Abstract
In a short time, single-cell platforms have become the norm in many fields of research, including multiple myeloma (MM). In fact, the large amount of cellular heterogeneity in MM makes single-cell platforms particularly attractive because bulk assessments can miss valuable information about cellular subpopulations and cell-to-cell interactions. The decreasing cost and increasing accessibility of single-cell platform, combined with breakthroughs in obtaining multiomics data for the same cell and innovative computational programs for analyzing data, have allowed single-cell studies to make important insights into MM pathogenesis; yet, there is still much to be done. In this review, we will first focus on the types of single-cell profiling and the considerations for designing a single-cell profiling experiment. Then, we will discuss what have learned from single-cell profiling about myeloma clonal evolution, transcriptional reprogramming, and drug resistance, and about the MM microenvironment during precursor and advanced disease.
Introduction
Multiple myeloma (MM), similar to a number of other malignancies, is associated with significant intra- and intertumor heterogeneity. This heterogeneity can be further exacerbated by the many different ways in which tumor cells interact with the bone marrow (BM) microenvironment. As such, methods that investigate cells in bulk can miss valuable information about subpopulations and rarer interactions. Single-cell profiling technologies can now assess at high resolution the genome, transcriptome, and epigenome of individual cells, thus allowing us to recognize cell identity, state, and activity, particularly when combining multiple technologies. These multiomics strategies have led to more thorough studies on the evolution of cancer, activity of subclones, response to treatment, and bodily consequences of the disease. Therefore, these technologies, either alone or combined, create an invaluable data deluge for understanding MM cells and their microenvironment. This is an emerging field with only a few landmark studies in MM. In this perspective, we will focus on peer-reviewed publications that performed single-cell profiling using next-generation sequencing or cytometry by time of flight (CyTOF) and the considerations for designing single-cell profiling experiments, as well as what we have learned so far by applying these techniques to the study of MM and its BM microenvironment.
Considerations for single-cell profiling
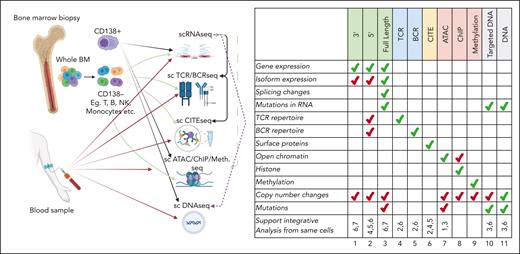
When designing a profiling experiment, it is important to consider how the sample will be acquired and which technologies will assess it. Furthermore, the target cell types will determine the sample processing and whether any cell sorting will need to be performed. For example, if the target tissue is BM MM or plasma cells (PCs) and/or the general microenvironment, then BM aspirate would be used either unseparated or after purifying various cell populations, especially MM or PCs using CD138 as a marker; however, additional markers such as the presence of CD38 or CD45 or the absence of CD56 or CD19 may also be used.1,2 If a specific microenvironment cell is a target, such as T cells, then the appropriate markers would be used, such as CD3+ and CD4+ or CD8+ cells (Figure 1). Cell integrity and quality are crucial requirements for successful single-cell experiments. Therefore, careful experimental planning, acquiring the required samples, and defining the goals affect the outcome of each study. The general rule of thumb is to keep conditions comparable instead of trying to remove the artifacts later via computational methods. If possible, using fresh samples processed within 2 hours can provide the best results, and this can be extended to 24 to 48 hours for multilocation studies. However, sampling times until cryopreservation is a driver of technical variability, ubiquitous across cell types, donors, protocols, and disease status. Still, cryopreserved samples such as dimethyl sulfoxide–stored samples can be used if the samples need to be stored long term. Such methods have no, or limited, impact on sequencing if the samples are stored properly and viable cells are selected before library preparation.3,4 However, storing samples increases the percentage of mitochondrial reads and contamination by background ambient reads.5 Only by careful application of computational corrections, cell culture, and storage adjustments can we design reliable retrospective or prospective studies.6
Summary of single-cell platforms used in MM research, and primary and secondary data use for each platform. Cartoon on the associated single-cell platforms with sample collection sites and how single-cell platforms have been used. Table on the right shows the primary (green) and secondary (red) use for each platform. Bottom row shows whether a platform can be matched with other platforms for integrative analysis.
Summary of single-cell platforms used in MM research, and primary and secondary data use for each platform. Cartoon on the associated single-cell platforms with sample collection sites and how single-cell platforms have been used. Table on the right shows the primary (green) and secondary (red) use for each platform. Bottom row shows whether a platform can be matched with other platforms for integrative analysis.
Although single-cell methods have improved significantly, there are still a few key aspects of sample collection and processing that affect data quality.7,8 The first of these is the number of cells. Any decision on sample amount and cell sorting should carefully consider the expected frequency and number of cells, aiming for a cell count sufficient for the proposed experiments. Cell counts that are too low may result in the failure of the profiling or the inability to reach statistical significance. Too many cells can result in overcrowding of the platform. Second, the condition of the sample can affect data quality. Early studies based on older protocols required fresh samples, which necessitated tight coordination between tissue acquisition and processing, posing a challenge in clinical settings. Newer protocols have been developed for viably stored cells, particularly for nuclei from frozen samples.9 Yet, these nuclei produce lower yields, and, for specific cells, it is harder to preprocess. Single-cell genomic studies are very sensitive to these and many subtler technical issues that can lead to batch effects, even when samples are processed by the same equipment at the same time. Computational methods have been developed to correct for technical differences between experiments10-13; however, processing and handling all samples with the same protocol is still a key factor in minimizing such effects on study results, reducing the probability of false-positive or false-negative findings.
The next consideration is picking the profiling technology. Among all the different platforms, single-cell RNA sequencing (scRNA-seq) is by far the most used.14,15 In fact, in MM research, 25 of 27 publications we found in our literature search used scRNA-seq in their study (Table 1). The gene expression data from scRNA-seq can infer cell identity, phenotype, and activity. In many cancer studies, scRNA-seq data have also been used for copy number alteration (CNA) detection. In general, scRNA-seq platforms can be divided into full-length transcript analysis vs digital counting of 3′ or 5′ transcript ends.16 The choice of sequencing method should be dictated by the goal of the experiment; for example, to prioritize retention of sequence information vs cost-effectiveness.17 Full-length transcripts allow for the detection of splice variants, single-nucleotide variants, indels, and fusion transcripts. However, only 30% of mutations are expressed in MM,18 and therefore, careful consideration needs to be made before secondary use, specifically for mutation detection. In contrast, 3′ or 5′ scRNA-seq approaches capture information from many more cells, require less hands-on time for library preparation, have increased ease of access, and cost less.
List of single-cell genomics papers published since 2016
| PMID . | Authors . | Cell types evaluated with single-cell genomics . | Single-cell platforms used . | Time point . | Tags . |
|---|---|---|---|---|---|
| 36026513102 | Dhodapkar et al, 2023 | BMMC | scRNA, scTCR, scCITE, CyTOF | Before and after anti-BCMA treatment | Microenvironment, MM cells, CART |
| 3670283491 | Pilcher et al, 2023 | CD138− | scRNA | Diagnosis | Microenvironment, rapid progression |
| 3637920890 | Sklavenitis-Pistofidis et al, 2022 | CD138− BMMC, PB | scRNA, scTCR, CyTOF | Baseline, cycle 9, end of the treatment | Immunotherapy, microenvironment, immune normalization |
| 3639663198 | Boiarsky et al, 2022 | CD138+ | scRNA | MGUS, SMM, MM, NPC | Transcriptional signature, progression |
| 36252999103 | Kehl et al, 2022 | BMMC, PB | scRNA, scCITE | Microenvironment, IgE, T cells | |
| 36163550104 | Gao et al, 2022 | CD138+ | scRNA | CNV | |
| 3634330666 | Lannes et al, 2022 | CD138+ | scDNA | SMM, NDMM | CNV, shallow seq |
| 3613128292 | Liang et al, 2022 | BMMC | scRNA | NDMM, RRMM | Clonal evolution, microenvironment, CNA |
| 36122410105 | Welters et al, 2022 | BM T cells | scRNA, scCITE, scVDJ | NDMM | T cells, clonal expansion |
| 36969740106 | Yao et al, 2022 | CD138− | scRNA, scCITE, CyTOF | NDMM | Microenvironment, classification by progression time |
| 3597711183 | Schinke et al, 2022 | CD138− | scRNA, scTCR | MGUS, SMM, NDMM | Microenvironment, disease progression |
| 3599208469 | Masuda et al, 2022 | CD138+ | scRNA | NDMM, RRMM | Drug-response, longitudinal samples |
| 3526457570 | Kumar et al, 2022 | CD138+ | scRNA | HMCL | Drug resistance |
| 3514507765 | Merz et al, 2022 | CD138+ (BM and OL) | scRNA | NDMM, RRMM | Spatial heterogeneity |
| 35646666107 | Kumar et al, 2022 | CD138+, HMCL | CyTOF | MM | PI, IMID, drug resistance |
| 3451830971 | Waldschmidt et al, 2021 | CD138+ | scRNA | RRMM | BRAF, drug resistance |
| 3484518887 | Tirier et al, 2021 | CD138+, CD138− | scRNA | RRMM | Drug response, microenvironment |
| 34732728108 | Croucher et al, 2021 | BM of mice | scRNA | Early, intermediate, and active MM | Vκ∗MYC mice, CNV |
| 3467539067 | Frede et al, 2021 | CD138+, CD45+ | scRNA, scATAC | RRMM, NPC | Transcriptional reprogramming under treatment, immunotherapeutic vulnerabilities, integrative analysis |
| 3413371868 | Alameda et al, 2021 | PCs from SLO, PB, and BM | scRNA | MGUS, AL, MM | Transcriptional rewiring, AL, PC malignancy comparison |
| 33602683109 | Cho et al, 2021 | CD56+ NK cells | scRNA | NDMM | Antibody-dependent cellular cytotoxicity, NK cells, daratumumab |
| 3401712286 | de Jong et al, 2021 | Nonhematopoietic microenvironment, CD38+ and CD38– immune microenvironments | scRNA | NDMM, NPC | Stromal cells, microenvironment, iMSC |
| 3396318257 | Liu et al, 2021 | BMMC | scRNA | SMM, NDMM, RRMM | Longitudinal samples, clonal evolution, microenvironment, integrative analysis |
| 3361936972 | Cohen et al, 2021 | CD138+ | scRNA | NDMM, RRMM, NPC | Longitudinal samples, therapeutic resistance |
| 3355851160 | Samur et al, 2021 | BMMC | scRNA | RRMM | CART, immunotherapy resistance, longitudinal samples, microenvironment |
| 3361936861 | Da Via et al, 2021 | BMMC | scRNA | RRMM | CART, immunotherapy resistance, longitudinal samples |
| 3269340659 | Goicoechea et al, 2021 | 8-color EuroFlow NGF antibody panel | scRNA | NDMM | Standard vs high-risk MM, transcriptome |
| 3229909364 | Maia et al, 2020 | CD34+ HPCs | Targeted scDNA | NDMM | Myelodysplastic syndrome, SNVs |
| 3159503963 | Garcés et al, 2020 | 8-color EuroFlow NGF antibody panel | scRNA | NDMM | Circulating cells, BM vs PB, transcriptome |
| 3340950181 | Zavidij et al, 2020 | CD138−, CD45+ | scRNA | MGUS, SMM, NDMM, NPC | Microenvironment, precursor conditions |
| 31988495110 | Teh et al, 2020 | CD138+, HMCL | CyTOF | Before and after treatment | Resistance, synergistic drug combination |
| 3101325482 | Bailur et al, 2019 | BMMC, CD138− | scRNA, CyTOF | MGUS,NDMM, NPC | T cells, myeloid cells, microenvironment |
| 30607001111 | Jang et al, 2019 | CD138+ | scRNA | MGUS, SMM, NDMM, RRMM | Disease progression, transcriptomic signatures |
| 31462637112 | Kourelis et al, 2019 | BMMC, PB | CyTOF | MM, MGUS, SMM, AL | Microenvironment, immune escape, PB vs BM |
| 3052332858 | Ledergor et al, 2018 | CD138+ | scRNA | MGUS, SMM, NDMM, AL, NPC | Circulating cells, BM vs PB, heterogeneity, subclones, CNA, BCR |
| 2780728262 | Lohr et al, 2016 | CD138+ | scRNA, targeted scDNA | MGUS, NDMM | Circulating cells, BM vs PB, heterogeneity, subclones, CNA, SNV |
| 25711758113 | Hansmann et al, 2015 | PB | CyTOF | MM, MGUS, AL | PBMC, immune cells |
| PMID . | Authors . | Cell types evaluated with single-cell genomics . | Single-cell platforms used . | Time point . | Tags . |
|---|---|---|---|---|---|
| 36026513102 | Dhodapkar et al, 2023 | BMMC | scRNA, scTCR, scCITE, CyTOF | Before and after anti-BCMA treatment | Microenvironment, MM cells, CART |
| 3670283491 | Pilcher et al, 2023 | CD138− | scRNA | Diagnosis | Microenvironment, rapid progression |
| 3637920890 | Sklavenitis-Pistofidis et al, 2022 | CD138− BMMC, PB | scRNA, scTCR, CyTOF | Baseline, cycle 9, end of the treatment | Immunotherapy, microenvironment, immune normalization |
| 3639663198 | Boiarsky et al, 2022 | CD138+ | scRNA | MGUS, SMM, MM, NPC | Transcriptional signature, progression |
| 36252999103 | Kehl et al, 2022 | BMMC, PB | scRNA, scCITE | Microenvironment, IgE, T cells | |
| 36163550104 | Gao et al, 2022 | CD138+ | scRNA | CNV | |
| 3634330666 | Lannes et al, 2022 | CD138+ | scDNA | SMM, NDMM | CNV, shallow seq |
| 3613128292 | Liang et al, 2022 | BMMC | scRNA | NDMM, RRMM | Clonal evolution, microenvironment, CNA |
| 36122410105 | Welters et al, 2022 | BM T cells | scRNA, scCITE, scVDJ | NDMM | T cells, clonal expansion |
| 36969740106 | Yao et al, 2022 | CD138− | scRNA, scCITE, CyTOF | NDMM | Microenvironment, classification by progression time |
| 3597711183 | Schinke et al, 2022 | CD138− | scRNA, scTCR | MGUS, SMM, NDMM | Microenvironment, disease progression |
| 3599208469 | Masuda et al, 2022 | CD138+ | scRNA | NDMM, RRMM | Drug-response, longitudinal samples |
| 3526457570 | Kumar et al, 2022 | CD138+ | scRNA | HMCL | Drug resistance |
| 3514507765 | Merz et al, 2022 | CD138+ (BM and OL) | scRNA | NDMM, RRMM | Spatial heterogeneity |
| 35646666107 | Kumar et al, 2022 | CD138+, HMCL | CyTOF | MM | PI, IMID, drug resistance |
| 3451830971 | Waldschmidt et al, 2021 | CD138+ | scRNA | RRMM | BRAF, drug resistance |
| 3484518887 | Tirier et al, 2021 | CD138+, CD138− | scRNA | RRMM | Drug response, microenvironment |
| 34732728108 | Croucher et al, 2021 | BM of mice | scRNA | Early, intermediate, and active MM | Vκ∗MYC mice, CNV |
| 3467539067 | Frede et al, 2021 | CD138+, CD45+ | scRNA, scATAC | RRMM, NPC | Transcriptional reprogramming under treatment, immunotherapeutic vulnerabilities, integrative analysis |
| 3413371868 | Alameda et al, 2021 | PCs from SLO, PB, and BM | scRNA | MGUS, AL, MM | Transcriptional rewiring, AL, PC malignancy comparison |
| 33602683109 | Cho et al, 2021 | CD56+ NK cells | scRNA | NDMM | Antibody-dependent cellular cytotoxicity, NK cells, daratumumab |
| 3401712286 | de Jong et al, 2021 | Nonhematopoietic microenvironment, CD38+ and CD38– immune microenvironments | scRNA | NDMM, NPC | Stromal cells, microenvironment, iMSC |
| 3396318257 | Liu et al, 2021 | BMMC | scRNA | SMM, NDMM, RRMM | Longitudinal samples, clonal evolution, microenvironment, integrative analysis |
| 3361936972 | Cohen et al, 2021 | CD138+ | scRNA | NDMM, RRMM, NPC | Longitudinal samples, therapeutic resistance |
| 3355851160 | Samur et al, 2021 | BMMC | scRNA | RRMM | CART, immunotherapy resistance, longitudinal samples, microenvironment |
| 3361936861 | Da Via et al, 2021 | BMMC | scRNA | RRMM | CART, immunotherapy resistance, longitudinal samples |
| 3269340659 | Goicoechea et al, 2021 | 8-color EuroFlow NGF antibody panel | scRNA | NDMM | Standard vs high-risk MM, transcriptome |
| 3229909364 | Maia et al, 2020 | CD34+ HPCs | Targeted scDNA | NDMM | Myelodysplastic syndrome, SNVs |
| 3159503963 | Garcés et al, 2020 | 8-color EuroFlow NGF antibody panel | scRNA | NDMM | Circulating cells, BM vs PB, transcriptome |
| 3340950181 | Zavidij et al, 2020 | CD138−, CD45+ | scRNA | MGUS, SMM, NDMM, NPC | Microenvironment, precursor conditions |
| 31988495110 | Teh et al, 2020 | CD138+, HMCL | CyTOF | Before and after treatment | Resistance, synergistic drug combination |
| 3101325482 | Bailur et al, 2019 | BMMC, CD138− | scRNA, CyTOF | MGUS,NDMM, NPC | T cells, myeloid cells, microenvironment |
| 30607001111 | Jang et al, 2019 | CD138+ | scRNA | MGUS, SMM, NDMM, RRMM | Disease progression, transcriptomic signatures |
| 31462637112 | Kourelis et al, 2019 | BMMC, PB | CyTOF | MM, MGUS, SMM, AL | Microenvironment, immune escape, PB vs BM |
| 3052332858 | Ledergor et al, 2018 | CD138+ | scRNA | MGUS, SMM, NDMM, AL, NPC | Circulating cells, BM vs PB, heterogeneity, subclones, CNA, BCR |
| 2780728262 | Lohr et al, 2016 | CD138+ | scRNA, targeted scDNA | MGUS, NDMM | Circulating cells, BM vs PB, heterogeneity, subclones, CNA, SNV |
| 25711758113 | Hansmann et al, 2015 | PB | CyTOF | MM, MGUS, AL | PBMC, immune cells |
Papers are ordered chronologically, and cell types analyzed with single-cell platforms are listed. Tags are globally categorizing the papers based on the research question addressed.
BMMC, bone marrow mononuclear cells; CART, chimeric antigen receptor T-cell therapy; CNV, copy number variant; HMCL, human myeloma cell lines; IgE, immunoglobulin E; IMID, immunomodulatory derivatives; iMSC, inflammatory mesenchymal stromal cells; NDMM, newly-diagnosed MM; NGF, next-generation flow; NPC, normal plasma cell; OL, osteolytic lesion; PMID, PubMed identification number; SLO, secondary lymphoid organ.
To assess cell surface protein expression, scRNA-seq is often combined with additional quantification methods such as single-cell T-cell receptor (TCR) or B-cell receptor (BCR) sequencing and cellular indexing of transcriptomes and epitopes (CITE).19-22 Particularly in MM, understanding the T-cell and B-cell repertoires is critical because they affect the response to therapy and disease progression. Single-cell TCR/BCR sequencing techniques provide the full-length immunoglobulin sequence, including isotypes and TCR α/β. In combination with scRNA-seq, it can track T-cell clonal expansion in steady state and in disease, and track changes in the transcriptome of the same clone over the disease course and with treatment. Oftentimes, scRNA-seq and TCR/BCR sequencing methods are combined with epitope detection protocols to fine-tune the cellular annotations. The recent focus on immunotherapies has highlighted the use of TCR and BCR sequencing platforms. However, their long-term use on larger data sets will provide actual information about their use in research and clinical settings other than for detecting minimal residual disease.
A popular alternative to next-generation sequencing (NGS) platforms to study immune cells is CyTOF. CyTOF provides clear advantages over scRNA as a powerful approach for making high-dimensional measurements of protein expression in immune cells23 and analyzing many more cells than most NGS platforms, which allows the detection of rare cell populations. However, it is limited to selected markers. Therefore, careful design is needed, and identifying novel makers is limited to what is profiled. NGS platforms aim to overcome such issues by combining CITE-seq studies with scRNA-seq studies. However, there is still a trade-off in the number of cells analyzed. How to integrate such platforms should be considered carefully before starting any study.
In addition, the epigenome can be assessed on a single-cell level using assay for transposase-accessible chromatin (ATAC) with sequencing protocols.24 scATAC with sequencing determines chromatin accessibility across the genome, which suggests the enabled or disabled regulatory functions of genomic regions. There are also methods to evaluate the binding of DNA-associated proteins, such as single-cell Cleavage Under Targets and Tagmentation,25 Drop–chromatin immunoprecipitation (ChIP)/scChIP sequencing26 and antibody-guided chromatin tagmentation sequencing,27 as well as single-cell methylation sequencing,28 but such technologies have not been widely used in MM research. These methods, however, hold great promise, because such approaches have been extremely helpful in evaluating the resistance or sensitivity to certain conditions or treatment in other cancers.23,28-33 In prostate cancer, integrated scATAC with sequencing and scRNA-seq identified associations between distinct chromatin landscapes, enzalutamide treatment, resistance, and alternative transcriptional programs.29 In breast cancer, scChIP sequencing found a subset of cells within untreated drug-sensitive tumors that shared a chromatin signature with resistant cells, which was undetectable using bulk approaches.30 Another study took the integrative approach a step further by using single-cell triple omics to reconstruct genetic lineages and follow their epigenomic and transcriptomic dynamics.33 scATAC and methylation sequencing can also be used in copy number variant detection and phylogenetic tree construction. In some cases, RNA levels were also inferred from epigenomic data, although this remains somewhat experimental (Figure 1). More recently, studies mainly focusing on the spatial organization of BM samples have been presented. Such technologies would bring further insight and new knowledge; however, they would also require additional protocols to preserve the spatial organization of the sample.
Particularly relevant to cancer is scDNA sequencing, which is most frequently used to evaluate clonal evolution and clonal selection. As opposed to transcriptome and epigenome solutions, scDNA-sequencing methods have been slower to develop. Some early platforms are not available now and others still suffer significantly from low cellular inputs and high cost. In other hematological diseases, amplicon sequencing (ultra-deep sequencing of polymerase chain reaction products) is a highly targeted approach that enables researchers to analyze genetic variation in specific genomic regions. Although these techniques can capture thousands of cells, they only investigate a predetermined list of targets. In MM, very few genes have been associated with outcome and risk, which hinders the design of a specific targeted approach. Other methods that do not require targeted capture need higher sample inputs. In either respect, the hands-on time for library preparation is a challenge, and both methods require some form of DNA amplification, which introduces bias. In addition, whole-genome sequencing methods are often used for detecting clonal changes by tracking CNAs. Single-nucleotide variation and indel detection often requires >8× to 15× coverage at a minimum, which is expensive and still leads to missing data because of dropout rates and uneven coverage.
Computational workflows to analyze single-cell genomics data are also more challenging than bulk sequencing platforms. Several single-cell genomics platforms provide in-house pipelines such as cell-ranger or tapestri. However, there are many open-source solutions developed for similar purposes. To date, there are >1000 tools developed for scRNA-seq platforms, and there are public resources in which computational tools are often listed such as scrna-tools.org. Although the benchmarking of particular tools is often a challenge and understanding the data and the methods is critical for benchmarking, studies that combine multiple data sets with simulation studies and possible ground truths validated in the wet lab provide useful insights to analysis.10,34-38 Most workflows and downstream analysis tools are developed by using R and Python. Therefore, understanding these programming languages would be helpful for customizing analysis, optimizing analytical codes, and for visualization. Preprocessing raw data before downstream analysis also requires high-throughput computing environments, and the required hardware to handle samples almost grows logarithmically with increasing sample size and cell numbers. Initial steps are more parallelizable and potentially accessible using standard central processing units (CPUs) and graphics processing units (GPUs), whereas downstream analyses are more heavy on random-access memory (RAM) consumption.34,39 There are also several workflows that were originally created for bulk genomics data, such as BWA40 and Salmon41 that have been adapted to single-cell genomics. Creating the best data possible and setting up the right workflow with appropriate quality control steps have a major impact on the outcomes of the analysis.
Delineating multiple myeloma, 1 cell at a time
All genomic studies published in the last decade have concluded that MM cells are widely heterogeneous.42-51 This was first seen in bulk sequencing studies, in which mutations caused noticeable noise in the signal. Yet, because bulk sequencing combines the genomes of different cells, using it to sensitively interrogate mutation rates heavily depends on sequencing depth and regions captured.52 Thus, it is incredibly difficult to build evolutionary trees and assess subclone rates using bulk data. The outlook worsens for bulk RNA-seq, because it averages the transcriptomes of different cells, making it difficult to detect changes in gene expression within particular cell types and states, as well as limiting the number of detectable mutations.18 In fact, in bulk data, the correlation between CNAs and gene expression is limited.53 As single-cell profiling looks at individual cells, it provides an approach to sensitively find, track, and interrogate cell types, states, and clones.54
Clonal evolution
Intratumor heterogeneity is considered a major driving factor for clonal selection and the cause of resistant clone growth.55,56 Liu et al combined scRNA-seq with various genomics platforms to evaluate clonal evolution and the effect of the tumor microenvironment by using longitudinal samples from patients at various stages.57 This approach allowed the authors to distinguish between somatic mutations in PCs vs microenvironment cells, and to evaluate coexisting mutations in the same cell. They investigated how PCs evolve from smoldering MM (SMM) to active MM to relapse by integrating somatic alterations (such as CNAs), cell-lineage marker gene expression (like B-cell maturation antigen [BCMA], SLAMF7, or CD138), and differential gene expression. They found that PC subpopulations are stable during the transition from precancer to diagnosis but are dynamically gained or lost between diagnosis and relapse.57 These cell subpopulations were involved in distinct activities, with 1 group focused on translation, 1 having potentially different interactions with the immune microenvironment, 1 focused on proliferation, and an occasional subgroup that appeared to be senescent.57 In addition, Ledergor et al58 also used scRNA-seq on samples from 40 patients with MM and 11 healthy donors, finding extensive subclonal structures for >30% of the patients with MM. In individuals who are asymptomatic with early disease and in those with minimal residual disease after treatment, rare tumor PCs were detected with molecular characteristics similar to those of active myeloma cells. The PETHEMA/GEM Cooperative Group study using scRNA-seq revealed a similar number of transcriptional clones in patients with newly diagnosed MM that had standard- or high-risk disease.59
Using single-cell methods, 2 studies have reported clonal selection after chimeric antigen receptor–T cell therapy.60,61 One study collected longitudinal samples from a patient before and after anti-BCMA chimeric antigen receptor–T cell therapy, at relapse, and after retreatment, and used scRNA-seq to assess cell state and infer copy number variants. A tiny subclone, which made up <5% of MM cells was observed with del 16p (BCMA locus) at the time of initial therapy. This clone became the dominant clone after treatment, with mutation in the second allele, resulting in biallelic loss of the target antigen (deletion and point mutation combined), suggesting positive selection and evolution. Similarly, another study61 evaluated serial samples from another patient treated with BCMA-targeting therapy and found that biallelic deletion of BCMA before treatment caused resistance and clonal selection. In both studies, scRNA-seq was supported by additional genomic profiling.
In addition to using scRNA-seq, a few studies have used targeted or whole-genome scDNA sequencing or scRNA to evaluate clonal evolution and the differences between BM PCs and circulating tumor cells (CTCs). Lohr et al62 used single-cell targeted sequencing and scRNA-seq to demonstrate that, even at the single-cell level, there is high concordance between single-nucleotide variants and CNAs between CTCs and the BM, and that CTCs can be used to track clonal evolution. This is important, because collecting serial samples from the BM is often impractical for patients and care providers. A study comparing transcriptomic differences between BM cells and CTCs at single-cell resolution also reported a high correlation in transcriptomes.63 Another study used targeted single-cell mutation profiling to evaluate the correlation between clonal hematopoiesis and myelodysplastic syndrome in patients with MM,64 finding that patients with MDS had a significantly higher rate of clonal hematopoiesis and that these clones had a median allelic frequency of 8%. In terms of evaluating tumor cells in different parts of the body, scRNA-seq data from paired osteolytic bone lesions and BM samples have now revealed spatial variability in tumor cell composition.65 The PCs of osteolytic lesions, compared with those of the BM, showed upregulated genes associated with myeloma bone disease and downregulated Wnt-signaling.
Clonal selection in the clinical setting was recently well documented using single-cell shallow whole-genome DNA sequencing.66 In 28% of patients at diagnosis, a very low-frequency high-risk subclone was identified by single-cell analysis but it was not detected by the conventional method (below its detection limit). This clone became detectable by the conventional method at relapse after therapy, with myeloma being relabeled as high-risk in these patients. The clinical impact of low-level high-risk clones detectable only by single-cell sequencing was confirmed by looking at a large data set of patients with MM. Patients who had high-risk events detected only at relapse had similar outcomes as patients in whom such events were detected at diagnosis. This study highlighted the need to use more sensitive approaches, like scDNA sequencing, to detect risk features in very small cell populations.
Transcriptional reprogramming
In addition to genomic heterogeneity, there is transcriptional heterogeneity and flexibility in myeloma. Using single-cell transcriptome and chromatin accessibility profiling, the coexistence of distinct transcriptional states has been reported in MM. Furthermore, MM treatments promote transcriptional reprogramming and differential enhancer recruitment while simultaneously reducing developmental potential.67 Treatment also generated a distinct complement of actionable immunotherapy targets. Transcriptional rewiring was also evaluated in normal PCs and 2 distinct PC disorders, MM and light-chain amyloidosis (AL).68 A transcriptional atlas of normal PC development in secondary lymphoid organs, peripheral blood (PB), and the BM was created to compare against PCs from AL, MM, and monoclonal gammopathy of undetermined significance (MGUS). The study showed that AL shares greater similarity to secondary lymphoid organ PCs, whereas MM is transcriptionally closer to PB PCs and newborn BM PCs. However, patients with AL and MM cells enriched in immature transcriptional programs had inferior survival.
Drug resistance studies
Single-cell profiling is a powerful tool to understand the impact of treatment at a cellular level. Using samples from a patient who was exposed to proteasome inhibitor (PI), an immunomodulatory agent, and anti-CD38 monoclonal antibody treatment, Masuda et al found that a population of cells that emerged during lenalidomide treatment disappeared after PI treatment.69 These cells had higher expression of PELI2, which was responsible for PI-induced cell death in an in vitro assay. To understand drug sensitivity and resistance, scRNA-seq of a variety of MM cell lines has been used to predict the combination of drugs that would be most effective, based on the percent of cells expressing the target genes, and single-cell proteomics has been used to evaluate the effect of treatment on cellular phenotype.70
To understand the outgrowth of resistant cells after therapy, full-length scRNA-seq on CD138+ cells from 3 patients with BRAF-mutated myeloma, before and after treatment with a BRAF/MEK inhibitor was performed.71 Patients had rapid cellular state changes in response to treatment, first on a transcriptional level with widespread enhancer remodeling, followed by the outgrowth of detectable, genetically discernible drug-resistant clones. These resistant cells relied on increased oxidative phosphorylation for their energy needs, yet at a single-cell level, the magnitude of the effect varied across patients and was inversely correlated with the activation of MAPK. The molecular dynamics of drug resistance has been studied using longitudinal scRNA-seq in a single-arm clinical trial. Samples from 41 patients with primary refractory or early relapsed disease were compared with samples from 11 healthy individuals and 15 patients with newly diagnosed MM. A gene expression signature that defined a subset of patients with primary-refractory MM, including perturbation in mitochondrial stress genes, the ER and UPR pathways, and proteasome machinery was reported.72 Nonresponders had downregulated PC function and upregulated proteasome and mitochondrial stress genes. Importantly, using scRNA-seq, they found clones that were sensitive or resistant to treatment, with poor responders tending to have large cellular populations of a resistant clone, either before or after treatment. Moreover, peptidylprolyl isomerase A was identified as a potential target for resistant MM, supported by a CRISPR screening. These studies provide the insight to now consider appropriate combinations of agents that can prevent or delay the development of such resistance. It should be noted that some of these studies draw their conclusions from a small sample size. Given that MM has high heterogeneity, larger studies are required, or further functional studies are needed, to validate and target some of these resistance mechanisms.
Investigating the microenvironment with single-cell strategies
The BM microenvironment has a major influence on oncogenesis in a variety of B-cell malignancies including MM, in which malignant PCs have bidirectional interactions with the BM elements.73,74 The multiple and complex interactions between MM cells and various BM cell types, including B, T, natural killer (NK), and myeloid cells, contribute to PC growth, proliferation, and drug resistance.75 As such, identifying the role of the various actors of the MM microenvironment in myelomagenesis and myeloma progression is essential.76,77 Single-cell sequencing methods have been used to characterize the immune ecosystem and microenvironment in various cancer types78-80 and, more recently, in MM. These applications have identified the changes in the immune microenvironment that are associated with the progression from early precursors (MGUS and SMM) to advanced myeloma, the impact of the microenvironment on MM cell behavior, and the impact of therapy.
Microenvironment and precursor disease
Changes in the immune system occur early during the MGUS stage, with an increase in CD16+ monocytes, NK, T cells, and precursors,81 as observed in a single-cell transcriptomic analysis of 19 000 CD45+/CD138− cells from the microenvironment of patients with MGUS (n = 5 patients), low-risk SMM (n = 3), high-risk SMM (n = 8), newly diagnosed MM (n = 7), and from 9 healthy donors. NK cells were more numerous in the early stages and associated with altered chemokine receptor expression and progressive loss of granzyme K+ memory cytotoxic T cells at myeloma progression.81 Changes in NK cell populations in patients with MGUS vs healthy donors have been reported, suggesting an early impact of the disease process on the immune microenvironment.82 This study also reported the enrichment of terminally differentiated T cells and stem-like T-cell factor 1–high memory T cells in MGUS, which, respectively, increased and decreased in size during the progression to MM. The loss of T-cell factor 1–positive memory T cells in MM may lead to the inability to maintain protective immunity over time and the loss of immune surveillance. These initial results of immune composition using scRNA-seq were validated with mass cytometry. Similar observations were also reported by other groups showing varying compositions of cell types over the disease course.57 To understand the T-cell repertoire and kinetics in depth, the usual scRNA-seq approach was expanded to include TCR sequencing.83 This combined investigation showed that newly-diagnosed MM (NDMM) was associated with a decrease in both memory and naïve CD4 T cells and an increase in CD8+ effector T cells, regulatory T cells, and CD14/CD16 monocytes.83 The analysis showed an enrichment of nonclonal memory B cells and no change in the clonality of T cells between MGUS, SMM, and MM.
Microenvironment and advanced disease
The BM microenvironment and stromal cells play a crucial role in MM pathogenesis by promoting MM cell growth, survival, and drug resistance.84 Interactions of stroma and MM cells have been connected to changes in chromatin states and transcriptomic programs, which led to therapeutic resistance, accelerated disease dissemination, and impaired long-term survival.85 However, the various treatments and more aggressive clones of advanced disease can also affect these compartments. A study comparing 13 patients with NDMM, and 7 healthy controls identified interferon-responsive effector T cell and CD8+ stem cell memory T-cell populations as potential sources of stromal cell–activating cytokines. Importantly, this proinflammatory signaling was not abrogated by efficient antimyeloma therapy.86 Another study reported single-cell transcriptomic data from 406 946 CD38− cells from 20 patients with relapsed/refractory MM (RRMM) and healthy donors. It showed an upregulation of inflammatory cytokines, with an accumulation of PD1+ γδ T cells and tumor-associated macrophages. Importantly, the most pronounced changes were observed during treatment with immunomodulatory imide drugs, with an increase in plasmacytoid dendritic cells, confirming a potential role for directly targeting plasmacytoid dendritic cells in MM.87,88 The impact of myeloma subtype has also been studied with scRNA-seq in 20 patients with RRMM before and after treatment. Subclones carrying high-risk features like 1q gain had a specific transcriptomic signature and frequently expanded during treatment. Different from the previously discussed works, integrated data from the BM microenvironment showed that CD14+ monocytes are a major driver of inflammation in MM and that RRMM cells contribute to generating an immunosuppressive microenvironment by upregulating inflammatory cytokines and closely interacting with the myeloid compartment.87 A recent study presented at the American Society of Hematology’s 2022 annual meeting focused on the spatial organization of tumor growth as well as immune infiltration in MM. It showed that T-cell entry depends on target recognition and costimulation and suggests a novel role for tumor-associated dendritic cells (DCs) in regulating the entry of neoantigen-specific T cells and the effector phase of the cancer immunity cycle.89 Single-cell profiling from a recent phase 2 study in which patients with high-risk SMM received elotuzumab, lenalidomide, and dexamethasone showed that posttherapy immune normalization and a higher abundance of GZMK+ cytotoxic T cells are associated with better progression-free survival. Critically, it also showed that blood-based immune profiling can reflect immune alterations observed in the BM.90 Another multicenter study recently published made a similar observation for active MM. The authors reported that patients who rapidly progressed had significantly higher enrichment of GZMK+ and TIGIT+ exhausted CD8+ T cells, with a significantly higher enrichment of M2 tolerogenic macrophages and activation of proproliferative signaling pathways, such as BAFF, CCL, and IL16.91
The compilation of the data suggests that a bidirectional interactive relationship between clonal evolution and the tumor microenvironment exists. The tumor cells affect the composition and state of the microenvironment, which in turn, provides growth and survival advantage to MM cells. Thus, the evolution process is not only selected by, but also shapes, the microenvironment. This microenvironment reprogramming showed significant patient specificity, which tracks with the tumor heterogeneity of patients.92
Future of single-cell applications in MM
Myeloma cells are highly heterogeneous42,43,93 and the immune cells surrounding them are in a highly complex and constantly changing structure.94 Within such a dynamic and complex structure, longitudinal studies are necessary to evaluate MM and its microenvironment, yet, because of the inherent variation between patients, we still need a well-established large data set to begin to understand the cellular states at diagnosis and at relapse from specific intervention. A recent pilot study investigating the microenvironment using scRNA-seq, CyTOF, and CITE-seq on the same 18 patients found high concordance between the technologies for identifying cell-type abundance.95 Another data set recently published showed high concordance between BM and PB samples90 (Figure 2). In addition to the microenvironment, some studies96-98 have started to report multiomic efforts in MM cells. Combining larger data sets with multiomics and longitudinal samples will provide conclusive evidence with granular details about tumor cells and microenvironmental interactions, including better subgroup comparisons. More importantly, with the rise of multiomic data sets, we will be able to integrate data from the same cell from various platforms.23,99 Such methods will increase the resolution we get from data but cost control will be a significant contributor to the success of these efforts.100
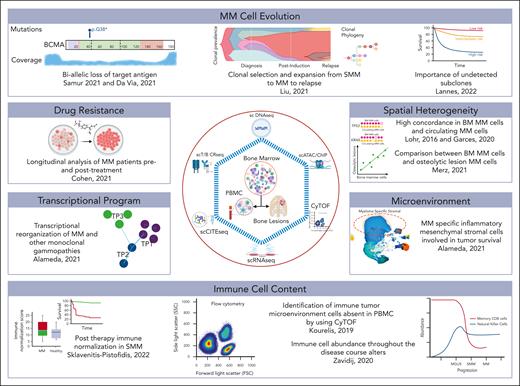
Overall summary of research concepts using cells from the BM, blood circulation, or lesions outside the BM, and single-cell platforms to address various hypotheses.
Overall summary of research concepts using cells from the BM, blood circulation, or lesions outside the BM, and single-cell platforms to address various hypotheses.
Emerging single-cell profiling strategies will likely provide additional information on MM. The spatial transcriptomic scRNA-seq technologies will likely contribute to an even better characterization of the intercellular crosstalk between myeloma cells and specific cell populations from the BM microenvironment.101 In addition, in the near future, we expect to see epigenetic changes profiled alongside RNA. Other single-cell platforms, such as single-cell perturb sequencing and single-cell barcoding platforms will allow evaluation of the selection and accumulation of somatic events, and we can expect to see single-cell methods in translational research settings with potential applications in clinical practice and personalized medicine.
Conclusions
Just like with any other cancer, research using single-cell platforms has become almost the norm in MM research (Figure 2). Decreasing cost, increasing accessibility, breakthroughs in obtaining multiomics data for the same cell, and innovative computational programs for analyzing data have made single-cell platforms increasingly attractive. As such, the future is bright in MM research with the availability of single-cell methodologies, with many insights already discovered and more awaited.
Acknowledgments
This work was supported by National Institutes of Health (NIH) Research Program Project grant P01-155258 (N.C.M. and M.K.S.) and NIH National Cancer Institute grant 5P50 CA100707 (N.C.M. and M.K.S.), Department of Veterans Affairs Merit Review Award I01BX001584-01 (N.C.M.), and Rogers Family Foundation grant (N.C.M. and M.K.S.).
Authorship
Contribution: M.K.S., R.S., and N.C.M. reviewed the literature, prepared the visuals and tables, and wrote the manuscript.
Conflict-of-interest disclosure: N.C.M. is a consultant for BMS, Janssen, Oncopep, Amgen, Pfizer, Karyopharm, Legend, NextRNA, Raqia, AbbVie, Takeda, and GSK; and on the board of directors of Oncopep. The remaining authors declare no competing financial interests.
Correspondence: Mehmet Kemal Samur, Dana-Farber Cancer Institute, 450 Brookline Ave, Boston, MA 02215; e-mail: mehmet_samur@dfci.harvard.edu; and Nikhil C. Munshi, Dana-Farber Cancer Institute, 450 Brookline Ave, Boston, MA 02215; e-mail: nikhil_munshi@dfci.harvard.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal