Key Points
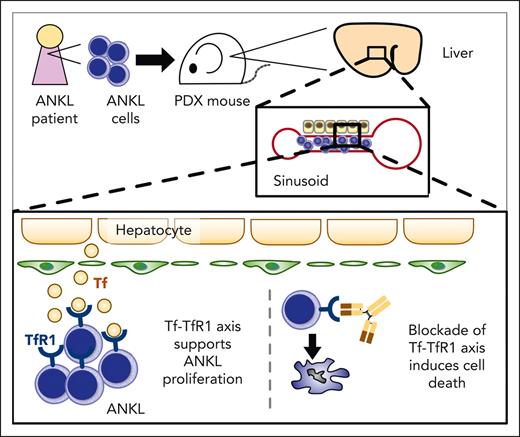
The liver, a noncanonical hematopoietic organ in adults, serves as a principal niche for ANKL.
Targeting TfR1 with monoclonal antibody PPMX-T003 is a promising strategy for treating ANKL.
Abstract
Aggressive natural killer cell leukemia (ANKL) is a rare lymphoid neoplasm frequently associated with Epstein-Barr virus, with a disastrously poor prognosis. Owing to the lack of samples from patients with ANKL and relevant murine models, comprehensive investigation of its pathogenesis including the tumor microenvironment (TME) has been hindered. Here we established 3 xenograft mice derived from patients with ANKL (PDXs), which enabled extensive analysis of tumor cells and their TME. ANKL cells primarily engrafted and proliferated in the hepatic sinusoid. Hepatic ANKL cells were characterized by an enriched Myc-pathway and proliferated faster than those in other organs. Interactome analyses and in vivo CRISPR-Cas9 analyses revealed transferrin (Tf)–transferrin receptor 1 (TfR1) axis as a potential molecular interaction between the liver and ANKL. ANKL cells were rather vulnerable to iron deprivation. PPMX-T003, a humanized anti-TfR1 monoclonal antibody, showed remarkable therapeutic efficacy in a preclinical setting using ANKL-PDXs. These findings indicate that the liver, a noncanonical hematopoietic organ in adults, serves as a principal niche for ANKL and the inhibition of the Tf-TfR1 axis is a promising therapeutic strategy for ANKL.
Introduction
Aggressive natural killer cell leukemia (ANKL) is a rare form of NK-cell neoplasm with an extremely poor prognosis.1 The term ANKL was first proposed in 1986 and categorized as a distinct entity since 2001 in the World Health Organization Classification.2,3 ANKL is known to be more prevalent among Asians than in other ethnic populations.1 However, a growing number of White or Latin American cases has been reported.3 According to a recent epidemiological study, 76.4% of ANKL cases in the US–affected White populations.4 Clinically, ANKL cells commonly exist in the bone marrow (BM), peripheral blood (PB), liver, and spleen. Although Epstein-Barr virus (EBV) reactivation was observed in most ANKL cases,1 little is known about the pathogenesis of ANKL and no standard of care has been established.3,5
Several studies have shown that ANKL and extranodal NK/T-cell lymphoma, another NK-cell malignancy, have common genomic abnormalities in DDX3X, STAT3, and JAK/STAT-Myc pathway–related genes.6-8 Thus, these 2 NK-cell malignancies cannot be distinguished by the relevant genetic backgrounds.
Recently, the tumor microenvironment (TME) has been suggested as a crucial factor in the tumorigenesis of various neoplasms.9-11 For instance, in hematological malignancies, our previous studies showed that EBV–reactivated B-cell lymphoma adjusts the TME through extracellular vesicles for their survival and progression.12,13 With this background, we decided to focus on the communication of ANKL cells with their TME. In this study, to assess the ANKL pathophysiology in vivo, we established xenograft mice derived from patients with ANKL (PDXs), which reflect the pathophysiology of the human body.14-16
We found that ANKL cells initially engraft and proliferate in liver sinusoids, with dependence on the transferrin (Tf)–transferrin receptor 1 (TfR1) interaction. Moreover, we demonstrated the therapeutic potential of an anti-TfR1 inhibitory antibody, PPMX-T003, in ANKL.
Methods
Human primary ANKL samples and tissue specimens
ANKL tissue samples were obtained from patients for diagnosis or autopsy. Informed consent was obtained for their use in research, in accordance with the Declaration of Helsinki. This study was approved by the institutional review board of Tokai University, Tokyo, Japan.
Mouse strains and PDX establishment
Female NOD/Sci-scid, IL2-Rgnull (NOG) mice (In-Vivo Science, Tokyo, Japan) were used for all in vivo experiments. All mice were housed at the animal facility of Tokai University in a specific pathogen-free area and received humane care. PDXs were established by injecting PB mononuclear cells or BM cells taken from patients, through the tail vein. PB mononuclear cells and BM cells were isolated by density gradient centrifugation using Lymphoprep (STEMCELL Technologies, Vancouver, Canada), resuspended with phosphate-buffered saline, and immediately injected into mice without further enrichment or culture. The ANKL3 cells were obtained from a previously reported PDX.17 In total, samples from 11 patients with T/NK-cell leukemia were used and PDXs were successfully obtained from 6 of these. Three PDXs were excluded from the analysis because the final diagnosis of the patients was not ANKL. Tumor cells were collected from the liver, BM, or spleen and transplanted into other NOG mice. To select liver-derived cells, ANKL cells were collected from the PDX liver and transplanted into a mouse. After 3 to 4 weeks, cells were collected from this PDX liver and transplanted into another mouse. This procedure was repeated 10 times in total. Spleen-derived ANKL cells were selected in the same manner. These procedures were inspired by the selection of brain metastatic breast cancer cell lines.18 Unless specifically stated, 1 million cells were injected per mouse. All animal experiments were approved by the Animal Experimentation Committee of Tokai University.
Leukemic cell collection from the liver, BM, spleen, and PB of mice
To isolate leukemic cells from the liver, the whole liver was minced and mechanically disrupted; cells were harvested and incubated in Hanks balanced salt solution containing collagenase A and DNase I (Sigma-Aldrich) for 20 minutes at 37°C, followed by further purification with Percoll (Cytiva). Bilateral femurs and tibias were excised and flushed to collect BM cells. The spleen was chopped with scissors and gently mashed to prepare a single-cell suspension. To collect PB, 0.5 mL of blood was drawn from the heart of a deeply anesthetized mouse. Red blood cells (RBCs) were lysed (BD Bioscience) and samples were subjected to the subsequent procedure.
Generation of Cas9-expressing ANKL cells and knockout with sgRNA
ANKL cells were collected from the PDX liver, transduced with FUGW-Cas9-Venus vector (modified from lentiCas9-Blast, Addgene), and then injected into mice. ANKL cells were collected after 3 to 4 weeks, sorted based on Venus expression, and serially injected into mice. This collecting-sorting-injection process was performed twice to obtain pure Venus+ ANKL (ANKL-Cas9) cells. ANKL-Cas9 cells were then frozen for future experiments. TFRC-deficient ANKL cells were established by transducing ANKL-Cas9 cells with a single-guide RNA (sgRNA) targeting TFRC (sgTFRC) using a CSII-sgRNA-mOrange vector.
Prediction of molecular interaction
Data sets for predicting molecular interactions in human were derived from the Human Reference Protein Interactome Mapping Project (HuRI; http://www.interactome-atlas.org/).19 Transcripts per million (TPM) values of each gene in ANKL (TPMANKL) and the human liver (TPMLiver) were derived from our bulk RNA-sequencing (RNA-seq) data and Genotype-Tissue Expression (GTEx) project V7 (https://gtexportal.org/home/), respectively. Interaction scores were calculated and ranked by the geometric means of TPMLiver and TPMANKL.
Treatment of ANKL-PDXs with PPMX-T003
ANKL-PDXs were intravenously treated with anti-TfR1 antibody (PPMX-T003; Perseus Proteomics Inc, Tokyo, Japan) according to the indicated schema. In the survival analysis, ANKL-PDXs were intravenously treated with phosphate-buffered saline or 10 mg/kg of PPMX-T003. In an advanced-disease model, treatment was started on day 14, when mice began losing weight.
Additional methods
The details of additional methods including cell lines, antibodies/reagents, flow cytometry, in vivo imaging system (IVIS), microscopic analysis, sgRNA sequence, bulk and single-cell RNA-seq (scRNA-seq), intracellular ferrous staining, and statistical analyses are provided in the supplemental Methods, available on the Blood website.
Results
ANKL-PDXs reproduce human clinical features
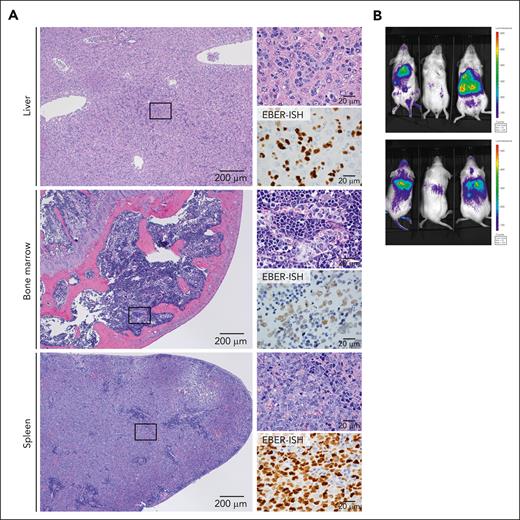
We successfully established 3 PDX strains (ANKL1, ANKL3, and ANKL5) using samples from patients who were EBV-positive (supplemental Figure 1A). All PDXs showed tumor infiltration into the liver, BM, and spleen (Figure 1A; supplemental Figure 1B). Remarkably, in the liver, tumor cells were localized in the sinusoidal region in all strains and in the periportal region in 1 strain (ANKL3). In the BM and spleen, ANKL cells were engrafted in the sinusoidal region (ANKL1 and ANKL5) or interstitial regions (ANKL3) of the BM and diffusely in the red pulp of the spleen (ANKL1, 3, 5). The pathology of the PDXs recapitulated that of patient specimens of the liver, spleen, or BM reported in previous studies.20,21 The PDXs reproduced the immunophenotype observed in the patient, including surface CD3− with flow cytometry, CD3ε+ with immunohistochemistry, and CD56+ as well (supplemental Figure 2A-C). Macroscopically, IVIS analysis using luciferase-transduced ANKL1 or ANKL3 (ANKL1-Luc and ANKL3-Luc) cells revealed exclusive strong bioluminescent signals in the region corresponding to the liver, spleen, and BM (Figure 1B; supplemental Figure 2D). Altogether, these results suggested that PDXs recapitulated the human pathology.
ANKL-PDXs reproduce human clinical features. (A) Hematoxylin and eosin (H&E) staining (left: low-power field; upper-right: high-power field) and EBV-encoded RNA in situ hybridization (EBER-ISH) (lower-right) of the liver, BM, and spleen sections derived from an ANKL1 PDX mouse. Representative image from 3 independent experiments. Scale bar, 200 μm (low-power field) and 20 μm (high-power field). (B) Bioluminescent signal detected using the IVIS in ANKL1 PDX mice (n = 3). The upper and lower panels show the supine and prone positions, respectively.
ANKL-PDXs reproduce human clinical features. (A) Hematoxylin and eosin (H&E) staining (left: low-power field; upper-right: high-power field) and EBV-encoded RNA in situ hybridization (EBER-ISH) (lower-right) of the liver, BM, and spleen sections derived from an ANKL1 PDX mouse. Representative image from 3 independent experiments. Scale bar, 200 μm (low-power field) and 20 μm (high-power field). (B) Bioluminescent signal detected using the IVIS in ANKL1 PDX mice (n = 3). The upper and lower panels show the supine and prone positions, respectively.
ANKL cells primarily proliferate in the liver
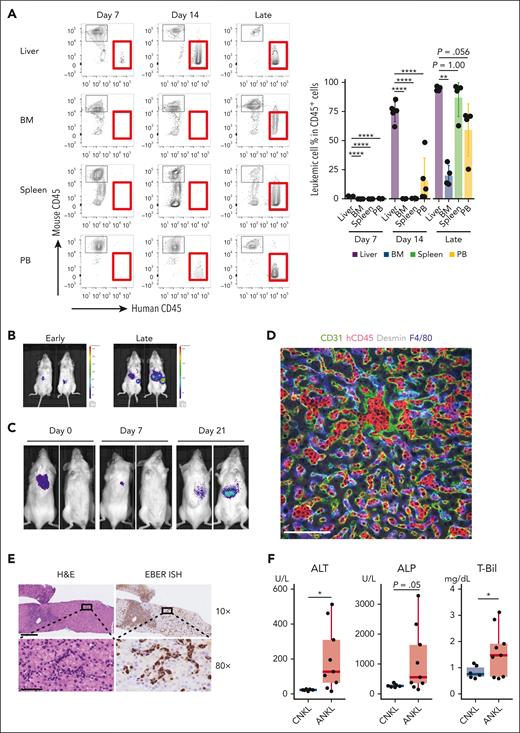
To investigate where ANKL cells primarily engraft and proliferate, the serial pathology after transplantation was evaluated. At 1 week after transplantation, ANKL cells were observed almost exclusively in the liver but not in other organs (Figure 2A; supplemental Figure 3A). The leukemic cells continued to proliferate mainly in the liver until 3 to 4 weeks after transplantation. Finally, leukemia progressed systemically, and mice became weak. Even the injection of splenic ANKL1-Luc or ANKL3-Luc cells resulted in liver engraftment and subsequent dissemination to other organs (Figure 2B; supplemental Figure 3B). In contrast, THP-1, a cell line derived from myeloid leukemia, preferentially engrafted to the BM (supplemental Figure 3C). Carboxyfluorescein diacetate succinimidyl ester–labeled ANKL1 cells in mice exclusively divided in the liver within 3 days after transplantation, even though they had been collected from the spleen (supplemental Figure 3D). To exclude the possibility that this result was caused by the accessibility of tail-vein injection to the liver, leukemic progression was examined after subcutaneous injection of ANKL1-Luc cells into the posterior cervical region. Bioluminescent signals in the neck regions were detected immediately after injection, but gradually disappeared and reappeared in the liver region in 2 out of 3 mice (Figure 2C; supplemental Figure 3E).
ANKL cells primarily proliferate in the liver. (A) The percentage of leukemic cells in each organ of PDX mice analyzed by flow cytometry at days 7 and 14 and 3 to 4 weeks when mice already became weak (late) after splenic ANKL1 cell injection at a dose of 1 × 103 cells (n = 4-5). (B) IVIS imaging of an ANKL1 PDX mouse at an early (1-2 weeks after transplantation) and late (3-4 weeks when mice already got weakened) phase. (C) IVIS imaging at days 0, 7, and 21 after hepatic ANKL1 cell subcutaneous injection at a dose of 1 × 107 cells. Representative image from 3 biological replicates. (D) Expression of CD31, hCD45, F4/80, and desmin in the liver of an ANKL1 PDX mouse. Data are representative of 2 experiments. Scale bar, 100 μm. (E) H&E staining (left) and EBER-ISH (right) for a liver section from a biopsy sample of a patient with newly diagnosed ANKL. Upper and lower images show the ×10 and ×80 original magnification, respectively. Scale bar, 300 μm (×10) and 50 μm (×80). (F) Comparison of liver function between patients with chronic NK-cell lymphocytosis (CNKL) and those with ANKL. Clinical data of alanine transaminase (ALT, left), alkaline phosphatase (ALP, middle), and total bilirubin (T-Bil, right) from the previous study were reanalyzed.22 ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
ANKL cells primarily proliferate in the liver. (A) The percentage of leukemic cells in each organ of PDX mice analyzed by flow cytometry at days 7 and 14 and 3 to 4 weeks when mice already became weak (late) after splenic ANKL1 cell injection at a dose of 1 × 103 cells (n = 4-5). (B) IVIS imaging of an ANKL1 PDX mouse at an early (1-2 weeks after transplantation) and late (3-4 weeks when mice already got weakened) phase. (C) IVIS imaging at days 0, 7, and 21 after hepatic ANKL1 cell subcutaneous injection at a dose of 1 × 107 cells. Representative image from 3 biological replicates. (D) Expression of CD31, hCD45, F4/80, and desmin in the liver of an ANKL1 PDX mouse. Data are representative of 2 experiments. Scale bar, 100 μm. (E) H&E staining (left) and EBER-ISH (right) for a liver section from a biopsy sample of a patient with newly diagnosed ANKL. Upper and lower images show the ×10 and ×80 original magnification, respectively. Scale bar, 300 μm (×10) and 50 μm (×80). (F) Comparison of liver function between patients with chronic NK-cell lymphocytosis (CNKL) and those with ANKL. Clinical data of alanine transaminase (ALT, left), alkaline phosphatase (ALP, middle), and total bilirubin (T-Bil, right) from the previous study were reanalyzed.22 ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Microscopically, leukemic cells were first engrafted in the sinusoidal area (supplemental Figure 4A) and robustly proliferated solely in the sinusoid (ANKL1) or in both the periportal and sinusoidal areas (ANKL3), as confirmed by confocal microscopy results showing leukemic cells in the sinusoidal vasculature, surrounded by endothelial cells, hepatic stellate cells, and macrophages (Figure 2D; supplemental Figures 4B-C and 5).
These results were also examined in humans. The liver specimen of a patient newly diagnosed with ANKL already showed leukemic infiltration of periportal and sinusoidal regions (Figure 2E), whereas the BM examination of the patient failed to reveal a definitive diagnosis. The liver sections of 2 autopsy cases also showed sinusoidal liver infiltration by ANKL cells (supplemental Figure 4D). Moreover, reanalysis of liver enzymes shown from a previous study comparing ANKL with chronic NK-cell lymphocytosis22 indicated markedly worse liver function in ANKL than in chronic NK-cell lymphocytosis (Figure 2F). Taken together, liver sinusoids serve as the primary niche for ANKL, where infiltrating leukemic cells can lead to liver dysfunction.
Liver-derived ANKL cells acquire a more aggressive phenotype characterized by an enriched Myc pathway
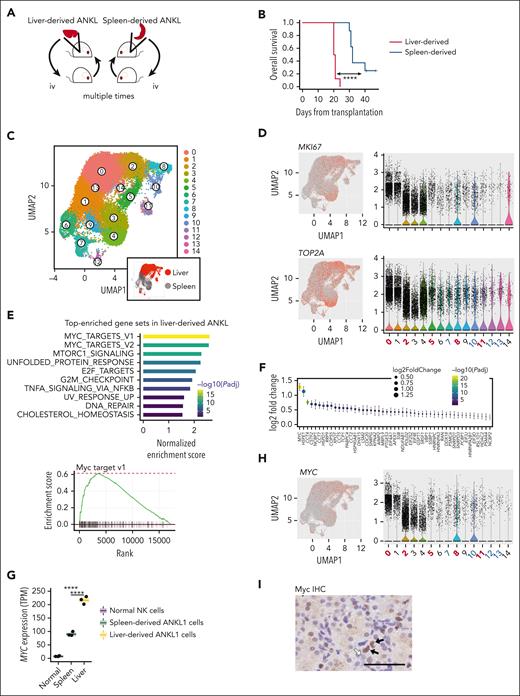
To further clarify the specific features of hepatic ANKL cells, we selected liver- or spleen-derived ANKL cells (Figure 3A). Mice bearing liver-derived ANKL1 or ANKL3 cells succumbed earlier than those bearing spleen-derived ANKL cells because of the rapid growth of leukemic cells (Figure 3B; supplemental Figure 6A). This suggests that liver-derived ANKL cells have a more aggressive phenotype than that of spleen-derived ANKL cells. We then evaluated the transcriptomic profile of liver- and spleen-derived ANKL1 cells using RNA-seq. Principal component analysis revealed distinct transcriptomic profiles between liver-derived ANKL1 cells, spleen-derived ANKL1 cells, and normal NK cells, even though the difference between liver- and spleen-derived ANKL1 cells was smaller than the difference between ANKL1 cells and normal NK cells (supplemental Figure 6B). We then explored the enrichment of cell-type signature among the top 100 differentially expressed genes using Metascape (supplemental Figure 6C).23 We found that “liver NK or NK/T cells” was the top-enriched cell signature, suggesting that liver-derived ANKL1 cells share some features with normal liver-resident NK-type cells (supplemental Figure 6D).24
Liver-derived ANKL cells acquire a more aggressive phenotype characterized by an enriched Myc pathway. (A) Schema for generating organ–derived ANKL cells. To select liver-derived cells, ANKL cells were collected from the PDX liver and transplanted into a mouse. After 3 to 4 weeks, cells were again collected from the liver of this mouse and transplanted into another mouse. We repeated the procedure 10 times in total. Spleen-derived ANKL cells were selected in the same way from PDX spleens. (B) Survival curve of PDX mice transplanted with 1 × 103 liver- or spleen-derived ANKL1 cells (n = 8 in each group). (C) Merged uniform manifold approximation and projection (UMAP) of liver-derived ANKL1 cells (17 039 cells) and spleen-derived ANKL cells (14 034 cells) (left). Liver-derived cells were isolated from the liver of PDX transplanted with liver-derived ANKL1 cells and spleen-derived cells were isolated from the spleen of PDX transplanted with spleen-derived ANKL1 cells (n = 1 for each cell type). Liver-derived ANKL1 cells are highlighted in red (right lower). Clusters 0, 2, 5, 8, and 11 are mainly composed of liver-derived cells; clusters 1, 3, 4, 6, 9, and 14 are spleen-derived cells; clusters 7, 10, 12, and 13 are mixed with liver- and spleen-derived cells. (D) Expression levels of typical cell cycle marker genes, MKI67 and TOP2A, were overlaid on a UMAP representation (left) and their violin plots are shown on the right. (E) The top 10 enriched gene sets listed by GSEA using hallmark gene sets of the Molecular Signature Database in liver-derived ANKL1 cells compared with those in spleen-derived ANKL1 cells (left). Enrichment plot of hallmark Myc targets v1 (right). (F) Comparison of significantly differentiated leading-edge genes in the hallmark Myc target v1 between liver- and spleen-derived ANKL1 cells. (G) Gene expression levels of MYC in normal NK cells, spleen-derived ANKL cells, and liver-derived ANKL cells. (H) The expression level of MYC was overlaid on a UMAP representation (left) and its violin plot is shown on the right. (I) Immunohistochemistry (IHC) of Myc in the liver section of a patient with newly diagnosed ANKL revealing variable expression of Myc among ANKL cells (black arrow indicates expression of Myc). Scale bar, 50 μm. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Liver-derived ANKL cells acquire a more aggressive phenotype characterized by an enriched Myc pathway. (A) Schema for generating organ–derived ANKL cells. To select liver-derived cells, ANKL cells were collected from the PDX liver and transplanted into a mouse. After 3 to 4 weeks, cells were again collected from the liver of this mouse and transplanted into another mouse. We repeated the procedure 10 times in total. Spleen-derived ANKL cells were selected in the same way from PDX spleens. (B) Survival curve of PDX mice transplanted with 1 × 103 liver- or spleen-derived ANKL1 cells (n = 8 in each group). (C) Merged uniform manifold approximation and projection (UMAP) of liver-derived ANKL1 cells (17 039 cells) and spleen-derived ANKL cells (14 034 cells) (left). Liver-derived cells were isolated from the liver of PDX transplanted with liver-derived ANKL1 cells and spleen-derived cells were isolated from the spleen of PDX transplanted with spleen-derived ANKL1 cells (n = 1 for each cell type). Liver-derived ANKL1 cells are highlighted in red (right lower). Clusters 0, 2, 5, 8, and 11 are mainly composed of liver-derived cells; clusters 1, 3, 4, 6, 9, and 14 are spleen-derived cells; clusters 7, 10, 12, and 13 are mixed with liver- and spleen-derived cells. (D) Expression levels of typical cell cycle marker genes, MKI67 and TOP2A, were overlaid on a UMAP representation (left) and their violin plots are shown on the right. (E) The top 10 enriched gene sets listed by GSEA using hallmark gene sets of the Molecular Signature Database in liver-derived ANKL1 cells compared with those in spleen-derived ANKL1 cells (left). Enrichment plot of hallmark Myc targets v1 (right). (F) Comparison of significantly differentiated leading-edge genes in the hallmark Myc target v1 between liver- and spleen-derived ANKL1 cells. (G) Gene expression levels of MYC in normal NK cells, spleen-derived ANKL cells, and liver-derived ANKL cells. (H) The expression level of MYC was overlaid on a UMAP representation (left) and its violin plot is shown on the right. (I) Immunohistochemistry (IHC) of Myc in the liver section of a patient with newly diagnosed ANKL revealing variable expression of Myc among ANKL cells (black arrow indicates expression of Myc). Scale bar, 50 μm. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
To detect the distinct cellular populations between liver- and spleen-derived ANKL1 cells, these cells were subjected to scRNA-seq. Unbiased clustering of ANKL1 cells revealed that liver- and spleen-derived ANKL1 cells were separated into different clusters almost without any overlap (Figure 3C). The expression of typical cell cycle marker genes, MKI67 and TOP2A, were mainly localized in the clusters of liver-derived ANKL1 (Figure 3D), again indicating the aggressive features of liver-derived ANKL cells.
To determine the molecular mechanisms underlying the aggressiveness of liver-derived ANKL cells, we further analyzed the bulk RNA-seq data comparing liver- and spleen-derived ANKL cells. Gene-set enrichment analysis (GSEA) showed that hallmark Myc targets was the most enriched pathway in the liver-derived ANKL compared with that in the spleen-derived ANKL cells (Figure 3E; supplemental Table 1). Indeed, expression of MYC, the highest-ranked gene among the leading-edge genes of the Myc pathway (Figure 3F), was higher in liver-derived ANKL1 cells than in spleen-derived ANKL1 cells or normal NK cells (Figure 3G). The scRNA-seq results confirmed the higher MYC expression in the clusters of liver-derived ANKL1 cells compared with that in the spleen-derived ANKL1 cells (Figure 3H). We also confirmed Myc expression in the liver biopsy specimen of a patient with newly diagnosed ANKL (Figure 3I). The variable expression of Myc among ANKL cells was compatible with the expression data in the scRNA-seq (Figure 3H). Overall, MYC is likely responsible for the aggressiveness of liver-derived ANKL1 cells.
Furthermore, to clarify the molecular mechanism separately in tumor cells and the liver niche, the transcripts obtained from the bulk liver tissues of healthy and PDX mice were separated into those derived from ANKL (human) and the liver niche (mouse) in silico using CASTIN (CAncer-STromal INteractome analysis) (supplemental Figure 6E).25 Principal component analysis demonstrated different transcriptomic profiles of the liver niche between healthy and late-phase PDX mice (supplemental Figure 6F). GSEA of these profiles revealed that the liver niche in the late phase was enriched with inflammation-related pathways (supplemental Figure 6G).
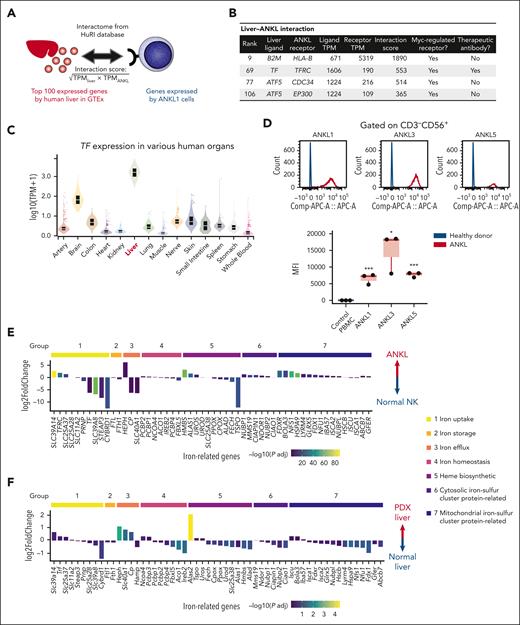
Tf and transferrin receptor interaction as a potential therapeutic target in ANKL
To investigate the therapeutically targetable molecular interactions between the liver and ANKL cells, we extracted the top 100 genes expressed in the normal human liver from the GTEx, searched the ligand-receptor interactions between these genes and the genes expressed by ANKL1 cells using HuRI (Figure 4A),19 and then ranked the liver-ANKL interaction. Because the above results as well as the previous study suggested the upregulated Myc-pathway as a critical feature of ANKL,8 we extracted Myc-regulated ANKL receptors from the interaction using TRRUST,26 and then considered their targetability using Thera-SAbDab27 (Figure 4B). Consequently, Tf–TfR1 (also known as CD71) was identified as a potential targetable interaction between the liver and ANKL.
Transferrin and transferrin receptor interaction as a potential therapeutic target in ANKL. (A) Schema for calculating the interaction score. The top 100 expressed genes in the human liver were extracted from the GTEx database. The interactome is based on the HuRI. (B) Liver-ANKL interaction associated with Myc regulation. (C) TF gene expression among major human tissues according to the GTEx data. (D) Transferrin receptor 1 (TfR1) expression in normal NK cells derived from healthy donor, ANKL1, ANKL3, and ANKL5 cells analyzed by flow cytometry. (E) Expression profile of iron-related genes in ANKL1-PDX compared with that in normal NK cells. Genes are classified into the following groups: group 1, iron uptake; group 2, iron storage; group 3, iron efflux; group 4, iron homeostasis; group 5, heme biosynthesis; group 6, cytosolic iron-sulfur cluster protein assemble; and group 7, mitochondrial iron-sulfur cluster protein biogenesis or export. (F) Expression profile of iron-related genes in the hepatic niche of PDX mice compared with that in the normal liver. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Transferrin and transferrin receptor interaction as a potential therapeutic target in ANKL. (A) Schema for calculating the interaction score. The top 100 expressed genes in the human liver were extracted from the GTEx database. The interactome is based on the HuRI. (B) Liver-ANKL interaction associated with Myc regulation. (C) TF gene expression among major human tissues according to the GTEx data. (D) Transferrin receptor 1 (TfR1) expression in normal NK cells derived from healthy donor, ANKL1, ANKL3, and ANKL5 cells analyzed by flow cytometry. (E) Expression profile of iron-related genes in ANKL1-PDX compared with that in normal NK cells. Genes are classified into the following groups: group 1, iron uptake; group 2, iron storage; group 3, iron efflux; group 4, iron homeostasis; group 5, heme biosynthesis; group 6, cytosolic iron-sulfur cluster protein assemble; and group 7, mitochondrial iron-sulfur cluster protein biogenesis or export. (F) Expression profile of iron-related genes in the hepatic niche of PDX mice compared with that in the normal liver. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
TfR1 is a type 2 transmembrane glycoprotein that binds Tf and plays a crucial role in iron uptake by cells. Several cellular functions such as oxygen transfer, mitochondrial function, and DNA synthesis or repair rely on iron.28,29 Furthermore, TfR1 is known to be overexpressed on various cancer cells including hematological malignancies and has thus been investigated as a therapeutic target.29 The liver expresses the highest level of TF among major human tissues based on the GTEx data (Figure 4C). The livers of PDXs also expressed a much higher level of Trf compared with that in the BM or spleen (supplemental Figure 7A). In addition, ANKL cells expressed higher levels of TfR1 mRNA as well as protein than those in the NK cells from healthy donors (Figure 4D; supplemental Figure 7B). The scRNA-seq data also indicated that more liver-derived ANKL1 cells tended to express TFRC than spleen-derived ANKL1 cells (supplemental Figure 7C). The expression profile of iron-related genes in ANKL1-PDXs was compared with that in normal NK cells based on the iron homeostasis pathway (PW:0000590) in RGD (Rat Genome Database) (Figure 4E).30 In addition to TFRC, SLC39A14, which encodes the metal cation symporter ZIP14, was significantly upregulated in the iron-uptake pathway. Furthermore, SLC40A1, encoding the sole iron exporter ferroportin, was significantly downregulated in the iron-efflux pathway in ANKL1 cells. These results demonstrated that iron homeostasis in ANKL cells is heavily biased toward iron uptake. Intriguingly, the mitochondrial iron-sulfur cluster protein biogenesis pathway was strongly upregulated in ANKL1 cells. In addition, liver-derived ANKL1 cells also favored mitochondrial iron-sulfur cluster protein biogenesis compared with spleen-derived ANKL1 cells, although this difference was smaller than that between ANKL1 cells and normal NK cells (supplemental Figure 7D). Notably, examining the expression pattern of iron-related genes in the hepatic niche revealed that Hamp, which encodes the iron master regulator hepcidin to store iron in the cells during inflammation, was not elevated despite the inflammatory signature of the hepatic niche (Figure 4F). Furthermore, the expression of Slc40a1 was significantly higher in the liver of PDXs than that of normal mice. These data suggest that the PDX liver supplies more iron for leukemic cells despite its inflammatory signature.
Taken together, these results indicate that ANKL cells have an enhanced iron utilization pathway and the Tf-TfR1 axis appears to be a therapeutic target.
ANKL cells are vulnerable to the inhibition of TfR1 function
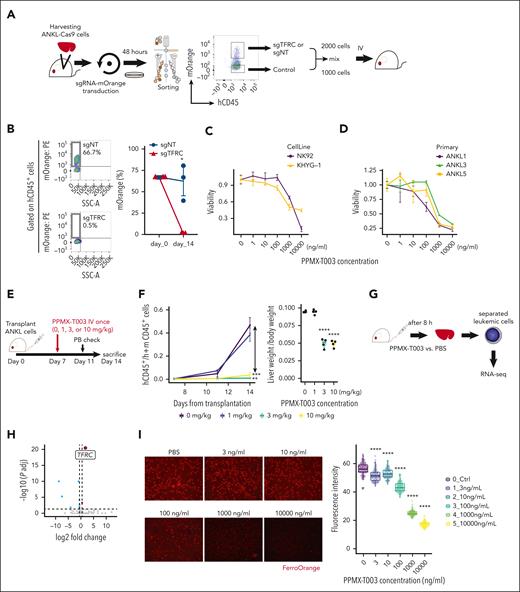
To examine the therapeutic effectiveness of targeting the Tf-TfR1 axis, we deleted TfR1 on ANKL cells with the CRISPR-Cas9 system in vitro, using the NK92 cell line expressing Cas9 (NK92-Cas9). At 1 week after sgRNA-mOrange transduction, the number of NK92-Cas9 transduced sgTFRC (NK92-sgTFRC) was much fewer than that of cells transduced with nontarget sgRNA (NK92-sgNT) (supplemental Figure 8A) under a successful CRISPR-Cas9 system, in which TfR1 expression in the remaining NK92-sgTFRC was much weaker than that in NK92-sgNT (supplemental Figure 8B). Next, to elucidate the necessity of TfR1 for the survival and proliferation of primary ANKL cells, direct in vivo CRISPR-Cas9 analysis was performed owing to their inability to survive and proliferate ex vivo (supplemental Figure 8C). The impact of transducing sgTFRC into ANKL1-Cas9 (ANKL1-sgTFRC) or ANKL3-Cas9 (ANKL3-sgTFRC) cells on tumor formation in vivo was compared with that of sgNT or other sgRNAs. Consequently, the burden of ANKL-sgTFRC cells in the PDX liver was markedly reduced compared with that of ANKL-sgNT cells (supplemental Figure 8D). This reduction was comparable with that of sgMYC and much stronger than that of sgRNAs targeting CXCR3 or ITGB7 (supplemental Figure 8D). The in vivo survival fitness of ANKL-sgTFRC was remarkably reduced, as shown by a competitive assay in which a 2:1 ratio of the ANKL1-sgRNA-mOrange cells and control ANKL1 cells resulted in almost complete disappearance of ANKL1-sgTFRC from mice that received transplantation after 14 days, whereas ANKL1-sgNT cells maintained their ratio to control ANKL1 cells (Figure 5A-B). These results suggest that TfR1 is essential for the survival and/or proliferation of ANKL.
ANKL cells are vulnerable to the inhibition of TfR1 function. (A) Schema of the in vivo competitive assay using the CRISPR-Cas9 system. ANKL1-Cas9 cells are harvested from PDX mice and then transduced with sgRNA-mOrange using a lentiviral vector. After 48 hours, hCD45+mOrange+ and hCD45+mOrange− cells are sorted, mixed at a ratio of 2000 to 1000 cells, and transplanted into mice. (B) The percentage of mOrange+ cells in the liver at day 14 after transplantation are compared between sgNT and sgTFRC transduced ANKL1-Cas9 cells (n = 3). (C) Viability of NK-cell leukemia cell lines after treatment with various concentrations of PPMX-T003 for 96 hours in vitro, evaluated by propidium iodide and hCD45 staining and flow cytometry. (D) Viability of primary ANKL cells obtained from ANKL1, ANKL3, and ANKL5 PDX mice after treatment with various concentrations of PPMX-T003 for 48 hours in vitro. (E) Schema for testing the efficacy of PPMX-T003 in vivo using ANKL-PDX mice. (F) Comparing the ratio of human CD45+ cells to mouse CD45+ cells in PB on days 11 and 14 after transplantation among ANKL1-PDX mice treated with 0, 1, 3, and 10 mg/kg of PPMX-T003 (left). Comparing the liver weight per body weight among 0, 1, 3, and 10 mg/kg of PPMX-T003 treatments in ANKL1-PDX mice (right) (n = 4). (G) Schema for RNA-seq to analyze the global gene expression changes in ANKL1 cells upon PPMX-T003 or phosphate-buffered saline (PBS) treatment in vivo (n = 3). (H) Volcano plot of RNA-seq data. (I) Intracellular ferrous staining of ANKL3 cells 48 hours after treatment with PPMX-T003 in vitro, stained with FerroOrange, and analyzed using confocal microscopy. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
ANKL cells are vulnerable to the inhibition of TfR1 function. (A) Schema of the in vivo competitive assay using the CRISPR-Cas9 system. ANKL1-Cas9 cells are harvested from PDX mice and then transduced with sgRNA-mOrange using a lentiviral vector. After 48 hours, hCD45+mOrange+ and hCD45+mOrange− cells are sorted, mixed at a ratio of 2000 to 1000 cells, and transplanted into mice. (B) The percentage of mOrange+ cells in the liver at day 14 after transplantation are compared between sgNT and sgTFRC transduced ANKL1-Cas9 cells (n = 3). (C) Viability of NK-cell leukemia cell lines after treatment with various concentrations of PPMX-T003 for 96 hours in vitro, evaluated by propidium iodide and hCD45 staining and flow cytometry. (D) Viability of primary ANKL cells obtained from ANKL1, ANKL3, and ANKL5 PDX mice after treatment with various concentrations of PPMX-T003 for 48 hours in vitro. (E) Schema for testing the efficacy of PPMX-T003 in vivo using ANKL-PDX mice. (F) Comparing the ratio of human CD45+ cells to mouse CD45+ cells in PB on days 11 and 14 after transplantation among ANKL1-PDX mice treated with 0, 1, 3, and 10 mg/kg of PPMX-T003 (left). Comparing the liver weight per body weight among 0, 1, 3, and 10 mg/kg of PPMX-T003 treatments in ANKL1-PDX mice (right) (n = 4). (G) Schema for RNA-seq to analyze the global gene expression changes in ANKL1 cells upon PPMX-T003 or phosphate-buffered saline (PBS) treatment in vivo (n = 3). (H) Volcano plot of RNA-seq data. (I) Intracellular ferrous staining of ANKL3 cells 48 hours after treatment with PPMX-T003 in vitro, stained with FerroOrange, and analyzed using confocal microscopy. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Furthermore, the Tf-TfR1 axis was interrupted using the anti-TfR1 antibody (PPMX-T003, also known as JST-TFR09) in ANKL. PPMX-T003 has been reported to suppress the growth of various cancer cell lines in vitro and can eradicate several types of hematological malignancies in xenograft murine models by blocking cellular iron uptake.31-33 The viability of various NK-cell leukemia cell lines and PDX-derived primary ANKL cells was reduced after treatment using PPMX-T003 in vitro, with the following IC50 values: NK92, 1155 ng/mL; KHYG-1, 242 ng/mL; ANKL1, 117 ng/mL; ANKL3, 754 ng/mL; and ANKL5, 262 ng/mL (Figure 5C-D). The cytotoxic effect of PPMX-T003 was also confirmed using a 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide or MTT assay (supplemental Figure 8E). Notably, in ANKL-PDXs, PPMX-T003 with no more than 3 and 1 mg/kg fully suppressed the leukemic growth of ANKL1 and ANKL3, respectively (Figure 5E-F; supplemental Figure 8F). Because the Tf-TfR1 axis also plays an important role in RBC development, we examined the toxicity of PPMX-T003 on human erythroblasts using cord blood CD34+ cells. PPMX-T003 showed dose-dependent growth inhibition of erythroblasts with a maximum inhibition rate of ∼75% compared with that of nontreated erythroblasts (supplemental Figure 8G).
The global gene expression changes in ANKL1 cells treated with PPMX-T003 determined using RNA-seq revealed TFRC as the top differentially expressed gene (Figure 5G-H). As a previous study reported that anti-TfR1 antibody treatment led to paradoxical TfR1 upregulation in hematological malignant cells owing to rapid iron deprivation,34 PPMX-T003 could efficiently deplete intracellular iron as indicated by a significant decrease in the intracellular ferrous ion (Fe2+) concentration of ANKL3 cells after treating with PPMX-T003 in vitro (Figure 5I). Furthermore, GSEA revealed that many of cellular biological processes of ANKL1 were downregulated after treatment with PPMX-T003 (supplemental Figure 8H). Collectively, abrogation of the Tf-TfR1 axis inhibits ANKL progression by reducing the intracellular iron pool, followed by downregulation of the cellular biological processes.
Inhibition of the Tf-TfR1 axis using anti-TfR1 antibody attenuates ANKL
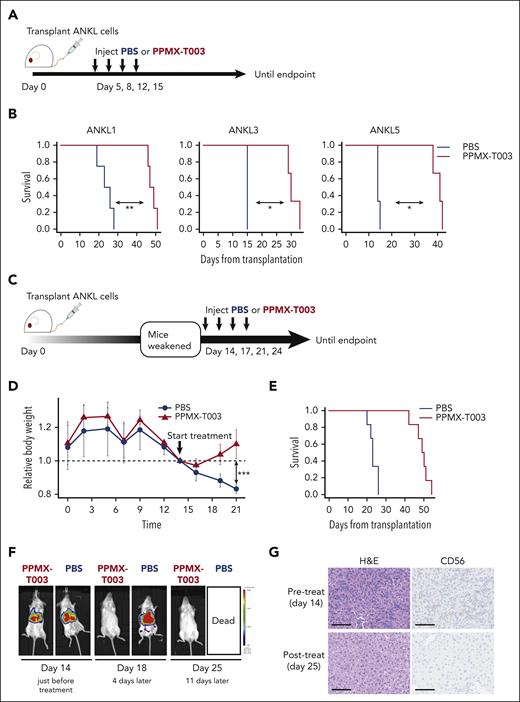
Finally, the therapeutic efficacy of PPMX-T003 for ANKL was investigated in a preclinical setting of PDXs. We intravenously administered PPMX-T003 to ANKL1, ANKL3, and ANKL5 PDX mice twice weekly, starting from day 5 after leukemic cell transplantation (Figure 6A). Mice treated with the vehicle began to develop ANKL and succumbed within 2 (ANKL3 and ANKL5) or several weeks (ANKL1), whereas those treated with PPMX-T003 did not succumb for more than one and a half months. PPMX-T003 significantly prolonged the survival of all PDX strains compared with that of the vehicle, suggesting that the drug may provide survival benefits to patients with ANKL (Figure 6B). In the clinic, the leukemic burden is already high at diagnosis; therefore, we assessed the therapeutic efficacy of PPMX-T003 in PDXs with advanced disease where they had already begun losing weight at day 14 (Figure 6C). ANKL1-PDXs treated with vehicle continued losing weight and died soon after a week or so, whereas those treated with PPMX-T003 recovered their weight within a few days and further showed prolonged survival to >1 month (Figure 6D-E). IVIS indicated that PPMX-T003 eradicated leukemia from the liver, whereas some lesions remained in the spleen and BM (Figure 6F; supplemental Figure 9A). Microscopically, in ANKL1 and ANKL3 PDXs treated with PPMX-T003, the leukemic lesions completely disappeared from the liver (Figure 6G; supplemental Figure 9B).
Inhibition of the Tf-TfR1 axis using anti-TfR1 antibody attenuates ANKL. (A) Schema for assessing the therapeutic efficacy of PPMX-T003 for ANKL in a preclinical setting. ANKL-PDX mice were treated with PBS or 10 mg/kg of PPMX-T003 on days 5, 8, 12, and 15 after transplantation of 1 × 106 ANKL cells and monitored until the end point. (B) Survival curve of ANKL1, ANKL3, and ANKL5 PDX mice treated with PBS or PPMX-T003 (n = 3-4 in each group). (C) Schema for evaluating the efficacy of PPMX-T003 treatment in PDX mice with advanced disease. ANKL-PDX mice were treated with PBS or 10 mg/kg of PPMX-T003 on days 14, 17, 21, and 24 after transplantation of 1 × 106 ANKL1 cells and monitored until the end point. (D) Relative weight changes of mice during the course of the experiment. Weights were corrected for body weight on day 14. (E) Survival curve of ANKL1-PDX mice treated with PBS or PPMX-T003 (n = 6). (F) IVIS analysis of ANKL1-PDX mice treated with 10 mg/kg of PPMX-T003 or PBS on days 14, 17, 21, and 24. Representative image from 3 biological replicates. (G) H&E staining (left) and CD56 IHC (right) for a liver section from ANKL1-PDX mice before and after the PPMX-T003 treatment. Scale bar, 100 μm. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Inhibition of the Tf-TfR1 axis using anti-TfR1 antibody attenuates ANKL. (A) Schema for assessing the therapeutic efficacy of PPMX-T003 for ANKL in a preclinical setting. ANKL-PDX mice were treated with PBS or 10 mg/kg of PPMX-T003 on days 5, 8, 12, and 15 after transplantation of 1 × 106 ANKL cells and monitored until the end point. (B) Survival curve of ANKL1, ANKL3, and ANKL5 PDX mice treated with PBS or PPMX-T003 (n = 3-4 in each group). (C) Schema for evaluating the efficacy of PPMX-T003 treatment in PDX mice with advanced disease. ANKL-PDX mice were treated with PBS or 10 mg/kg of PPMX-T003 on days 14, 17, 21, and 24 after transplantation of 1 × 106 ANKL1 cells and monitored until the end point. (D) Relative weight changes of mice during the course of the experiment. Weights were corrected for body weight on day 14. (E) Survival curve of ANKL1-PDX mice treated with PBS or PPMX-T003 (n = 6). (F) IVIS analysis of ANKL1-PDX mice treated with 10 mg/kg of PPMX-T003 or PBS on days 14, 17, 21, and 24. Representative image from 3 biological replicates. (G) H&E staining (left) and CD56 IHC (right) for a liver section from ANKL1-PDX mice before and after the PPMX-T003 treatment. Scale bar, 100 μm. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
On the basis of the results of this study, blockade of the Tf-TfR1 axis using PPMX-T003 shows promising therapeutic efficacy for ANKL.
Discussion
In this study, we established ANKL-PDXs, and our results indicated that ANKL cells primarily engraft and proliferate in the liver sinusoids. Next, we analyzed the transcriptome of ANKL cells at both bulk and single-cell resolution and determined that liver-derived ANKL cells showed over-activation of Myc-associated genes. Among these genes, we discovered TfR1 as a novel therapeutic target against ANKL. Furthermore, we demonstrated the therapeutic potential of an anti-TfR1 antibody, PPMX-T003, against ANKL in vivo.
Although the principal niche of ANKL has been assumed to be BM, some patients with ANKL show absence of BM infiltration.21,35,36 Consistent with this clinical observation, our data strongly suggest that ANKL cells prefer the liver to the BM as a proliferative niche.
Recently, 2 different groups reported that 20% to 50% of patients with ANKL showed activation of the JAK/STAT pathway.7,8 NK-cell tumors are sensitive to inhibitors of the JAK/STAT pathway in vitro.7 However, our interactome analyses could not identify a significant relationship between JAK/STAT-related molecules and the liver TME (supplemental Table 1). Inhibition of the JAK/STAT pathway may suppress ANKL proliferation independently of PPMX-T003. Thus, the synergistic effects of their combined use against ANKL need to be evaluated further.
The liver plays a central role in regulating iron homeostasis under inflammatory conditions.28,37 Although the liver restricts iron availability during inflammation,38 ANKL cells received iron from the liver TME in ANKL-PDXs under inflammatory conditions (Figure 4F; supplemental Figure 6G). We thus assumed that ANKL cells may alter the expression of iron-related genes in the liver to receive sufficient amounts of iron for their survival and growth through a yet unknown mechanism.
Previous studies have investigated the potential of TfR1-targeting treatment against various neoplasms.29 In particular, PPMX-T003 has been tested and has shown antineoplastic potential against various hematological malignancies such as adult T-cell leukemia/lymphoma, acute myeloid leukemia, and polycythemia vera.31-33 Furthermore, a phase 1 study using PPMX-T003 in patients with polycythemia vera has already been started (NCT05074550). From this background, we can immediately start another phase 1 study of PPMX-T003 for patients with ANKL based on this study.
Although ANKL-PDXs are useful for investigating unknown pathogenesis as well as evaluating novel drugs in preclinical studies, several limitations exist in this study. We should consider that the selection pressure on tumor cells can differ between humans and immunocompromised mice. However, our ANKL-PDXs still recapitulated the clinical characteristics of human ANKL. Another crucial limitation is that we are unable to confirm our findings regarding whether ANKL primarily proliferates in human liver sinusoids, as most patients are diagnosed after the systemic infiltration of ANKL cells. Notably, clinical observations of severe hepatic injury are consistent with our findings. In addition, although PPMX-T003 eradicated ANKL cells from the liver and prolonged PDX survival, some ANKL cells remained in the spleen and BM even after PPMX-T003 treatment and the PDX mice eventually died (supplemental Figure 9A). The biology of ANKL in the spleen and BM TME needs to be further investigated to identify additional therapeutic targets that can work synergistically with TfR1 blockade. Regarding the clinical application of PPMX-T003, the tolerability and efficacy of PPMX-T003 needs to be evaluated further in patients, although PPMX-T003 has shown a tolerable safety profile in cynomolgus monkeys.32 In fact, PPMX-T003 induced growth inhibition of human erythroblasts in our analysis (supplemental Figure 8G). However, it is important to note that anemia can be managed by transfusion of RBCs in the clinic, as is done for other anticancer drugs. The upcoming phase 1 trial of PPMX-T003, specifically targeting patients with ANKL, will address this problem.
In summary, ANKL cells initially engraft and proliferate in liver sinusoids with dependence on the Tf-TfR1 axis. We also found that PPMX-T003, an anti-TfR1 antibody, showed therapeutic potential against ANKL. Overall, the model established in this study provides a strong basis for understanding ANKL.
Acknowledgments
The authors thank Masako Takamatsu, Natsumi Kurosaki, Etsuko Nagashima, Masatoshi Kakizaki, Hideki Nakasone, and members of the Department of Innovative Medical Science at Tokai University, Division of Hematology at Jichi Medical University Saitama Medical Center, and 3rd Department of Internal Medicine at Yamagata University Faculty of Medicine for their assistance, advice, and helpful discussions. The authors also thank the Support Center for Medical Research and Education at Tokai University for providing technical assistance and Lilin Zhang and Fumiko Nomura at Perseus Proteomics Inc for providing data regarding the toxicity of PPMX-T003. The authors thank Editage (www.editage.com) for English-language editing.
This work was supported by grants-in-aid for scientific research from the Japan Society for the Promotion of Science (JP20H03716 and JP20K21550 [A.K.]; 20J14747 [K.K.]), AMED (JPAS2414005Q and JP17im0110705h0004 [T.M.]), P-CREATE (20cm0106275 [A.K.]), PRIME (10gm5910004 [A.K.]), and the Research Program on Hepatitis (23fk0210133s0101 [A.K.]) from the Japan Agency for Medical Research and Development; JST-CREST (JPMJCR19H5 [A.K.]) from the Japan Science and Technology Agency; a Jichi Medical University Graduate Student Research Award 2021 (K.K.); the Japan Leukemia Research Fund (K.K.); and a Tokai University Tokuda Memorial Cancer/Genome Basic Research Grant for Young Investigators (Y. Miyatake).
Authorship
Contribution: K.K., R.Y., and A.K. conceptualized the study; K.K., R.Y., Y. Miyatake, H. Higuchi, T.M., S.N., S. Iwabuchi, S.H., K.-I.I., and A.K. designed the methodology; K.K., R.Y., Y. Miyatake, and A.K. validated the study; K.K., R.Y., R.I., Y. Murakami, S. Iwabuchi, and S.H. conducted formal analysis; K.K., R.Y., Y. Miyatake, J.C., H. Higuchi, R.I., Y. Murakami, N.N., and A.K. conducted investigation; H.M., T.I., A.I., S. Iida, N.F., H. Harigae, Y.F., N.T., H.W., F.I., H.N., H.T., T.M., K.-I.I., N.N., and K.I. were responsible for resources; K.K., R.Y., Y. Miyatake, S. Iwabuchi, S.H., and A.K. curated the data; K.K., R.Y., Y. Miyatake, and A.K. wrote the original draft; all the authors reviewed and edited the manuscript; K.K., R.Y., Y. Miyatake, S. Iwabuchi, S.H., and A.K. performed visualization; K.I., Y.K., K.A., and A.K. supervised; K.K. and A.K. administrated the project; and K.K., Y. Miyatake, T.M., and A.K. were responsible for funding acquisition.
Conflict-of-interest disclosure: T.M. was an employee and is a board member and chief technology officer of Perseus Proteomics Inc and has stock options of Perseus Proteomics Inc. The remaining authors declare no competing financial interests.
Correspondence: Ai Kotani, Department of Innovative Medical Science, Tokai University School of Medicine, 143 Shimokasuya, Isehara, Kanagawa 259-1193, Japan; e-mail: aikotani@k-lab.jp.
References
Author notes
∗K.K., R.Y., and Y. Miyatake contributed equally to this study.
Data are available on request from the corresponding author, Ai Kotani (aikotani@k-lab.jp).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal