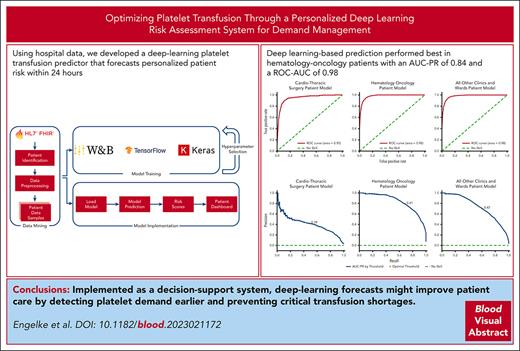
Using hospital data, we developed a deep-learning platelet transfusion predictor that forecasts personalized patient risk within 24 hours.
Deep learning–based prediction performed best in patients with hematologic cancers with an AUC-PR of 0.84 and an ROC-AUC of 0.98.
Visual Abstract
Platelet demand management (PDM) is a resource-consuming task for physicians and transfusion managers of large hospitals. Inpatient numbers and institutional standards play significant roles in PDM. However, reliance on these factors alone commonly results in platelet shortages. Using data from multiple sources, we developed, validated, tested, and implemented a patient-specific approach to support PDM that uses a deep learning–based risk score to forecast platelet transfusions for each hospitalized patient in the next 24 hours. The models were developed using retrospective electronic health record data of 34 809 patients treated between 2017 and 2022. Static and time-dependent features included demographics, diagnoses, procedures, blood counts, past transfusions, hematotoxic medications, and hospitalization duration. Using an expanding window approach, we created a training and live-prediction pipeline with a 30-day input and 24-hour forecast. Hyperparameter tuning determined the best validation area under the precision-recall curve (AUC-PR) score for long short-term memory deep learning models, which were then tested on independent data sets from the same hospital. The model tailored for hematology and oncology patients exhibited the best performance (AUC-PR, 0.84; area under the receiver operating characteristic curve [ROC-AUC], 0.98), followed by a multispecialty model covering all other patients (AUC-PR, 0.73). The model specific to cardiothoracic surgery had the lowest performance (AUC-PR, 0.42), likely because of unexpected intrasurgery bleedings. To our knowledge, this is the first deep learning–based platelet transfusion predictor enabling individualized 24-hour risk assessments at high AUC-PR. Implemented as a decision-support system, deep-learning forecasts might improve patient care by detecting platelet demand earlier and preventing critical transfusion shortages.
Introduction
Platelet demand management is an important aspect of patient care. Identifying and treating patients who need platelet transfusions requires careful planning to ensure the right amount of platelet concentrates (PCs) are available at the desired time. This task can be challenging and time-consuming, particularly in hospital settings, in which a lack of human resources can negatively affect patient care.
Moreover, because of the short shelf-life of PCs and fluctuating demand, platelet inventory management is subject to a delicate balance between high expiration rates and product shortages. Despite increasing methods available to reduce platelet wastage, high spoilage rates remain a global problem exacerbated by increasing demand.1 New developments in platelet preparation, storage, and other novel approaches may help address this problem. The systematic review "Is platelet expiring out of date?" by Flint et al2 provides valuable insight into platelet inventory management by evaluating existing methods to reduce platelet expiration rates and improve product availability. It was found that although complex methods, such as operational research and simulation, had the lowest predicted wastage, they may be difficult to implement at a typical blood bank.2 More recent studies use methods that include machine learning3-5 (ML), statistical time series forecasting,3-5 and peak demand analysis.6 When trying to predict global demand, these studies were able to reduce wastage, yet it was found that using more complex methods, such as long short-term memory (LSTM), did not lead to notably fewer outdated PCs. Additionally, Perelman et al6 showed more platelet demand peaks than other blood components over 10 years. This suggests a need for specific strategies to manage peak platelet demand. In contrast to previous studies that addressed the problem from a global perspective, our study, to the best of our knowledge, is the first to examine platelet demand from an individualized viewpoint, opening a new dimension for further research and potential solutions.
A clinical decision-support system framework described in recent health care studies can potentially increase efficiency and improve patient care.7,8 Such systems outline instances in which a predefined probability threshold for a specific event is crossed. Recognizing their potential applicability to transfusion medicine, we opted for an approach that leverages the latest advances in artificial intelligence. A supervised deep learning (DL) architecture was chosen to solve the supervised binary classification problem. A supervised binary classification problem refers to a task in which a ML model is trained to predict 1 of 2 possible outcomes. In our study, the 2 possible outcomes (referred to as “labels”) are “the patient will need a PC transfusion” and “the patient will not need a PC transfusion.” DL is a type of ML that uses artificial neural networks to learn and make decisions.9 Supervised learning is the most common type of learning, in which the label is known in advance.9
This study aimed to create a personalized support system for PC demand management by designing a DL model that computes a risk score. The risk score, a decimal between 0 and 1, expresses the likelihood a patient will need a platelet transfusion in the next 24 hours. A higher score indicates a greater likelihood that a transfusion will be needed. By calculating a risk score for individual patients, physicians can optimize platelet transfusion timing, improve resource allocation, and improve health care outcomes. We used data from various clinical information technology systems to develop and train this model.
Methods
Statement of ethics
This study was conducted according to the Declaration of Helsinki and approved by the Ethics Committee of the Medical Faculty of the University of Duisburg-Essen (approval number 20-9386-BO). Because of the retrospective nature of the study, the requirement of written informed consent was waived for individual patients when data sets were anonymized for model development.
Methodological overview
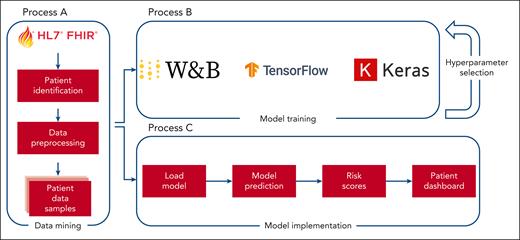
The model construction and deployment processes are outlined in Figure 1. An identical data extraction procedure was used for both model training and deployment, albeit with different parameters. Specifically, clinical data from January 2017 to December 2022 was used for model training along with the corresponding labels. Conversely, for the practical application of the model, prospective data are collected to facilitate future predictions about patients currently admitted to the hospital.
Model development and deployment overview to create predictive models capable of creating real-time predictions. Process A transforms patient data into a format suitable for training and prediction tasks. Process B uses the output of process A to generate training samples that are then used to develop machine learning models. Process C uses the same output from process A to create samples derived from current hospital patients, enabling immediate predictions through the use of the trained models generated by process B. W&B, Weights & Biases.
Model development and deployment overview to create predictive models capable of creating real-time predictions. Process A transforms patient data into a format suitable for training and prediction tasks. Process B uses the output of process A to generate training samples that are then used to develop machine learning models. Process C uses the same output from process A to create samples derived from current hospital patients, enabling immediate predictions through the use of the trained models generated by process B. W&B, Weights & Biases.
Data mining process
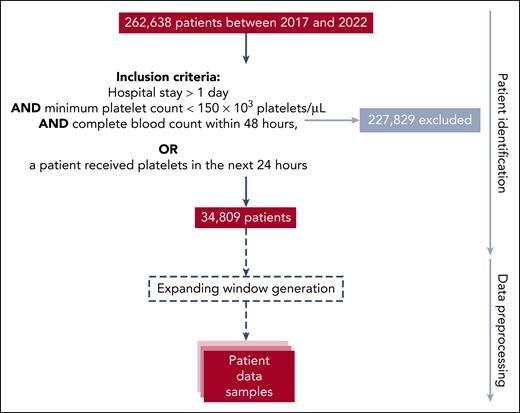
This retrospective study was conducted over 5 years at the Essen University Hospital, a tertiary care academic referral center in Germany. The cohort data were obtained from the hospital's internal Health Level Seven International Fast Healthcare Interoperability Resources (FHIR)10 server. FHIR is a standard server for storing information and exchanging electronic health data, such as records generated during the clinical routine. FHIR-PYrate11 was used to query the FHIR server and obtain relevant patient information, including patient stay, laboratory results, diagnosed conditions, prior platelet transfusions, medication intake, received procedures, sex, and age, in a standardized way. Figure 2 shows the process from identifying valid patients to the transformation into valid samples.
Study cohort selection and data preprocessing. The data mining process is illustrated as a top-down flow diagram. The entire cohort comprised 262 638 patients, and the identification process returned 34 809 patients for the scope of this study. During data preprocessing, 437 791 samples were generated, which were further split into training, validation, and test sets on a per-patient level. Each extracted resource had to pass validation processes to ensure the raw data were consistent. The validation processes included comparing the total number of available data in FHIR by resource to the downloaded resources and manual spot checks between source systems and the extracted data set.
Study cohort selection and data preprocessing. The data mining process is illustrated as a top-down flow diagram. The entire cohort comprised 262 638 patients, and the identification process returned 34 809 patients for the scope of this study. During data preprocessing, 437 791 samples were generated, which were further split into training, validation, and test sets on a per-patient level. Each extracted resource had to pass validation processes to ensure the raw data were consistent. The validation processes included comparing the total number of available data in FHIR by resource to the downloaded resources and manual spot checks between source systems and the extracted data set.
Inclusion criteria for the model were established to capture pertinent cases while still ensuring the relevance of the data to control data imbalance without losing relevant samples. The criterion “hospital stay >1 day” was established to include patients under stable medical monitoring for a sufficient duration to yield data. The requirement “minimum platelet count <150 × 103 platelets per μL” per sample ensured those at risk for thrombocytopenia-related bleeding incidents, according to the reference values12 and the hemotherapeutic guidelines,13,14 were primarily considered, directing the model's focus. In addition, a “complete blood count within 48 hours” was necessary to obtain valid samples. Patients who had received platelet transfusions within 24 hours, regardless of other criteria, were included in the study because of their relevance to the objective of the models.
Primarily, PCs are used to prevent and treat platelet-related bleeding. The indication for a PC transfusion depends predominantly on the bleeding risk, clinical symptoms, underlying disease, and platelet count.13,14 Although acute hemorrhage and the risk of bleeding could not be foreseen with the current data architecture, the diseases and the platelet count of such cases were considered.
The foundation for generating training samples was defined by the start and end of a patient’s stay at the hospital. Each sliding window was sampled with a frequency of 12 hours, and each day was translated to 2 data points. These time spans were filled with laboratory test results, conditions diagnosed, platelet transfusions administered, medications received, and procedures underwent, all of which translated into features. A complete list of the selected features generated from medications, procedures, and conditions can be found in supplemental Tables 1 to 4 (available on the Blood website). The features were selected according to the recommendation of the hemotherapeutic guidelines13,14 and several medical expert interviews.
Data were then used to generate training samples and labels. Each patient's data point was further transformed into expanding data windows. Expanding window generation is a method used to generate a sequence of overlapping windows of a fixed lower bound and a variable upper bound size, starting with a small window and increasing the window size with each iteration.15,16 The stepping size between each expansion was 1 day. To ensure all input sequences in a batch have the same length, the data were reduced to the last 60 data points (30 days) of a window. If the expanding window length consisted of fewer than 60 data points, padding tokens were appended to the beginning of the sequence to exclude data cells without any information. The trainable input data varied from 1 to 30 days. The platelet count between 2 platelet observations was linearly interpolated. A binary label was formed for each training sample, identifying whether a patient will need a PC in the next 24 hours. The first label represents no transfusion, and the second represents ≥1 PCs.
Platelet classification and study design
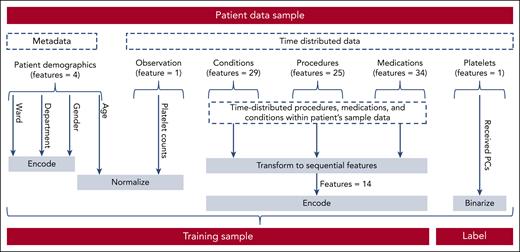
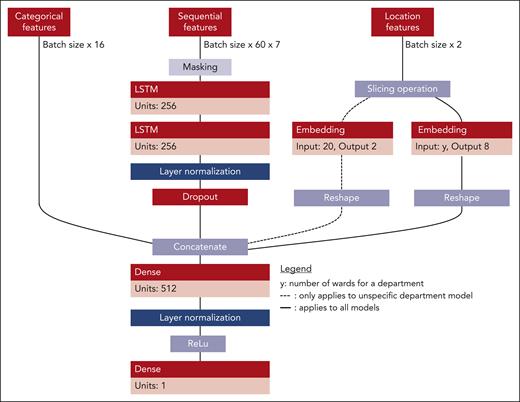
The extracted data were fed into a final transformation process to generate valid training samples (Figure 3). The transformed data were divided into 2 groups, 80% for training and 20% for testing, on a per-patient level. One-fifth of the training data set were used as validation data. Each patient could only appear in one of the splits. The data were heavily skewed toward the first class, because most patients received no PC transfusions (4% positive labels). Most transfusions were consumed by either the hematology and oncology department or the cardiothoracic surgery department. Given the relevance of these distinct departments for the entire hospital's transfusion volume, we trained department-specific models for these 2 departments and a third multispecialty model for the remaining cohort across the entire hospital. The training samples consisted of 3 input types that represented the input space of the model's architecture. The first input type, sequential features, consists of tensor elements representing the generated time series with a sample size of batch size 60 × 7. Batch size refers to the number of training examples or sequences processed together in each forward and backward pass during the training process. The number 60 represents the length of each sequence (the number of time steps) in each example. The 7 features of each time series element included the platelet count, a cosine and sine representation of the day of the week, and Boolean variables indicating whether the platelet count was measured or interpolated, whether a PC was given, whether a time slot was a holiday in the hospital's state, and whether it took place on the weekend. Second, the model incorporated location features, uniquely identifying a patient's last institute and ward. Each is presented as a set of elements within the sample size of batch size ×2. The first feature represented the encoded department, and the second represented the encoded ward. When training the department-specific models, the department-encoding feature was disregarded. Finally, categorical features were represented by a sample size of batch size ×16. These features include data from past procedures, diagnosed conditions, and medications (supplemental Tables 1-4) within the last 30 days as well as sex and age. The last layer of each model is a dense layer, which contains an activation function and reduces the dimension of the feature space to 1. An activation function is a mathematical operation applied to the output of a neural network layer that introduces nonlinearity. In this case, a sigmoid function is used as the activation function to map the output between 0 and 1 for binary classification.17Figure 4 shows an overview of the model's architecture.
Data sampling from raw patient samples to trainable samples. Before using the patient data samples retrieved from the data mining process for training, each sample had to pass another preprocessing step. First, metadata, such as department, specific ward, and sex, had to be encoded, and age was normalized. Next, the received PCs per time window were encoded. Then, categorical features of the time-distributed data were transformed into a one-hot encoding (see supplemental Figures 1-4 for a complete list of conditions, medications, and procedures). Finally, the observations were normalized. This was done for every sample before the data were batched.
Data sampling from raw patient samples to trainable samples. Before using the patient data samples retrieved from the data mining process for training, each sample had to pass another preprocessing step. First, metadata, such as department, specific ward, and sex, had to be encoded, and age was normalized. Next, the received PCs per time window were encoded. Then, categorical features of the time-distributed data were transformed into a one-hot encoding (see supplemental Figures 1-4 for a complete list of conditions, medications, and procedures). Finally, the observations were normalized. This was done for every sample before the data were batched.
Architecture of the LSTM–based multi-input single output model. The first input type is a tensor representing the generated time series, with 7 features for each time series element, including platelet count, day of the week (represented by cosine and sine), and Boolean variables indicating measurements, PC given, holiday, and weekend. The second input type is location features uniquely identifying a patient's last institute and ward. The third input type is categorical features generated from past procedures, diagnosed conditions, and given medications within the last 30 days. The sample size for each input type is batch size 60 × 7, batch size ×2, and batch size ×16, respectively. The institute encoding feature is disregarded when training department-specific models. The model's final output, generated through the sigmoid activation function in the last dense layer, represents the likelihood that a patient will need a platelet transfusion.
Architecture of the LSTM–based multi-input single output model. The first input type is a tensor representing the generated time series, with 7 features for each time series element, including platelet count, day of the week (represented by cosine and sine), and Boolean variables indicating measurements, PC given, holiday, and weekend. The second input type is location features uniquely identifying a patient's last institute and ward. The third input type is categorical features generated from past procedures, diagnosed conditions, and given medications within the last 30 days. The sample size for each input type is batch size 60 × 7, batch size ×2, and batch size ×16, respectively. The institute encoding feature is disregarded when training department-specific models. The model's final output, generated through the sigmoid activation function in the last dense layer, represents the likelihood that a patient will need a platelet transfusion.
For the DL models, TensorFlow18 with Keras19 and Weights and Biases20 were used for training and optimization. As optimizers, Adam21 and AdamW22 were chosen. The weight decay regularization for the AdamW optimizer was chosen as a 10th of the learning rate.22 To accelerate the hyperparameter search, 2 algorithms were used. First, early stopping was used to track the validation precision-recall curve.23 The early stopping patience was set to 10 epochs, and weights were restored for the best run. Second, the hyperband method was used to stop runs,24 which performed poorly compared with the previous validation precision-recall scores. Each model ran for 30 epochs if not stopped early because of early stopping or the hyperband method. As a search strategy, Bayesian optimization was chosen, and the goal was to maximize the best area under the precision-recall curve (AUC-PR) score.25 Additional metrics reported are Matthews correlation coefficient score, precision, specificity, and sensitivity. Optimizing the multispecialty model was achieved using the hyperparameter search spaces shown in supplemental Table 5, for which 300 models were generated. Likewise, the department-specific models were optimized using the same hyperparameter search spaces shown in supplemental Table 5 but with a larger sample of 600 models because of its relatively lower computational complexity. To further refine the models, sharpness-aware minimization without mini-batch splitting was used to improve further generalization and robustness to label noise in terms of model training convergence.26 Therefore, another 100 models were trained for each model type using the hyperparameter spaces in supplemental Table 5. After identifying the best model configuration, a fivefold cross-validation27 was conducted, creating an ensemble model that averaged the risk scores for each domain. Cross validation is a prevalent method in ML that involves dividing the data set into a specific number of subsets based on the cohort. A separate model is then trained for each of these splits. To ensure the reliability of the test data set, a bootstrapping method with a fold of 1000 and a confidence interval of 0.95 was reported. Bootstrapping is a common resampling technique, in which samples are selected with replacements from the original data set. As a result, some samples may be repeated several times, whereas others may not be included at all.28
After the best-performing model was found, the threshold for interpreting probabilities to class labels needed to be determined. Because the data set was highly imbalanced toward the class of patient samples receiving no transfusions, the precision-recall curve was calculated across the number of occurring thresholds within the data set using scikit-learn.29 The optimal threshold was determined by calculating the maximum F1 score for each threshold on the best-performing models, which was motivated by the unbalanced nature of our data set in which the minority class (true label) had significant importance.30 To understand the developed models, we used LIME,31 a well-established method to understand the relative feature contribution of specific features within the prediction process. Specifically, we calculated the relative feature contributions for each sample in our data set and subsequently estimated the overall contribution of features by averaging over all samples.
To compare the DL approach with classical ML methods, we trained XGBoost32 with a Random Forest33 (RF) classifier to compare the DL approach with classical ML methods. Weights and Biases20 was used for model training and optimization. Hyperparameter settings, model performance, and implementation details can be found in supplemental Tables 6 and 7. Feature importance can be found in supplemental Table 1.
Results
Patient characteristics
After the processing steps shown in Figure 2, the study group comprised 34 809 individuals hospitalized between 2017 and 2022. The age of the patients ranged from 1 to 110 years (42% female patients), with a median of 64 years. The baseline characteristics of the training and test cohorts are detailed in Table 1.
Patient characteristics of the selected cohort
| Statistic . | Male training . | Male test . | Female training . | Female test . |
|---|---|---|---|---|
| Age range (minimum-maximum), y | 0-106 | 0-111 | 0-110 | 0-104 |
| Interquartile range, y | 55-78 | 54-77 | 43-77 | 41-76 |
| Median age, y | 67 | 67 | 64 | 63 |
| Standard deviation age, y | 20.99 | 21.17 | 23.05 | 23.31 |
| Total patients | 16 251 | 4 110 | 11 596 | 2 852 |
| Total proportions transfused | 23 951 | 5 177 | 15 201 | 3 478 |
| Total proportions transfused in hemato-oncology | 13 724 (57%) | 3 056 (59%) | 8 964 (59%) | 2 095 (60%) |
| Total proportions transfused in cardiothoracic | 4 582 (19%) | 1 024 (20%) | 2 408 (16%) | 564 (16%) |
| Total proportions transfused in multispecialty | 5 645 (24%) | 1 097 (21%) | 3 828 (25%) | 819 (24%) |
| Leukemia | 9 758 | 2312 | 6 990 | 2 113 |
| Leukemia remission | 1 154 | 267 | 1 030 | 263 |
| Lymphoma | 11 121 | 3 041 | 6 605 | 1 547 |
| Other malignancies | 2 426 | 558 | 1 406 | 419 |
| Anemia | 6 849 | 1 687 | 6 110 | 1 409 |
| Coagulopathies | 21 108 | 4 955 | 13 407 | 3 126 |
| Infections | 565 | 140 | 355 | 86 |
| Rheumatic diseases | 300 | 56 | 1 043 | 204 |
| Palliative care | 263 | 60 | 290 | 96 |
| Other therapies | 25 142 | 6 871 | 19 008 | 4 266 |
| Surgery interventions | 19 939 | 4 515 | 11 394 | 2 328 |
| Chemotherapy | 92 938 | 24 519 | 74 287 | 18 228 |
| Antiinfectives | 80 204 | 20 130 | 51 678 | 10 663 |
| Other drugs | 7 627 | 1 899 | 4 730 | 972 |
| Statistic . | Male training . | Male test . | Female training . | Female test . |
|---|---|---|---|---|
| Age range (minimum-maximum), y | 0-106 | 0-111 | 0-110 | 0-104 |
| Interquartile range, y | 55-78 | 54-77 | 43-77 | 41-76 |
| Median age, y | 67 | 67 | 64 | 63 |
| Standard deviation age, y | 20.99 | 21.17 | 23.05 | 23.31 |
| Total patients | 16 251 | 4 110 | 11 596 | 2 852 |
| Total proportions transfused | 23 951 | 5 177 | 15 201 | 3 478 |
| Total proportions transfused in hemato-oncology | 13 724 (57%) | 3 056 (59%) | 8 964 (59%) | 2 095 (60%) |
| Total proportions transfused in cardiothoracic | 4 582 (19%) | 1 024 (20%) | 2 408 (16%) | 564 (16%) |
| Total proportions transfused in multispecialty | 5 645 (24%) | 1 097 (21%) | 3 828 (25%) | 819 (24%) |
| Leukemia | 9 758 | 2312 | 6 990 | 2 113 |
| Leukemia remission | 1 154 | 267 | 1 030 | 263 |
| Lymphoma | 11 121 | 3 041 | 6 605 | 1 547 |
| Other malignancies | 2 426 | 558 | 1 406 | 419 |
| Anemia | 6 849 | 1 687 | 6 110 | 1 409 |
| Coagulopathies | 21 108 | 4 955 | 13 407 | 3 126 |
| Infections | 565 | 140 | 355 | 86 |
| Rheumatic diseases | 300 | 56 | 1 043 | 204 |
| Palliative care | 263 | 60 | 290 | 96 |
| Other therapies | 25 142 | 6 871 | 19 008 | 4 266 |
| Surgery interventions | 19 939 | 4 515 | 11 394 | 2 328 |
| Chemotherapy | 92 938 | 24 519 | 74 287 | 18 228 |
| Antiinfectives | 80 204 | 20 130 | 51 678 | 10 663 |
| Other drugs | 7 627 | 1 899 | 4 730 | 972 |
Overall, the data between the training and test set were evenly distributed, yet with sex-dependent heterogeneity, which we, therefore, included as a training feature. Age was not normally distributed within the data set. Within the 5-year study period, a mean of 49.493 PCs per year were transfused. When analyzed by clinical departments, 57% of PCs were applied in the hematology-oncology department, 19% in the cardiothoracic surgery department, and 24% in other departments. On an individual level, 31% of patients were associated with the cardiothoracic surgery department, another 31% came from hematology-oncology, and 37% were treated at other departments.
Model performance
The models showed differences in performance, as measured by the highest AUC-PR score. None of the RF models beat the performance scores of the DL models (refer to supplemental Table 7). Acknowledging the comparative limitations of RF models, our focus shifted to exploring DL models. On the validation set, the multispecialty model achieved a mean performance of 0.72 AUC-PR in 400 trained models, with the best model achieving a score of 0.77 AUC-PR (a 7% increase). The cardiothoracic model had a mean performance of 0.43 in 700 runs, with the best model achieving a score of 0.45 AUC-PR (a 5% increase). The hematology-oncology model had a mean performance of 0.81 in 700 runs, with the best model achieving a score of 0.84 AUC-PR (a 5% improvement). Table 2 summarizes the top-performing ensemble models across several metrics and reports the scores of the final chosen models, including the hematology-oncology model, which had the highest AUC-PR scores. The validation scores between each fold are presented in supplemental Material 4. The use of sharpness-aware minimization did not boost performance for any of the best-performing models. All models were further optimized on the validation set, analyzing the optimal threshold on the AUC-PR curve, as shown in Figure 5, for each domain. The ideal threshold for the cardiothoracic model was identified as 0.39. For the multispecialty models, it was identified as 0.47, and for the hematology-oncology model, it was 0.41. A risk score exceeding these thresholds indicates the patient has a high likelihood of needing a platelet transfusion. A high-performing model is signified by a sharp, rounded AUC-PR curve in the top right quadrant, indicating an optimal ratio of true positives to false negatives and suggesting a highly efficient classification system with minimal errors and high precision recall. The hematology-oncology (Figure 5E) model displayed higher AUC-PR scores than all other models across departments and wards (Figure 5F), whereas the cardiothoracic model fell short in performance (Figure 5D). The best-performing models differed significantly in their parameter structures. The hematology-oncology model with the highest AUC-PR scores was constructed with 2 677 985 trainable parameters. In comparison, the multispecialty model had 1 106 285 parameters, all of which were trainable, showing a substantial reduction in complexity. Lastly, the cardiothoracic model included 1 057 475 trainable and 1920 nontrainable parameters. These differences in model structures underscore the varying algorithmic needs of different specialties and indicate a one-size-fits-all model may not yield optimal results.
Model performance on validation and test data
| Model . | Validation set . | |||||
|---|---|---|---|---|---|---|
| AUC-PR . | MCC . | F1 score . | Precision . | Specificity . | Sensitivity . | |
| LSTM hemato-oncology | 0.88 | 0.77 | 0.79 | 0.80 | 0.98 | 0.77 |
| LSTM cardiothoracic | 0.55 | 0.52 | 0.53 | 0.56 | 0.99 | 0.50 |
| LSTM multispecialty | 0.78 | 0.68 | 0.69 | 0.73 | 0.99 | 0.66 |
| Model . | Validation set . | |||||
|---|---|---|---|---|---|---|
| AUC-PR . | MCC . | F1 score . | Precision . | Specificity . | Sensitivity . | |
| LSTM hemato-oncology | 0.88 | 0.77 | 0.79 | 0.80 | 0.98 | 0.77 |
| LSTM cardiothoracic | 0.55 | 0.52 | 0.53 | 0.56 | 0.99 | 0.50 |
| LSTM multispecialty | 0.78 | 0.68 | 0.69 | 0.73 | 0.99 | 0.66 |
| . | Test set . | |||||
|---|---|---|---|---|---|---|
| LSTM hemato-oncology | 0.84 (0.83-0.84) | 0.73 (0.72-0.73) | 0.74 (0.74-0.75) | 0.77 (0.76-0.77) | 0.98 (0.98-0.98) | 0.72 (0.71-0.73) |
| LSTM cardiothoracic | 0.41 (0.39-0.43) | 0.41 (0.40-0.43) | 0.43 (0.42-0.44) | 0.44 (0.43-0.45) | 0.99 (0.98-0.99) | 0.42 (0.4-0.43) |
| LSTM multispecialty | 0.73 (0.73-0.73) | 0.64 (0.64-0.64) | 0.65 (0.65-0.65) | 0.70 (0.69-0.70) | 0.99 (0.99-0.99) | 0.61 (0.61-0.62) |
| . | Test set . | |||||
|---|---|---|---|---|---|---|
| LSTM hemato-oncology | 0.84 (0.83-0.84) | 0.73 (0.72-0.73) | 0.74 (0.74-0.75) | 0.77 (0.76-0.77) | 0.98 (0.98-0.98) | 0.72 (0.71-0.73) |
| LSTM cardiothoracic | 0.41 (0.39-0.43) | 0.41 (0.40-0.43) | 0.43 (0.42-0.44) | 0.44 (0.43-0.45) | 0.99 (0.98-0.99) | 0.42 (0.4-0.43) |
| LSTM multispecialty | 0.73 (0.73-0.73) | 0.64 (0.64-0.64) | 0.65 (0.65-0.65) | 0.70 (0.69-0.70) | 0.99 (0.99-0.99) | 0.61 (0.61-0.62) |
The scores in parentheses represent the 0.95 confidence interval.
Performance analysis for best-performing LSTM models. The area under the receiver operating characteristic curve (ROC-AUC) demonstrated a score of 0.98 for both the hematology-oncology model (B) and the multispecialty model (C) and a score of 0.95 for the cardiothoracic model (A). The AUC-PR (D-F) is represented for each model type. The ideal threshold for the cardiothoracic model (D) was identified as 0.39 (0.38 for the test set), compared with 0.47 (0.42 for the test set) for the multispecialty models (F) and 0.41 (also 0.37 for the test set) for the hematology-oncology model (E). Confusion matrices for all the models that have been trained are shown (G-I). Note that the “true negative” class (referring to those patients who were accurately identified as not having received any PCs) was disproportionately represented in all data sets, and the multispecialty model included a disproportionately large sample size.
Performance analysis for best-performing LSTM models. The area under the receiver operating characteristic curve (ROC-AUC) demonstrated a score of 0.98 for both the hematology-oncology model (B) and the multispecialty model (C) and a score of 0.95 for the cardiothoracic model (A). The AUC-PR (D-F) is represented for each model type. The ideal threshold for the cardiothoracic model (D) was identified as 0.39 (0.38 for the test set), compared with 0.47 (0.42 for the test set) for the multispecialty models (F) and 0.41 (also 0.37 for the test set) for the hematology-oncology model (E). Confusion matrices for all the models that have been trained are shown (G-I). Note that the “true negative” class (referring to those patients who were accurately identified as not having received any PCs) was disproportionately represented in all data sets, and the multispecialty model included a disproportionately large sample size.
The best-performing model by domain was used based on the data pipeline delineated in Figure 1. To optimize the process, a scheduling system triggers the daily extraction of data and execution of the model implementation pipeline at the end of each day. Every patient meeting the eligibility criteria outlined in Figure 2 is subsequently assigned a risk score, representing the likelihood they will need a platelet transfusion in the next 24 hours. These risk scores are consequently dispatched to the hospital's in-house FHIR server, facilitating additional data application. Supplemental Figure 2 demonstrates a hypothetical application of this use case via the model implementation process. In this dashboard visualization, the blood symbol signifies patients at risk of requiring a PC transfusion. An example case involves using the model-based decision as a baseline when forecasting platelet demand for the following day among stationary patients.
Explainability
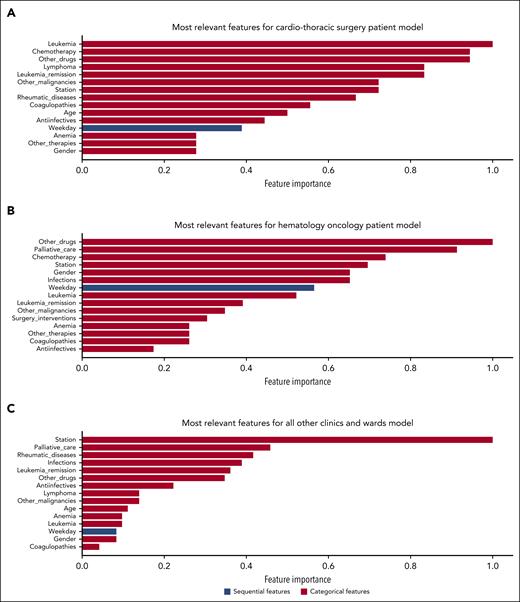
The most relevant features calculated on each test data set differed between clinical departments (Figure 6). Interestingly, the platelet count was not among the most relevant features contributing to the model’s prediction. The most relevant sequential feature was the weekday. This reflects the predominating impact of different operational practices between weekends and weekdays and operating room schedules. The model also integrated relevant clinical features, such as infections, malignant disorders, drug prescriptions with hematologic toxicity, and demographic features such as age and sex. It is important to note that although the importance of categorical and location features can be calculated reliably, caution is advised when assessing the overall importance of sequential features, because their contribution may vary over specific time points. For comprehensive interpretability, we examined the sequential features’ average contribution in our analysis.
Explainability analysis for best-performing LSTM models. The feature importance bar charts (A-C) show the most relevant features by model type divided into categorical and sequential features.
Explainability analysis for best-performing LSTM models. The feature importance bar charts (A-C) show the most relevant features by model type divided into categorical and sequential features.
Discussion
In this study, for the first time to our knowledge, we developed, tested, and implemented a patient-specific system to support platelet transfusion management at the individual level for each tertiary academic hospital patient. Using multimodal data, our DL models provided risk scores forecasting platelet transfusions within the subsequent 24 hours at high AUC-PR.
PCs pose challenges in inventory management owing to their perishability and limited shelf-life. Demand is often unpredictable, resulting in high expiration rates (typically between 10% and 20%) and occasional supply shortages.2 Over the past 4 decades, numerous strategies have been tested to address these issues. These strategies were analyzed in a systematic review2 that highlighted their complexity and applicability in typical blood bank scenarios. Despite such efforts, platelet expiration remains a global problem.34 Predicting individual patient platelet demand at the point of care is desirable both to improve total demand estimates and enable precise, individualized treatment plans.
In our prior research, we trained an XGBoost32 and RF33 model intending to predict patient-specific platelet demand in 3 days, focusing primarily on patients with extended hospital stays.35 This model relied on an array of electronic health records. However, it fell short in performance, and its utility for clinical implementation was curtailed because of a selection bias. Specifically, the model only incorporated patients who had stayed in the hospital for a minimum of 9 days, and data collection began only after the completion of the seventh day. This bias impeded the model's performance and significantly constrained its potential for routine clinical application because of its inability to generate accurate predictions for patients with shorter hospital stays.
These limitations have been fully addressed with a newly developed data mining process and with the DL approach for individualized platelet demand prediction. The marked improvement in reliability across a more diverse patient population, including a noteworthy ROC-PR score of 0.84 among patients with hematology-oncology conditions, is a crucial step in implementing data science tools in transfusion medicine. The substantial increase in the number of training samples and the cohort size (nearly 8500) using sequential and categorical data improved performance, supported realistic estimates, and enabled the development of department-specific models to overcome the constraints of prior models. The model for the hematology-oncology department demonstrated the best decision scores with an ROC-PR score of 0.84, a sensitivity of 0.72, and a precision of 0.77. This is likely because patients with hematology-oncology conditions are more prone to receive prophylactic platelet transfusions for thrombocytopenia, which is easier to predict than spontaneous bleeding. For patients undergoing cardiac surgery, emergency procedures, or intensive care, the event of acute bleeding and the need for a platelet transfusion is much more complex and challenging to predict. Overall, the trained models demonstrate high specificity but relative low sensitivity, as detailed in Table 2. This poses a potential challenge in clinical decision-making, because it may lead to false negatives and overlook patients who might benefit from a platelet transfusion.
The models possess the capability to aid health care professionals on several levels. A 24-hour forecast implemented as a decision-support tool within the hospital infrastructure can facilitate platelet management both for treating physicians and blood banks. Because these predictions reflect individual patient transfusion needs, clinicians are able to identify patients at high risk in advance. This may reduce the number of administered platelet products, because only patients needing platelet transfusions would be identified. Commercial solutions, such as GE HealthCare's Command Center, offer patient dashboards that could display the output of these models. Supplemental Figure 2 illustrates the mock integration of this use case. At the level of the blood banks, this tool can be leveraged to support the management of platelet donor activation, for example, at times of higher and lower than average platelet demand. Implementing an accurate forecasting system is critical to optimize blood bank inventory management, given the challenges faced by the health care system because of insufficient blood supply.36 The COVID-19 pandemic's recent blood supply crisis underscored the urgent necessity for improved forecasting methodologies, and a solution could boost pandemic preparedness.37 By combining traditional time series forecasting with our developed models, the response to the fluctuating demand can, therefore, be improved, at the cost of additional computational resources and increased complexity of the established systems. It may also optimize resource allocation between different blood banks by enabling the cooperative management of platelet shortages between different hospital corporations participating in the use of this tool. Although platelet transfer from donor centers to hospitals is common practice, its logistics can be streamlined.
The proposed solution is based on the FHIR standard, which provides the necessary features for easy interoperability across different health care settings. Thus, the model can be applied in different settings, including small regional hospitals and large tertiary centers, provided that the necessary data inputs are available. However, the applicability and effectiveness of the model may vary based on center characteristics. For example, centers without hematology-oncology services, in which the demand for platelets is driven mainly by surgical and critical care services, may face unique challenges not encountered in this study setting. Therefore, evaluating the model using local data is critical to ensure its validity and reliability before implementing this solution in a new setting. If the model's performance does not match the initial results, fine tuning according to the specific characteristics of local data could improve the model’s predictive ability and, ultimately, its utility in managing platelet demand. In this sense, the solution provides a flexible framework for platelet demand management that can be adapted to different health care settings according to specific needs and conditions.
The implementation of artificial intelligence–driven models in health care has some limitations. All of the presented models performed better in specificity than in sensitivity. Three limitations, in particular, affected the specificity scores of the cardiothoracic model: missing standardized data about future events, such as planned surgeries or medical devices/machines; data inconsistency; and errors caused by practitioners in their daily routines. We took data validity measures to ensure the completeness of data, yet not all errors could be avoided. Additionally, we did not incorporate the specific surgeon responsible for procedures into our feature set because of data privacy concerns. This omission made it difficult to predict prophylactic platelet transfusions for preventing bleeding during forthcoming surgical interventions. Finally, periprocedural acute bleeding as a complication of surgical intervention occurs unexpectedly and hence was difficult to predict. Efforts to improve data quality and quantity can ultimately pave the way for a more proactive and personalized approach to patient care, ensuring health care resources are used effectively and efficiently to meet the needs of each patient.
The use of our prediction model is limited by the inclusion criteria, which may exclude patients with incidents of acute bleeding and introduce bias into the model. Patients with a hospital stay of less than a day, no blood count, or a platelet count that never dipped below 150 × 103 platelets per μL were not considered, thus potentially missing cases with fatal outcomes, such as critical bleeding. However, it is important to note that the models have been primarily designed as supplementary tools for inpatient settings where these criteria can be reliably met.
Given their predictable behavior of patient platelet counts in stable clinical conditions, we used linear interpolation to fill gaps in platelet count observations. Although this approach may not fully capture abrupt shifts caused by unexpected events, its effect on model performance is negligible because the model differentiates between measured and interpolated data points. Moreover, the model relies on several other parameters, such as diagnoses, procedures, and medications. Future iterations of the model could benefit from more sophisticated imputation methods, such as those based on ML, which could provide a more nuanced estimate of platelet counts amidst unpredictable clinical events. Furthermore, a refined model that specifically targets the challenging task of predicting platelet demand for the cardiothoracic unit, primarily by incorporating variables such as impending surgical procedures and anticipated clinical procedures into the prediction algorithm, has the potential to overcome the limitations of the hematology-oncology model.
To the best of our knowledge, this is the first DL-based platelet transfusion predictor enabling individualized 24-hour risk assessments at high AUC-PR. It demonstrated that complex decisions about future clinical patient needs are possible by integrating data from different domains. Implemented as a decision-support system for platelet monitoring, DL forecasts might improve patient care by means of earlier detection of platelet demand and prevention of critical transfusion shortages.
Acknowledgments
The authors express their gratitude to all physicians and data managers at the various centers of the Essen University Hospital, whose data contributions have made this research achievable. The authors thank Anisa Kureishi for her language corrections on the manuscript as well as Paritosh Dhawale and Stefan Doll from GE HealthCare for their significant assistance with integrating the model's predictions into the patient dashboard.
This work was supported by funding that was based on a resolution of the German Bundestag by the Federal Government (ZMVI1-2519DAT713).
Authorship
Contribution: M.E. and G.B. acquired the data; M.E., F.N., P.A.H., C.S.S., V.P., R.H., and S.K. designed the study, analyzed the data, interpreted the results, and cowrote the manuscript; K.B., J.H., and A.T.T. contributed to write the manuscript and critically revised the manuscript; and all authors contributed clinical expertise, interpreted the results, and approved its final content.
Conflict-of-interest disclosure: A.T.T. reports consultancy fee from CSL Behring, Maat Pharma, Biomarin, and Onkowissen. The remaining authors declare no competing financial interests.
Correspondence: Peter A. Horn, Institute for Transfusion Medicine, University Hospital Essen, Hufelandstr 55, NRW 45147 Essen, Germany; email: peter.horn@uk-essen.de.
References
Author notes
The data supporting this study's findings are available upon reasonable request from the corresponding author, Peter Alexander Horn (peter.horn@uk-essen.de).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal