 samples), nilotinib for 2 days (n = 15 911 single cells; 2 samples), or nilotinib for 2 weeks (n = 11 449 single cells; 2 samples). (B) The percent of indicated clusters within CML LSK after treatment with vehicle, NIL for 2 days, or NIL for 2 weeks. (C) Gene set enrichment analysis subpopulations of gene sets (FDR < 0.05) comparing TKI-persistent with TKI-depleted clusters. Net enrichment score (NES), and statistical significance (FDR) are represented by color and size, respectively. (D-G) The SCENITH assay was performed on BM cells from CML mice treated with vehicle or NIL (TKI) for 2 days and 14 days (n = 5-6). (D) LTHSC, short-term HSC (STHSC), MPP-GM, and MPP-MKE frequency within CML LSK cells, assessed via flow cytometry, after treatment with TKI for 2 days (left) and 14 days (right). (E) Protein synthesis (Puro-MFI) within LSK subpopulations after 2 days (top) and 14 days (bottom) of TKI treatment. (F-G) Glucose dependence (F) and FAO/AAO capacity (G) within LSK subpopulations after 2 days (top) and 14 days (bottom) of TKI treatment. Significance values: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001. Results represent mean ± SEM of multiple replicates. FDR, false discovery rate; MK, megakaryocytic progenitors; TNF, tumor necrosis factor.
Enrichment of primitive CML stem cell subpopulations after TKI treatment. LSK cells were isolated from the BM of CML mice treated with vehicle or nilotinib (Nil) for 2 days or 2 weeks, and scRNA sequencing was performed using the 10x Genomics platform. Cluster identification was based on gene profiles. Clusters are color coded. (A) Uniform manifold approximation and projection (uMAP) display of scRNA sequencing from CML LSK cells treated with vehicle (n = 42 238 single cells, 4 samples), nilotinib for 2 days (n = 15 911 single cells; 2 samples), or nilotinib for 2 weeks (n = 11 449 single cells; 2 samples). (B) The percent of indicated clusters within CML LSK after treatment with vehicle, NIL for 2 days, or NIL for 2 weeks. (C) Gene set enrichment analysis subpopulations of gene sets (FDR < 0.05) comparing TKI-persistent with TKI-depleted clusters. Net enrichment score (NES), and statistical significance (FDR) are represented by color and size, respectively. (D-G) The SCENITH assay was performed on BM cells from CML mice treated with vehicle or NIL (TKI) for 2 days and 14 days (n = 5-6). (D) LTHSC, short-term HSC (STHSC), MPP-GM, and MPP-MKE frequency within CML LSK cells, assessed via flow cytometry, after treatment with TKI for 2 days (left) and 14 days (right). (E) Protein synthesis (Puro-MFI) within LSK subpopulations after 2 days (top) and 14 days (bottom) of TKI treatment. (F-G) Glucose dependence (F) and FAO/AAO capacity (G) within LSK subpopulations after 2 days (top) and 14 days (bottom) of TKI treatment. Significance values: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001. Results represent mean ± SEM of multiple replicates. FDR, false discovery rate; MK, megakaryocytic progenitors; TNF, tumor necrosis factor.
We also observed reduced proportions and numbers of MPP–granulocyte monocytes (GMs) and increased proportions of LTHSCs and MPP–megakaryocyte and erythrocytes (MPP-MKEs) after 2 weeks of TKI treatment, via flow cytometry, indicating similar trends to those of scRNA analysis (Figure 4D; supplemental Figure 6A). Using SCENITH, we showed that protein synthesis was significantly reduced in short-term HSCs and MPP-GMs, but not LTHSCs or MPP-MKEs, after 2 days and 2 weeks of TKI treatment (Figure 4E). FAO/amino acid oxidation capacity had significantly increased in LTHSCs and MPP-MKEs but not short-term HSCs and MPP-GMs after 2 weeks of TKI treatment (Figure 4F-G). Mitochondrial dependence and glycolytic capacity were not changed (supplemental Figure 6B-C). Analysis of coexisting healthy LSK cells within CML mice indicated the enrichment of healthy LTHSCs (supplemental Figure 7A), without significant changes in protein synthesis, mitochondrial dependence, glycolytic capacity, and substrate dependence after TKI treatment (supplemental Figure 7B-F).
We conclude that primitive CML subpopulations characterized by reduced metabolic signatures and enhanced HSCs and quiescence signatures are enriched after TKI treatment. OXPHOS, MYC, and E2F signatures are initially inhibited in TKI-persistent cells after TKI treatment but partially restored with continued treatment. LTHSCs also demonstrate altered substrate use for energy production, potentially contributing to maintenance of their metabolic activity and enrichment with continued TKI treatment.
Adaptive alterations in metabolic gene signatures in quiescent leukemia stem cells in response to TKI treatment
CML qHSCs, which are at the apex of the hematopoietic hierarchy, showed reduced expression of OXPHOS signatures after 2 days of TKI treatment, with recovery to levels similar to those in vehicle controls after 2 weeks of treatment (Figure 5A; supplemental Figure 8A). MYC and ribosome/translation gene signatures were also reduced after 2 days and partially restored after 2 weeks of treatment. In contrast, quiescence and HSC signatures were enriched and E2F signatures depleted after both 2 days and 2 weeks of treatment (Figure 5B; supplemental Figure 8B-D).
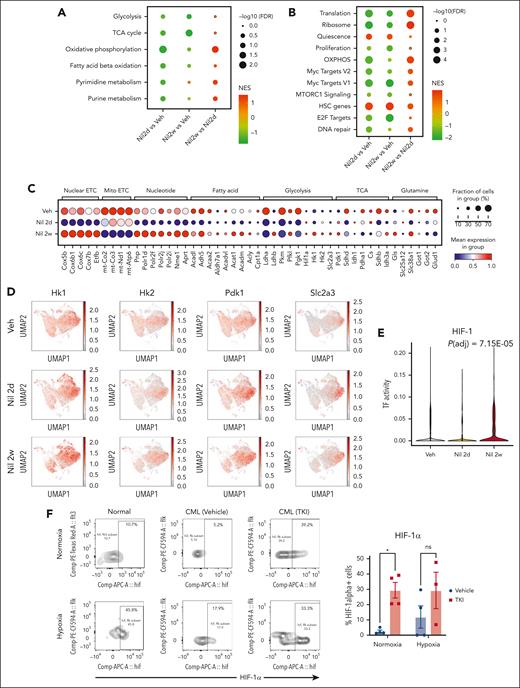
Alterations in metabolic gene signatures in primitive quiescent leukemia stem cells in response to TKI treatment. (A-B) Dot plots showing enrichment of metabolism-related gene sets (A) and hallmark gene sets (B) in the qHSC cluster, comparing cells treated with nilotinib for 2 days (Nil 2d) with vehicle (Veh) (left), with nilotinib for 2 weeks (Nil 2w) vs vehicle (middle), or with nilotinib for 2 weeks vs 2 days (right). NES and statistical significance (FDR) are represented by color and size, respectively. (C) Dot plot showing expression of genes within metabolic pathways in CML qHSC, treated with Veh, Nil 2d, and Nil 2w. The statistical significance (FDR) and fraction of cells expressing the gene are represented by color and size, respectively. (D) uMAP showing the expression of Hk1, Hk2, Pdk1, and Slc2a3 in CML LSK cells treated with vehicle, Nil 2d, or Nil 2w. (E) Enrichment of the HIF-1 regulon in CML qHSC treated with Nil 2d or Nil 2w compared with Veh, based on pySCENIC analysis. (F) The percentage of HIF-1α–expressing normal LTHSCs, CML LTHSCs treated with vehicle, and CML LTHSCs treated with Nil (TKI) for 2 weeks in normoxic or hypoxic conditions (n = 3-4). Significance values: ∗P < .05. Results represent mean ± SEM of multiple replicates.
Alterations in metabolic gene signatures in primitive quiescent leukemia stem cells in response to TKI treatment. (A-B) Dot plots showing enrichment of metabolism-related gene sets (A) and hallmark gene sets (B) in the qHSC cluster, comparing cells treated with nilotinib for 2 days (Nil 2d) with vehicle (Veh) (left), with nilotinib for 2 weeks (Nil 2w) vs vehicle (middle), or with nilotinib for 2 weeks vs 2 days (right). NES and statistical significance (FDR) are represented by color and size, respectively. (C) Dot plot showing expression of genes within metabolic pathways in CML qHSC, treated with Veh, Nil 2d, and Nil 2w. The statistical significance (FDR) and fraction of cells expressing the gene are represented by color and size, respectively. (D) uMAP showing the expression of Hk1, Hk2, Pdk1, and Slc2a3 in CML LSK cells treated with vehicle, Nil 2d, or Nil 2w. (E) Enrichment of the HIF-1 regulon in CML qHSC treated with Nil 2d or Nil 2w compared with Veh, based on pySCENIC analysis. (F) The percentage of HIF-1α–expressing normal LTHSCs, CML LTHSCs treated with vehicle, and CML LTHSCs treated with Nil (TKI) for 2 weeks in normoxic or hypoxic conditions (n = 3-4). Significance values: ∗P < .05. Results represent mean ± SEM of multiple replicates.
Analysis of genes contributing to OXPHOS signatures showed initially reduced but subsequently restored expression of nuclear-encoded, but not mitochondrially encoded, electron transport chain genes after 2 weeks of TKI treatment (Figure 5C). We also observed restored expression of specific genes representing critical steps in glycolysis (Hk1, Hk2, Pfkl, and Ldhb), the TCA cycle (Pdk1 and Idh3a), glutaminolysis (Gls), FAO (Acadl, Acadm, Acat1, and Acly), and glucose and glutamine transport (Slc38a1 and Slc2a3) after 2 weeks of TKI treatment. An increase in Pdk1 coupled with the decrease in Pdha1 is consistent with reduced glucose oxidation and increased lactate labeling observed upon metabolite analysis.
Several metabolic genes upregulated in qHSCs after 2 weeks of TKI treatment were classical HIF-1 transcription factor (TF) targets, including Hk1, Hk2, Pdk1, and Slc2a3 (Figure 5D). Hif1a transcript levels had also increased. Using pySCENIC analysis, we found that HIF-1 TF regulatory network (regulon) expression was increased within individual CML qHSCs after TKI treatment (Figure 5E).28 TKI treatment also altered other regulons important for metabolism, including increased MYC, nuclear respiratory factor 1 (NRF-1), CCAAT enhancer binding protein α, and TP53 regulon expression and reduced FOXO3 and STAT3 regulon expression (supplemental Figure 8E).
HIF-1 is a central regulator of cellular response to hypoxia but can be noncanonically upregulated in cancers. HIF-1 is a heterodimer composed of a constitutively expressed Hif-1β subunit, and a hypoxia-inducible Hif-1α subunit. HIF-1α protein levels measured using intracellular labeling and flow cytometry were significantly increased in CML LTHSCs after 2 weeks, (Figure 5F) but not 2 days (supplemental Figure 8F), of TKI treatment. In contrast to healthy LTHSCs, Hif-1α protein levels in TKI-treated CML LTHSCs were similar under hypoxic and normoxic conditions, suggestive of reduced sensitivity to degradation. Consistent with this, CML qHSCs showed reduced expression of the oxygen-sensitive Egln1, Egln2, and Egln3 proline hydroxylases, which mediate proteasomal degradation of HIF-1α29 after TKI treatment (supplemental Figure 8G). We conclude that TKI treatment increases Hif-1α levels and HIF-1 target gene expression in qHSCs.
HIF-1 inhibition depletes CML stem cells in combination with TKI treatment
We investigated the role of HIF-1 in maintenance of CML stem cells using echinomycin, a dimeric peptide antibiotic HIF-1 inhibitor (HIFi) that blocks the DNA binding of HIF-1.30 We treated a cohort of CML mice with vehicle, HIFi, nilotinib, or the combination for 2 weeks (Figure 6A). Nilotinib reversed PB neutrophilia and depleted leukemic stem and progenitor cells in the spleen but not in the BM. In contrast, compared with vehicle or nilotinib treatment alone, the combination of HIFi with nilotinib led to the significant depletion of CML BM stem and progenitor cells (Figure 6B-D; supplemental Figure 9A-D). BM cells from mice treated with HIFi, with or without nilotinib, demonstrated markedly reduced repopulating capacity via limiting dilution transplantation analysis (Figure 6E; supplemental Figure 9E). Mice receiving HIFi and nilotinib in combination also showed enhanced survival after completion of treatment compared with mice that received nilotinib alone (Figure 6F). These results indicate a critical role for HIF-1 in CML stem cell maintenance after TKI treatment.
HIF-1 inhibition depletes CML stem cells in combination with TKI treatment. (A) Experimental strategy. BM cells (2 × 106) from BCR-ABL transgenic mice (CD45.1/2) in which leukemia had been induced via tetracycline withdrawal were transplanted into CD45.1 recipient mice. Once mice had developed leukemia 6 or 8 weeks after transplantation, they were treated with vehicle, nilotinib (TKI, 50 mg/kg per day by oral gavage), HIFi (echinomycin 10 mg/kg, intraperitoneal for 5 consecutive days, with 2 days off times, 2 cycles), or the combination for 14 days. Mice were euthanized, and PB, the BM, and the spleen were harvested for analysis (n = 6-10). (B) Total neutrophils (based on differential count) in the PB are shown. (C) Total number of LTHSCs in the spleen are shown. (D) Total number of LTHSCs per 2 femurs and 2 tibiae (4 bones) are shown. (E) BM cells from CML mice treated with vehicle, nilotinib, HIFi, or combination for 2 weeks were transplanted in limiting-dilution transplantation (500 000 cells per mouse, n = 8; 1 000 000 cells per mouse, n = 6; and 2 000 000 cells per mouse, n = 6 each) into CD45.1 recipient mice together with competing normal CD45.2 cells (100 000 cells per mouse). Donor cell engraftment in the PB was evaluated after 16 weeks. (F) Kaplan-Meier plots of CML mice treated with vehicle, TKI, HIFi, or combination for 2 weeks and then followed-up for survival (n = 10 each). Log-rank test indicated significantly increased survival for combination- vs TKI-treated mice (P = .012). (G) Irradiated NRGS mice (400 cGy) received transplantation with human CML CD34+ BM cells (0.5×106 cells per mouse). After successful human cell engraftment was confirmed after 6 to 8 weeks, mice were treated with vehicle, nilotinib, HIFi, or combination for 14 days. Mice were euthanized and the BM was harvested for analysis (n = 6-8). (H-K) The absolute number of human CD45+ cells (H), human CD34+CD38+ cells (I), and human CD34+CD38−CD90+ cells (J) are shown. (K) The percent of human CD34+CD38−CD90+ cells expressing HIF-1α after vehicle and nilotinib treatment for 2 weeks are shown. Results represent mean ± SEM of multiple replicates. Significance values: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗ P < .0001. Hu, human; WT, wild-type.
HIF-1 inhibition depletes CML stem cells in combination with TKI treatment. (A) Experimental strategy. BM cells (2 × 106) from BCR-ABL transgenic mice (CD45.1/2) in which leukemia had been induced via tetracycline withdrawal were transplanted into CD45.1 recipient mice. Once mice had developed leukemia 6 or 8 weeks after transplantation, they were treated with vehicle, nilotinib (TKI, 50 mg/kg per day by oral gavage), HIFi (echinomycin 10 mg/kg, intraperitoneal for 5 consecutive days, with 2 days off times, 2 cycles), or the combination for 14 days. Mice were euthanized, and PB, the BM, and the spleen were harvested for analysis (n = 6-10). (B) Total neutrophils (based on differential count) in the PB are shown. (C) Total number of LTHSCs in the spleen are shown. (D) Total number of LTHSCs per 2 femurs and 2 tibiae (4 bones) are shown. (E) BM cells from CML mice treated with vehicle, nilotinib, HIFi, or combination for 2 weeks were transplanted in limiting-dilution transplantation (500 000 cells per mouse, n = 8; 1 000 000 cells per mouse, n = 6; and 2 000 000 cells per mouse, n = 6 each) into CD45.1 recipient mice together with competing normal CD45.2 cells (100 000 cells per mouse). Donor cell engraftment in the PB was evaluated after 16 weeks. (F) Kaplan-Meier plots of CML mice treated with vehicle, TKI, HIFi, or combination for 2 weeks and then followed-up for survival (n = 10 each). Log-rank test indicated significantly increased survival for combination- vs TKI-treated mice (P = .012). (G) Irradiated NRGS mice (400 cGy) received transplantation with human CML CD34+ BM cells (0.5×106 cells per mouse). After successful human cell engraftment was confirmed after 6 to 8 weeks, mice were treated with vehicle, nilotinib, HIFi, or combination for 14 days. Mice were euthanized and the BM was harvested for analysis (n = 6-8). (H-K) The absolute number of human CD45+ cells (H), human CD34+CD38+ cells (I), and human CD34+CD38−CD90+ cells (J) are shown. (K) The percent of human CD34+CD38−CD90+ cells expressing HIF-1α after vehicle and nilotinib treatment for 2 weeks are shown. Results represent mean ± SEM of multiple replicates. Significance values: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗ P < .0001. Hu, human; WT, wild-type.
To investigate the role of HIF-1 in maintaining human CML stem cells, CML CD34+ cells were xenografted in immunodeficient NRGS mice and mice treated with vehicle, HIFi, nilotinib, or combination for 2 weeks (Figure 6G). Nilotinib reduced human leukemic CD45+ cells, CD34+CD38+ progenitors, and CD34+CD38+CD90− MPPs, but not primitive CD34+CD38−CD90+ stem cells, in the BM (Figure 6H-J; supplemental Figure 9F-G). In contrast, HIFi by itself and in combination with nilotinib treatment caused a significant depletion of CD34+CD38−CD90+ stem cells in the BM (Figure 6J). Nilotinib increased HIF-1α protein levels in human CD34+CD38−CD90+ stem cells (Figure 6K). These results indicate that HIF-1 also plays an important role in maintaining human CML stem cells after TKI treatment.
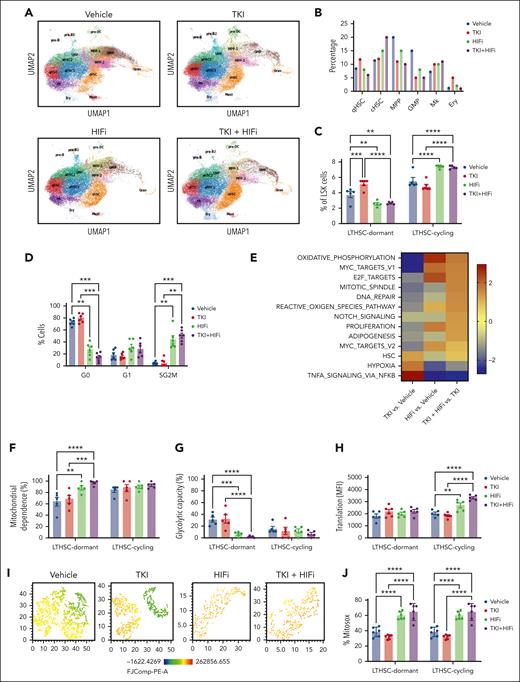
HIF-1 inhibition enhances mitochondrial activity, ROS, and cell cycling in TKI-treated CML stem cells
We performed scRNA sequencing analysis of LSK cells from CML mice treated with HIFi and nilotinib (Figure 7A). Combination with HIFi reduced qHSCs and increased cHSCs in nilotinib-treated cells (Figure 7B). Flow cytometry confirmed that the combination reduced dormant (CD34−) LTHSCs and increased cycling (CD34+) LTHSCs, compared with nilotinib alone (Figure 7C, Supplemental Figure 10A). Cell cycle analysis using Ki67-DAPI labeling showed that HIFi reduced quiescence and enhanced cycling of nilotinib-treated CML LTHSCs (Figure 7D). HIFi reduced the expression of HIF target genes (Hk1, Hk2, Slc2a3, and Pdk1) in nilotinib-treated qHSCs (Supplemental Figure 10B). Gene set enrichment analysis showed that combination treatment increased the expression of OXPHOS, MYC, cell cycling (proliferation G2M, E2F, and mitotic), and ROS signatures in qHSCs and reduced the expression of hypoxia and tumor necrosis factor signatures, compared with controls or nilotinib alone (Figure 7E). HIFi also increased nucleotide metabolism, pyruvate and glucose levels, FAO, and glutathione signatures (supplemental Figure 10C).
HIF-1 inhibition enhances mitochondrial activity, ROS, and cell cycling in TKI-treated CML stem cells. scRNA sequencing analysis was performed on LSK cells from CML mice treated with vehicle, nilotinib (TKI), echinomycin (HIFi), or the combination (TKI + HIFi). (A) uMAP display of CML LSK cells treated with vehicle (n = 12 368 single cells; 4 samples), TKI (n = 19 866 single cells; 2 samples), HIFi (n = 18 686 single cells; 4 samples), or the combination (n = 19 836 single cells; 4 samples). (B) The percentage of cells within individual clusters after treatment, as indicated. (C) Flow cytometry analysis of dormant LTHSCs (CD34−) and cycling LTHSCs (CD34+) after treatment, as indicated. (D) Cell cycle analysis of LTHSCs from CML mice treated with vehicle, TKI, HIFi, or the combination (n = 6 each) using Ki67-DAPI labeling. (E) NES of hallmark gene sets (FDR < 0.05) comparing combination-, TKI-, and vehicle-treated qHSCs. (F-H) Results of SCENITH analysis showing changes in protein synthesis (metabolic activity) (F), mitochondrial dependence (G), and glycolytic capacity (H) in cycling and dormant LTHSCs from mice receiving the indicated treatments (n = 5-6). (I) t-distributed stochastic neighbor embedding plot showing TMRM fluorescence in dormant LTHSCs from mice receiving the indicated treatments (n = 6, concatenated). (J) Mitochondrial ROS levels measured using Mitosox in dormant and cycling LTHSCs from mice receiving the indicated treatments (n = 6 each). Results represent mean ± SEM of multiple replicates. Significance values: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
HIF-1 inhibition enhances mitochondrial activity, ROS, and cell cycling in TKI-treated CML stem cells. scRNA sequencing analysis was performed on LSK cells from CML mice treated with vehicle, nilotinib (TKI), echinomycin (HIFi), or the combination (TKI + HIFi). (A) uMAP display of CML LSK cells treated with vehicle (n = 12 368 single cells; 4 samples), TKI (n = 19 866 single cells; 2 samples), HIFi (n = 18 686 single cells; 4 samples), or the combination (n = 19 836 single cells; 4 samples). (B) The percentage of cells within individual clusters after treatment, as indicated. (C) Flow cytometry analysis of dormant LTHSCs (CD34−) and cycling LTHSCs (CD34+) after treatment, as indicated. (D) Cell cycle analysis of LTHSCs from CML mice treated with vehicle, TKI, HIFi, or the combination (n = 6 each) using Ki67-DAPI labeling. (E) NES of hallmark gene sets (FDR < 0.05) comparing combination-, TKI-, and vehicle-treated qHSCs. (F-H) Results of SCENITH analysis showing changes in protein synthesis (metabolic activity) (F), mitochondrial dependence (G), and glycolytic capacity (H) in cycling and dormant LTHSCs from mice receiving the indicated treatments (n = 5-6). (I) t-distributed stochastic neighbor embedding plot showing TMRM fluorescence in dormant LTHSCs from mice receiving the indicated treatments (n = 6, concatenated). (J) Mitochondrial ROS levels measured using Mitosox in dormant and cycling LTHSCs from mice receiving the indicated treatments (n = 6 each). Results represent mean ± SEM of multiple replicates. Significance values: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Using the SCENITH assay, we found that, compared with vehicle or nilotinib alone, combination treatment increased mitochondrial dependence in dormant LTHSCs (Figure 7F-G), increased glucose dependence in dormant and cycling LTHSCs (supplemental Figure 10D-E), and increased protein synthesis in cycling LTHSCs (Figure 7H). TMRM fluorescence, a measure of mitochondrial membrane potential which is an essential component of OXPHOS, was increased in dormant and cycling LTHSCs via combination treatment compared with via vehicle or nilotinib alone (supplemental Figure 10F).31 Indeed, HIFi treatment eliminated TMRMlow cells within dormant LTHSCs (Figure 7I). ROS activity is linked to OXPHOS and plays an important role in LTHSC dormancy.32 HIFi treatment increased cytoplasmic and mitochondrial ROS, measured using 2',7'-dichlorodihydrofluorescein diacetate and Mitosox labeling respectively, in dormant and cycling LTHSCs (Figure 7J; supplemental Figure 10G). These results support the role of HIF-mediated inhibition of OXPHOS and ROS levels in maintaining dormancy, repopulating potential, and TKI insensitivity of CML stem cells.
Discussion
Resistance of CML stem cells to TKI treatment highlights the limits of curative potential of targeted therapies and the need for additional targeted approaches. To our knowledge, this study has made several novel findings related to metabolic adaptation of CML cells to TKI therapy. TKI treatment enriches primitive CML stem cell subsets with reduced metabolic gene expression. CML stem cells adapt to TKI treatment through altered substrate use and maintenance of OXPHOS. We show that enhanced activity of HIF-1, a key regulator of energy metabolism, after TKI treatment plays a critical role in inhibiting OXPHOS and ROS in primitive CML stem cells, maintaining stem cell dormancy, repopulating potential, and TKI insensitivity.
We show that TKI treatment induces dynamic changes in energy metabolism in CML-committed progenitors. TKI treatment initially inhibited OXPHOS, glycolysis, glutaminolysis, and TCA cycle activity in c-Kit+ CML-committed progenitor cells. However, glycolytic and glutaminolytic activity was restored, and glucose-mediated lactate generation, TCA cycle activity, and amino acid synthesis was increased with continued TKI treatment. We show that these metabolic changes are related to both the selection of distinct progenitor subpopulations and metabolic reprogramming within the selected subpopulations.
scRNA sequencing revealed that primitive CML stem cell subpopulations characterized by reduced OXPHOS and increased stem cell and quiescence signatures were enriched after TKI treatment. We have shown that TKI-sensitive CML cells are more reliant on OXPHOS compared with persistent cells.33 In normal hematopoiesis, qHSCs exhibit low OXPHOS levels but switch to a high OXPHOS state after activation associated with reduced self-renewal and enhanced myeloid differentiation.34,35 Primitive CML cells reportedly have upregulated OXPHOS.19 However, the most primitive CML qHSCs did not show increased OXPHOS compared with healthy qHSCs (data not shown). These metabolically inert features of CML qHSCs may contribute to difficulties in eliminating these cells. TKI treatment initially led to further inhibition of expression of metabolic gene signatures, but continued treatment led to the partial restoration of OXPHOS and MYC signatures in TKI-persistent subpopulations. Therefore, TKI treatment leads to both the selection of metabolically distinct primitive stem cell subpopulations and adaptive changes in persistent cells. Our results suggest that altered substrate use could contribute to maintenance of energy metabolism, which will be an important area for future investigation.
We identified an increased HIF-1 activity as a critical mechanism of CML stem cell adaptation to TKI treatment. Enhanced Hif-1α expression after TKI treatment may be related to reduced oxygen-induced degradation. HIF-1 functions as a primary regulator of hypoxic response by activating transcription of genes involved in energy metabolism, angiogenesis, and oxygen delivery. Hif-1α–deficient mice show the loss of HSC quiescence and depleted HSC numbers under stress.36 However, cell-intrinsic Hif-1α deletion does not affect HSC survival or regenerative capacity.37 BCR/ABL expression induces Hif-1α expression in cell lines.38 HIF-1 increases glycolysis and reduces proliferation in imatinib-resistant BCR-ABL lines.39 HIF-1 is also reported to support BCR-ABL–expressing LSK cells40 and clonogenicity of CML CD34+ cells during hypoxia,41 and to modify effects of TKI treatment.42
HIF-1 regulates the balance between oxidative and glycolytic metabolism by transactivating glucose transporters and glycolytic enzymes, diverting glucose-derived pyruvate to lactate, and switching nuclear-encoded cytochrome oxidase subunit composition to optimize respiratory efficiency.43-45 These effects are consistent with metabolic changes observed in quiescent CML stem cells after TKI treatment. HIF-1 inhibition reduced expression of known HIF-1 targeted glycolytic regulatory genes, increased mitochondrial activity and ROS generation, reduced quiescence, increased cycling, and reduced self-renewal and regenerating potential of CML stem cells. HIF1/2 regulation of glycolytic gene expression was not functionally relevant in leukemia cell lines.46 HIF-1α and HIF-2α play important roles in protecting HSC and AML cells from ROS.36,47,48 Our results support an important role for HIF-1–mediated inhibition of OXPHOS and ROS in maintaining leukemic stem cell dormancy, repopulating potential, and TKI insensitivity.
TKI treatment also leads to increased activity of other TFs with metabolic regulatory function, including MYC and NRF-1. MYC is a master regulator of metabolic reprogramming in human cancers and is required for CML development and maintenance.49,50 However, regulated thresholds of MYC abundance and activity are required for leukemia stem cell activity and drug resistance.51,52 CML stem cells with lower MYC target gene expression were enriched via TKI treatment. Sustained anabolism and proliferation in cells with higher MYC activity may confer increased TKI sensitivity. HIF-1 can counteract MYC transcriptional activity, and may contribute to maintenance of CML stem cell quiescence and survival with continued TKI treatment.53 Indeed, HIF-1 inhibition increased expression of MYC gene signatures in TKI-treated CML stem cells. We previously showed a role for Sirtuin1 (SIRT1) overexpression in CML progenitors in increased OXPHOS via modulation of PPAR-γ coactivator 1-α, an important coactivator of NRF-1.20 Increased NRF-1 activity could contribute to the restoration of OXPHOS by enhancing nuclear mitochondrial respiratory gene expression, and its interaction with HIF-1 in CML stem cells warrants further study.
We conclude that HIF-1 is a critical adaptive mechanism and dependency in CML stem cells after TKI treatment. Future studies to elucidate mechanisms of HIF-1 regulation in leukemia stem cells, including interactions with the BM microenvironment, will be of considerable interest. Our findings further support a vital role for HIF-1 signaling in tumorigenesis and resistance in cancers and emphasize the critical need for development of therapeutic HIFis.
Acknowledgments
The authors thank Vidya Sagar Hanumanthu and Shanrun Liu at the UAB Comprehensive Flow Cytometry Core for providing help with fluorescence-activated cell sorting and scRNA sequencing; the UAB Animal Resources Center for maintaining mice colonies; Maya Robinson and Amanda Mullens for processing human samples; and Mason Harris for assistance with animal studies.
This work was supported by National Cancer Institute, National Institutes of Health grants (R01 CA248794 and R01 CA172447) (R.B.).
Authorship
Contribution: S.Q., V.S., and R.B. conceptualized the study; S.Q., V.S., C.Y., J.L., B.K.C., H.L., S.A., A.R.W., D.K.C., S.D.F., A.J.P., and R.L. performed study investigations; S.Q., V.S., and R.B. performed analyses; R.B., V.M.D.-U., and J.W.L. provided resources; D.K.C. performed data curation; R.B., S.Q. and V.S. wrote the original draft; R.B., S.Q., V.S., M.B.G., A.J.P., A.R.W., R.S.W., V.M.D.-U., R.L., and J.W.L wrote, reviewed, and edited the manuscript; R.B. acquired funding; and R.B. supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ravi Bhatia, Division of Hematology & Oncology, Department of Medicine, University of Alabama at Birmingham, 1802 6th Ave South, North Pavilion, Room 2555C, Birmingham, AL 35294; e-mail: rbhatia@uabmc.edu.
References
Author notes
∗S.Q. and V.S. contributed equally to this study.
The scRNA sequencing data are deposited in The National Center for Biotechnology Information’s Gene Expression Omnibus database (accession number GSE207346).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![TKI treatment leads to dynamic alterations in energy metabolism in CML progenitors. Mice with CML were treated with nilotinib or vehicle for 2 days or 2 weeks, and BM c-Kit+ cells were selected and metabolomic profiling was performed. (A) The relative abundance of glycolytic intermediates after 2 days of nilotinib treatment is shown (n = 3-4). (B) The relative abundance of TCA cycle intermediates after 2 days of nilotinib treatment is shown (n = 3-4). (C) The ratio of ATP/ADP and GTP/GDP after 2 days of nilotinib treatment is shown (n = 3-4). (D) The relative abundance of glycolytic intermediates after 2 weeks of nilotinib treatment is shown (n =3). (E) The relative abundance of TCA cycle intermediates after 2 weeks of nilotinib treatment is shown (n = 3). (F) The ratio of ATP/ADP and GTP/GDP after 2 weeks of nilotinib treatment is shown (n = 3). (G) BM c-Kit+ cells were labeled with [U-13C6]glucose in vitro (n = 3). The percent labeling fraction of glycolytic end products (pyruvate, lactate, and alanine) and citric acid cycle intermediates (citrate/isocitrate, α-ketoglutarate, and malate) after 2 days and after 2 weeks of vehicle (Veh) or nilotinib (NIL) treatment is shown. The labeling fractions are corrected for 13C natural abundance. Significance values: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. Results represent mean ± SEM of multiple replicates. 3PG/2PG, 3-phosphoglyceric acid/2-phosphoglyceric acid; ADP, adenosine diphosphate; a-KG, α-ketoglutarate; F-1,6-BP, fructose 1,6-bisphosphate; G6P/F6P, glucose 6-phosphate/fructose 6-phosphate; GDP, guanosine diphosphate; GSH, glutathione; GTP, guanosine triphosphate; ns, not significant; PEP, phosphoenolpyruvic acid.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/142/6/10.1182_blood.2022018196/2/m_blood_bld-2022-018196-gr2.jpeg?Expires=1769099688&Signature=n6hjdMwfGyy0ClbljfawudatElYEE6l9AlWjpR-daGfj1NPOr8m180cK8Wm-zogYlfV4xMHIjFqG1Y50whuvWHu6girmz5ul0XyScisS1JsM11ptnFuj-n9CDsF1na7vQXfZ~GtT0l~eIDyec2TbHmXgTgN-cj8ojRU6ZZE1f7u9y4TkZ22EUHNEs6jfdnTBDGaG0uVa9Dc09EfeieRccT5hwoZi1yFO09hHQicZ7kWmeZ2i5uX10qICVaybPteAkBtpnvw3DNdxdHHLlRYxWyzoB~RFPN2jbUsYT2cmqiLyn~92jrC-8Su4uewFAitIAJzeo8v6aXzcql06cu-Agg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal