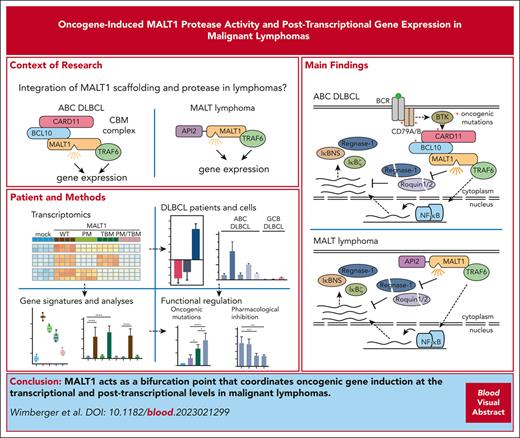
Key Points
MALT1 paracaspase acts as a bifurcation point for inducing transcriptional and posttranscriptional gene expression in malignant lymphomas.
MALT1 cleaves mRNA-destabilizing factors to enhance NF-κB–dependent and –independent gene induction in ABC DLBCL and MALT lymphoma.
Abstract
Constitutive mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1) activity drives survival of malignant lymphomas addicted to chronic B-cell receptor signaling, oncogenic CARD11, or the API2-MALT1 (also BIRC3::MALT1) fusion oncoprotein. Although MALT1 scaffolding induces NF-κB–dependent survival signaling, MALT1 protease function is thought to augment NF-κB activation by cleaving signaling mediators and transcriptional regulators in B-cell lymphomas. However, the pathological role of MALT1 protease function in lymphomagenesis is not well understood. Here, we show that TRAF6 controls MALT1-dependent activation of NF-κB transcriptional responses but is dispensable for MALT1 protease activation driven by oncogenic CARD11. To uncouple enzymatic and nonenzymatic functions of MALT1, we analyzed TRAF6-dependent and -independent as well as MALT1 protease–dependent gene expression profiles downstream of oncogenic CARD11 and API2-MALT1. The data suggest that by cleaving and inactivating the RNA binding proteins Regnase-1 and Roquin-1/2, MALT1 protease induces posttranscriptional upregulation of many genes including NFKBIZ/IκBζ, NFKBID/IκBNS, and ZC3H12A/Regnase-1 in activated B-cell–like diffuse large B-cell lymphoma (ABC DLBCL). We demonstrate that oncogene-driven MALT1 activity in ABC DLBCL cells regulates NFKBIZ and NFKBID induction on an mRNA level via releasing a brake imposed by Regnase-1 and Roquin-1/2. Furthermore, MALT1 protease drives posttranscriptional gene induction in the context of the API2-MALT1 fusion created by the recurrent t(11;18)(q21;q21) translocation in MALT lymphoma. Thus, MALT1 paracaspase acts as a bifurcation point for enhancing transcriptional and posttranscriptional gene expression in malignant lymphomas. Moreover, the identification of MALT1 protease–selective target genes provides specific biomarkers for the clinical evaluation of MALT1 inhibitors.
Introduction
Mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1), also known as paracaspase 1, is a human protease that emerged as a promising drug target, especially for treatment of hematologic malignancies.1,2 MALT1 is an integral subunit of the CARD11/CARMA1-BCL10-MALT1 (CBM) signalosome that initiates immune effector responses upon B-cell receptor (BCR) and T-cell receptor ligation on lymphocytes.3 MALT1 has a dual function in the CBM complex: by recruiting the E3 ligase TRAF6 to the CBM complex, MALT1 acts as a noncatalytic scaffold to induce Jun N-terminal kinase and canonical nuclear factor κB (NF-κB) signaling.4 Furthermore, the MALT1 protease is activated at the CBM complex and cleaves regulators of cell signaling (eg, A20, CYLD, and HOIL-1/RBCK1), transcription (RelB), and messenger RNA (mRNA) metabolism (eg, Regnase-1 and Roquin1/2).5 Although MALT1 protease activity is dispensable for initial antigenic stimulation, substrate cleavage modulates immune-cell functions.6-8
In the heterogeneous group of non-Hodgkin lymphomas (NHL), many B-cell lymphomas are characterized by oncogenic lesions affecting key components of the BCR pathway, which leads to the concomitant induction of NF-κB signaling and MALT1 protease activation.9 Diffuse large B-cell lymphoma (DLBCL) represents an aggressive lymphoid malignancy with the highest incident rate of all NHLs.10 Gene expression profiling defined 2 major subtypes: the germinal center B-cell–like (GCB) and the activated B-cell–like (ABC) DLBCL.11 ABC DLBCLs have an inferior prognosis and are characterized by chronic BCR signaling and recurrent oncogenic mutations in BCR mediators CD79A, CD79B, CARD11, and BCL10.12-15 Besides canonical NF-κB signaling, constitutive MALT1 protease activation contributes to growth and survival of BCR-addicted ABC DLBCLs.16-18 MALT1 acts downstream of Bruton tyrosine kinase (BTK) and therefore MALT1 inhibition represents an intriguing opportunity for patients with NHL exhibiting primary or secondary resistance to BTK inhibitors and in combinatorial treatment protocols.15,19-22 Several MALT1 inhibitors comprising different chemical entities have been developed23 and show effective and selective killing of MALT1-dependent ABC DLBCL cells in vitro and in xenografts.15,24-27 Clinical studies evaluating the safety and efficacy of MALT1 inhibition either in monotherapy or in combination with BTK inhibition have been started in patients with relapsed/refractory NHL (NCT03900598, NCT04876092, and NCT04657224). Beyond ABC DLBCL, MALT1 protease activation contributes to oncogenic transformation of MALT lymphoma. In MALT lymphoma the chromosomal translocation t(11;18)(q21;q21) leads to the expression of the API2-MALT1 fusion protein, which activates canonical and noncanonical NF-κB signaling.28,29 Aberrant MALT1 protease activation by the API2-MALT1 fusion drives noncanonical NF-κB signaling and represents a potential druggable vulnerability in MALT lymphomas.28
Despite the preclinical and clinical progress, it is not clear how MALT1 protease function contributes to growth and survival of lymphoma cells. MALT1 protease inhibition causes repression of NF-κB signature genes in ABC DLBCL cells,16,26 which has been attributed to cleavage of negative regulators of canonical NF-κB such as A20, CYLD, and RelB.17,30,31 However, MALT1 also cleaves HOIL-1, an essential component of linear ubiquitin assembly complex (LUBAC), which acts as a positive survival factor in ABC DLBCL cells.32,33 In T cells, MALT1-catalyzed cleavage and inactivation of Regnase-1 and Roquin-1/2 induces gene expression by releasing a brake of these RNA binding proteins (RBPs) on posttranscriptional induction of many transcripts.34,35 Here, we identify Regnase-1 and Roquin-1/2 as MALT1 substrates in lymphoma cells expressing oncogenic CARD11, CD79A/B, or API2-MALT1 fusion protein. Cleavage of RBPs lifts the posttranscriptional suppression and leads to high expression of distinct genes in ABC DLBCL. Thus, we provide evidence that MALT1 orchestrates oncogene–induced gene expression in malignant lymphomas through its noncatalytic and catalytic functions.
Methods
Cell lines, generation of knockouts, and inhibitor treatment
Cell lines were cultured and knockouts (KOs) were generated as previously described.4,36-38 For inhibitor treatment, cells were treated with the respective ibrutinib (20 nM), MLT-748 (2 µM), or S-mepazine (20 µM) for 18 hours before analysis. Protocols are available in supplemental Material and Methods (available on the Blood website).
Lentiviral transduction
Stable expression of wild-type (WT) or mutant CARD11, MALT1B, and API2-MALT1B as well as the NF-κB enhanced green fluorescent protein (EGFP) reporter was performed as previously described using lentiviral transduction, and NF-κB reporter activation was assessed by flow cytometry.36,39 Protocols are available in supplemental Material and Methods.
Preparation of cell lysates and western blotting
Cell lysates from 2 × 106 to 4 × 106 cells were prepared in coimmunoprecipitation or high-salt buffer and western blotting was performed as previously described.39 Protocols and antibodies are available in supplemental Material and Methods.
Transfection and posttranscriptional reporter assays
DLBCL cells were transfected with luciferase reporter genes containing NF-κB inhibitor zeta (NFKBIZ) 3′ untranslated region (UTR), NF-κB inhibitor delta (NFKBID) 3′ UTR, or no UTR (control) using electroporation. Cells were lysed 48 hours after transfection. Luciferase activity was assessed using a dual luciferase assay (Promega) in Centro LB960 (Berthold). Protocols are available in supplemental Material and Methods.
RNA isolation, complementary DNA synthesis, and quantitative real-time PCR
RNA was isolated, reverse transcribed, and quantitative real-time polymerase chain reaction (qRT-PCR) was performed on an LC480 (Roche) as previously described.40 Protocols and primers are available in supplemental Material and Methods.
RNA-seq and differential gene expression analyses
RNA sequencing (RNA-seq) was performed in paired-end mode (2 × 150 bases) in the Novaseq6000 sequencer (Illumina) with a depth of ≥30 × 106 reads per sample. The raw reads and MultiQC sequencing quality report were collected from the Helmholtz Center Munich Genomics Core Facility.41 RNA-seq analysis was performed as described in supplemental Material and Methods. Differential gene expression analysis in primary DLBCL samples was performed as described.42 Protocols are available in supplemental Material and Methods.
Results
TRAF6 bridges oncogenic CARD11 to canonical NF-κB activation in B cells
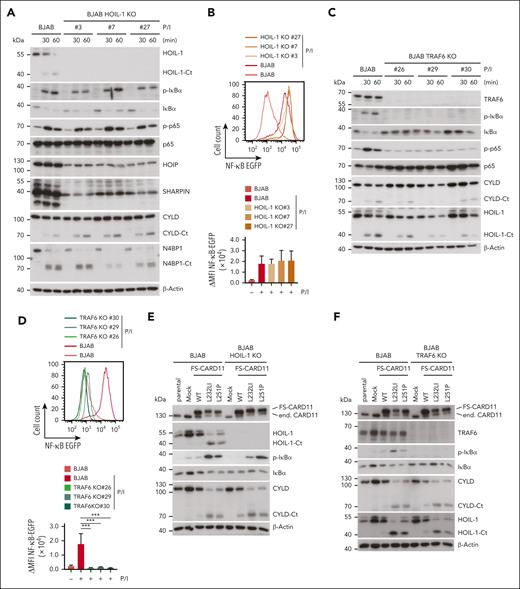
Conjugation of Met1-linked or Lys63-linked ubiquitin chains by LUBAC or TRAF6, respectively, has been suggested to contribute to canonical IKK/NF-κB activation downstream of the CBM complex in T and B cells.4,32,33,43,44 We aimed to determine which E3 ligase activity is primarily responsible for controlling NF-κB activation and signaling downstream of oncogenic CARD11. To this end, we knocked out either the LUBAC core subunit HOIL-1 or the E3 ligase TRAF6 in the GCB DLBCL cell line BJAB, which is characterized by low constitutive NF-κB activation and NF-κB–independent growth.45 As expected, HOIL-1 ablation caused destabilization of the entire LUBAC, as evident from a concomitant decrease in the expression of HOIP and SHARPIN (Figure 1A). However, HOIL-1 deficiency in 3 independent BJAB clones did not significantly impair inducible IκBα phosphorylation/degradation, p65 phosphorylation, and transcriptional activation of a NF-κB reporter gene in response to phorbol 12-myristate 13-acetate/ionomycin (P/I), which mimics BCR stimulation in driving CBM complex–dependent B-cell activation (Figure 1A-B). In contrast, TRAF6 ablation in BJAB cells abolished CBM-dependent NF-κB signaling and transcriptional activation (Figure 1C-D). Neither TRAF6 nor HOIL-1 deficiency affected inducible MALT1 protease activation, as revealed by equivalent substrate cleavage after P/I stimulation in parental and KO BJAB cells (Figure 1A,C).
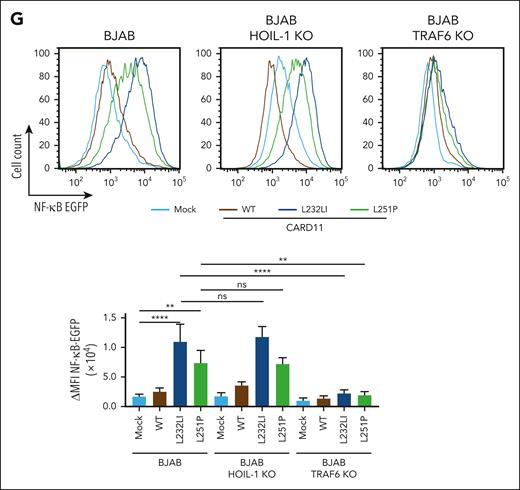
Requirements of TRAF6 and HOIL-1 for CBM and oncogenic CARD11 signaling in BJAB cells. (A,C) Western blot analysis of 3 HOIL-1 KO (A) and TRAF6 KO (C) clones after stimulation with P/I for the indicated time points to determine MALT1 substrate cleavage and NF-κB signaling. (B,D) Representative flow cytometric analyses (top panel) and quantification of changes in mean fluorescence intensity (ΔMFI, bottom panel) of the 6 × NF-κB–EGFP reporter in 3 HOIL-1 KO (B) and TRAF6 KO (D) BJAB cells after P/I stimulation (5 hours). n = 3 replicates, all error bars depict the mean ± standard deviation (SD), ordinary 1-way analysis of variance (ANOVA) with Tukey multiple comparisons, ∗∗∗P < .001. (E-F) Western blot analysis of overexpression of FS-CARD11 WT, L232LI, and L251P in HOIL-1 KO (E) and TRAF6 KO (F) BJAB cells to determine NF-κB signaling and MALT1 substrate cleavage. (G) Representative flow cytometric analyses (top panel) and quantification of ΔMFI (bottom panel) of the 6 × NF-κB–EGFP reporter in parental (left), HOIL-1 KO (middle), and TRAF6 KO (right) BJAB cells expressing CARD11 WT and mutants. N = 3 replicates, all error bars depict the mean ± SD, ordinary 1-way ANOVA with Tukey multiple comparisons; ns, not significant; ∗∗P < .01 and ∗∗∗∗P < .0001.
Requirements of TRAF6 and HOIL-1 for CBM and oncogenic CARD11 signaling in BJAB cells. (A,C) Western blot analysis of 3 HOIL-1 KO (A) and TRAF6 KO (C) clones after stimulation with P/I for the indicated time points to determine MALT1 substrate cleavage and NF-κB signaling. (B,D) Representative flow cytometric analyses (top panel) and quantification of changes in mean fluorescence intensity (ΔMFI, bottom panel) of the 6 × NF-κB–EGFP reporter in 3 HOIL-1 KO (B) and TRAF6 KO (D) BJAB cells after P/I stimulation (5 hours). n = 3 replicates, all error bars depict the mean ± standard deviation (SD), ordinary 1-way analysis of variance (ANOVA) with Tukey multiple comparisons, ∗∗∗P < .001. (E-F) Western blot analysis of overexpression of FS-CARD11 WT, L232LI, and L251P in HOIL-1 KO (E) and TRAF6 KO (F) BJAB cells to determine NF-κB signaling and MALT1 substrate cleavage. (G) Representative flow cytometric analyses (top panel) and quantification of ΔMFI (bottom panel) of the 6 × NF-κB–EGFP reporter in parental (left), HOIL-1 KO (middle), and TRAF6 KO (right) BJAB cells expressing CARD11 WT and mutants. N = 3 replicates, all error bars depict the mean ± SD, ordinary 1-way ANOVA with Tukey multiple comparisons; ns, not significant; ∗∗P < .01 and ∗∗∗∗P < .0001.
To determine which ligase is channeling oncogenic CARD11 to NF-κB signaling, we transduced CARD11 WT, L232LI, or L251P into parental, TRAF6 or HOIL-1 KO BJAB cells. Expression of both oncogenic variants induced constitutive NF-κB signaling and MALT1 protease activation in BJAB cells (Figure 1E-F). Although, NF-κB signaling and transcriptional activation of the NF-κB reporter induced by oncogenic CARD11 was intact in HOIL-1 KO BJAB cells, activation of the NF-κB signaling pathway was abolished in the absence of TRAF6 (Figure 1E-G). Neither TRAF6 nor HOIL-1 deficiency perturbed the ability of oncogenic CARD11 to trigger MALT1 catalyzed substrate cleavage (Figure 1E-F). Thus, in B cells, TRAF6 but not HOIL-1 and LUBAC is bridging oncogenic CARD11 to the canonical IKK/NF-κB pathway, whereas both ligases are dispensable for MALT1 protease activity.
Oncogenic CARD11 signaling controls TRAF6-dependent and -independent gene induction
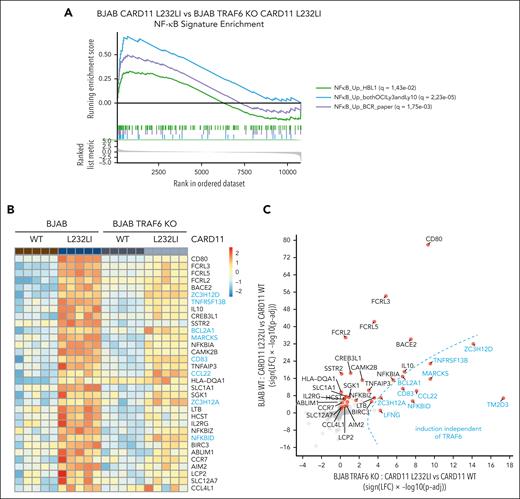
Because TRAF6 deficiency impaired NF-κB activation but not MALT1 protease activity, the analyses of TRAF6 KO BJAB cells allowed us to uncouple the pathological role of MALT1 protease and scaffolding function downstream of oncogenic CARD11. We used gene expression profiling by RNA-seq to compare the gene induction of oncogenic CARD11 in the presence or absence of TRAF6 in BJAB expressing CARD11 WT or L232LI. Individual samples from each group cluster in the principal component analysis (PCA) and gene set enrichment analysis revealed a significant induction of 3 NF-κB–dependent ABC DLBCL signature genes upon expression of CARD11 L232LI (supplemental Figure 1A-B). ABC DLBCL–derived gene signatures were still induced in TRAF6 KO BJAB cells (supplemental Figure 1C) but the gene set enrichment analysis score was significantly reduced compared with parental BJAB cells (Figure 2A). We compared gene induction by CARD11 L232LI in BJAB cells in the absence or presence of TRAF6, which allowed us to determine effects of TRAF6 in an unbiased manner (Figure 2B-C; supplemental Figure 1D-E). CARD11 L232LI induced upregulation of a variety of NF-κB target genes (eg, CD80, TNFAIP3, IL10, NFKBIZ, and NFKBIA) and induction of most genes was reduced in TRAF6-deficient cells, demonstrating the role of TRAF6 for gene induction downstream of oncogenic CARD11 (Figure 2B-C; supplemental Figure 1D-E). However, CARD11 L232LI significantly induced a subset of genes even in the absence of TRAF6 but, in general, at a lower level. To determine a potential contribution of MALT1 protease activity on CARD11 oncogenic gene expression in the absence of TRAF6, we treated BJAB cells with the potent and selective MALT1 inhibitor, MLT-748, which efficiently inhibited CARD11 L232LI–induced MALT1 substrate cleavage (supplemental Figure 2A).46 Despite overall weak gene induction, all top genes induced by CARD11 L232LI in TRAF6 KO cells were decreased upon MALT1 inhibition (supplemental Figure 2B). Although qRT-PCR showed that residual induction of CD80 and TNFAIP3 in TRAF6 KO cells was not significantly decreased by MALT1 inhibition, many TRAF6-controlled genes (eg, IL10, NFKBIZ, and BIRC3) were further downregulated by MALT1 inhibition, indicating a corequirement for optimal induction (Figure 2D; supplemental Figure 2C). A number of CARD11 L232LI–induced genes like NFKBID, ZC3H12A, and ZC3H12D were not relying on TRAF6 but were strongly dependent on MALT1 protease activity. Expression of NFKBID, ZC3H12A, and NFKBIZ is directly controlled on the posttranscriptional level by the MALT1 substrates Roquin-1/2 and Regnase-1.34,35 Regnase-1 and Roquin-1/2 are also cleaved by MALT1 independent of TRAF6 in CARD11 L232LI–expressing BJAB cells, which induced MALT1 protease–dependent expression of IκBNS p70 and p35 fragments, which are encoded by NFKBID (Figure 2E; supplemental Figure 2A).47 The data suggest that MALT1 protease controls TRAF6/NF-κB–dependent and –independent gene expression.
TRAF6-dependent and -independent gene expression profiling (GEP) of oncogenic CARD11. (A) Comparative gene set enrichment analysis of 3 ABC DLBCL–derived NF-κB gene signatures of CARD11 L232LI in parental vs TRAF6 KO BJAB cells. Three different NF-κB signatures were compared, with q values for the individual signatures shown in the figure. (B) Heat map of genes differentially expressed in parental and TRAF6 KO BJAB cells expressing CARD11 WT or L232LI (log fold change [LFC] > 1.3; adjusted P value [Padj] < .01). Genes are ordered based on the Padj values of parental CARD11 L232LI vs CARD11 WT. Red denotes higher, blue denotes lower expression on the color scale; n = 5 for each sample. Genes highlighted in blue are induced in the absence of TRAF6. (C) Signed Padj plot comparing gene induction by CARD11 L232LI in parental vs TRAF6 KO BJAB cells. Transcripts enriched in TRAF6 KO cells are depicted in blue. (D) Evaluation of the expression of selected transcripts by qRT-PCR in parental and TRAF6 KO BJAB cells transduced with CARD11 WT or CARD11 L232LI. Changes in transcript expression after MALT1 protease inhibition by MLT-748 (2 μM; 18 hours) in CARD11 L232LI–expressing TRAF6 KO BJAB cells. n = 5, all error bars depict the mean ± SD; left panel: ANOVA with Tukey multiple comparisons; right panel: unpaired Student t test; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001. (E) Western blot analysis of BJAB and TRAF6 KO BJAB cells expressing CARD11 WT and L232LI to determine cleavage of Regnase-1 and Roquin-1/2 and expression of IκBNS. Unspecific bands are marked with an asterisk.
TRAF6-dependent and -independent gene expression profiling (GEP) of oncogenic CARD11. (A) Comparative gene set enrichment analysis of 3 ABC DLBCL–derived NF-κB gene signatures of CARD11 L232LI in parental vs TRAF6 KO BJAB cells. Three different NF-κB signatures were compared, with q values for the individual signatures shown in the figure. (B) Heat map of genes differentially expressed in parental and TRAF6 KO BJAB cells expressing CARD11 WT or L232LI (log fold change [LFC] > 1.3; adjusted P value [Padj] < .01). Genes are ordered based on the Padj values of parental CARD11 L232LI vs CARD11 WT. Red denotes higher, blue denotes lower expression on the color scale; n = 5 for each sample. Genes highlighted in blue are induced in the absence of TRAF6. (C) Signed Padj plot comparing gene induction by CARD11 L232LI in parental vs TRAF6 KO BJAB cells. Transcripts enriched in TRAF6 KO cells are depicted in blue. (D) Evaluation of the expression of selected transcripts by qRT-PCR in parental and TRAF6 KO BJAB cells transduced with CARD11 WT or CARD11 L232LI. Changes in transcript expression after MALT1 protease inhibition by MLT-748 (2 μM; 18 hours) in CARD11 L232LI–expressing TRAF6 KO BJAB cells. n = 5, all error bars depict the mean ± SD; left panel: ANOVA with Tukey multiple comparisons; right panel: unpaired Student t test; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001. (E) Western blot analysis of BJAB and TRAF6 KO BJAB cells expressing CARD11 WT and L232LI to determine cleavage of Regnase-1 and Roquin-1/2 and expression of IκBNS. Unspecific bands are marked with an asterisk.
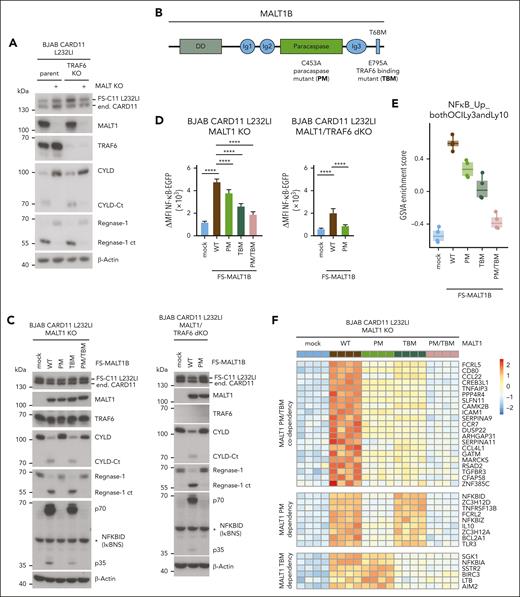
Oncogenic CARD11 drives gene induction via MALT1 scaffolding and protease functions
To decipher the exact contributions of MALT1 scaffold and protease functions for gene regulation, we deleted MALT1 in CARD11 L232LI–expressing parental or TRAF6 KO BJAB cells (Figure 3A). The MALT1 KO BJAB cells were reconstituted with MALT1B WT, paracaspase mutant (PM) C453A, TRAF6-binding mutant (TBM) E795A, or the combined PM/TBM (Figure 3B). Independent of TRAF6, CARD11 L232LI triggered MALT1-catalyzed substrate cleavage in BJAB cells expressing MALT1 WT or TBM but not catalytically inactive MALT1 PM or PM/TBM (Figure 3C). EGFP–NF-κB reporter gene induction was severely impaired by MALT1B TBM, because of its inability to recruit TRAF6 to the CBM complex (Figure 3D; supplemental Figure 3A).48 However, NF-κB activation was also significantly decreased by MALT1 PM and only the combined mutations in MALT1 PM/TBM nearly abolished NF-κB activation by oncogenic CARD11. Accordingly, in TRAF6/MALT1 double-KO BJAB cells, CARD11 L232LI induced substrate cleavage and weak NF-κB activation in MALT1 WT but not PM cells, confirming that MALT1 protease activity can augment NF-κB gene induction even in the absence of TRAF6 (Figure 3C-D).
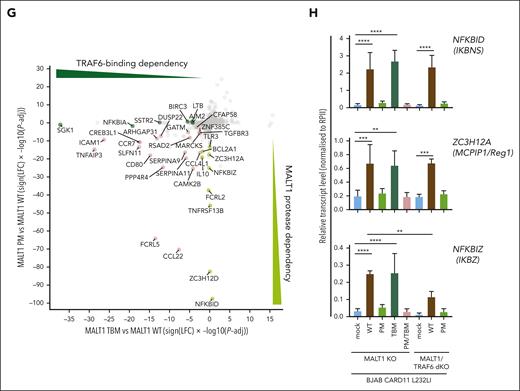
MALT1 scaffolding- and protease-dependent GEP by oncogenic CARD11. (A) Western blot analysis of MALT1 KO and MALT1/TRAF6 double-KO (dKO) BJAB cells expressing CARD11 L232LI. Effects of loss of MALT1 and/or TRAF6 on CYLD and Regnase-1 cleavage are shown. (B) Schematic depiction of MALT1B protein and domains. The MALT1 C453A PM and E795A TBM are shown. (C) Western blot analysis of MALT1 KO (left) and MALT1/TRAF6 dKO (right) BJAB expressing CARD11 L232LI reconstituted with mock vector, MALT1 WT, or mutants. Cleavage of MALT1 substrates CYLD and Regnase-1 and expression of NFKBID/IκBNS was determined. Unspecific bands are marked with an asterisk. (D) Quantification of ΔMFI of the 6 × NF-κB–EGFP reporter in MALT1 KO and MALT1/TRAF6 dKO BJAB cells reconstituted with mock vector, MALT1 WT, or mutant constructs. n = 4 replicates, all error bars depict the mean ± SD, ANOVA with Tukey multiple comparisons, ∗∗∗∗P < .0001. (E) Gene set variation analysis (GSVA) quantifying the effects of MALT1 WT and mutants on a published ABC DLBCL–derived NF-κB gene signature induced by CARD11 L232LI. (F) Heat map of differentially expressed genes in the MALT1 KO BJAB cells expressing CARD11 L232LI in dependency of reconstitution with mock vector, MALT1 WT, PM, TBM, or PM/TBM. Red denotes higher, blue denotes lower expression. n = 4 for each sample. (G) Signed Padj plot comparing gene induction by MALT1 PM and TBM compared with MALT1 WT reconstituted MALT1 KO BJAB cells expressing CARD11 L232LI. Genes in the top-left and the bottom-right corner are strongly TRAF6 binding or MALT1-protease dependent, respectively. Annotated are the genes from the heat map in panel F. (H) Relative transcript expression analysis of ZC3H12A, NFKBID, and NFKBIZ in dependency of the indicated reconstituted MALT1 mutants and MALT1 WT in MALT1 KO BJAB cells expressing CARD11 L232LI by qRT-PCR. n = 4, all error bars depict the mean ± SD, ANOVA with Tukey multiple comparisons; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001.
MALT1 scaffolding- and protease-dependent GEP by oncogenic CARD11. (A) Western blot analysis of MALT1 KO and MALT1/TRAF6 double-KO (dKO) BJAB cells expressing CARD11 L232LI. Effects of loss of MALT1 and/or TRAF6 on CYLD and Regnase-1 cleavage are shown. (B) Schematic depiction of MALT1B protein and domains. The MALT1 C453A PM and E795A TBM are shown. (C) Western blot analysis of MALT1 KO (left) and MALT1/TRAF6 dKO (right) BJAB expressing CARD11 L232LI reconstituted with mock vector, MALT1 WT, or mutants. Cleavage of MALT1 substrates CYLD and Regnase-1 and expression of NFKBID/IκBNS was determined. Unspecific bands are marked with an asterisk. (D) Quantification of ΔMFI of the 6 × NF-κB–EGFP reporter in MALT1 KO and MALT1/TRAF6 dKO BJAB cells reconstituted with mock vector, MALT1 WT, or mutant constructs. n = 4 replicates, all error bars depict the mean ± SD, ANOVA with Tukey multiple comparisons, ∗∗∗∗P < .0001. (E) Gene set variation analysis (GSVA) quantifying the effects of MALT1 WT and mutants on a published ABC DLBCL–derived NF-κB gene signature induced by CARD11 L232LI. (F) Heat map of differentially expressed genes in the MALT1 KO BJAB cells expressing CARD11 L232LI in dependency of reconstitution with mock vector, MALT1 WT, PM, TBM, or PM/TBM. Red denotes higher, blue denotes lower expression. n = 4 for each sample. (G) Signed Padj plot comparing gene induction by MALT1 PM and TBM compared with MALT1 WT reconstituted MALT1 KO BJAB cells expressing CARD11 L232LI. Genes in the top-left and the bottom-right corner are strongly TRAF6 binding or MALT1-protease dependent, respectively. Annotated are the genes from the heat map in panel F. (H) Relative transcript expression analysis of ZC3H12A, NFKBID, and NFKBIZ in dependency of the indicated reconstituted MALT1 mutants and MALT1 WT in MALT1 KO BJAB cells expressing CARD11 L232LI by qRT-PCR. n = 4, all error bars depict the mean ± SD, ANOVA with Tukey multiple comparisons; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001.
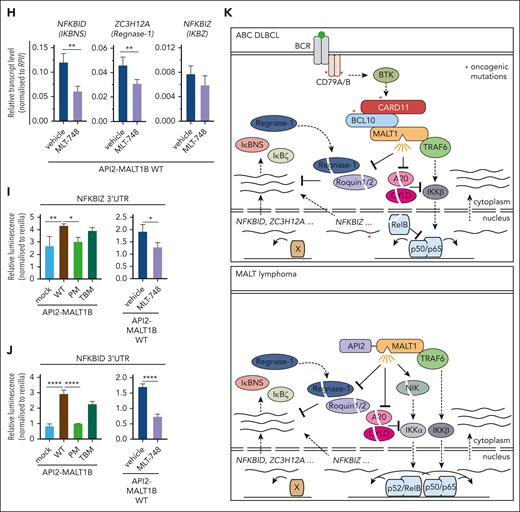
We performed transcriptome analyses in CARD11 L232LI MALT1 KO BJAB cells either transduced with mock vector (empty vector) or reconstituted with MALT1 WT and mutants. In PCA, samples cluster based on the genetic manipulations (supplemental Figure 3B). Gene set variation analyses revealed a strong enrichment of the ABC DLBCL–derived NF-κB gene signatures in MALT1 WT reconstituted cells (Figure 3E; supplemental Figure 3C). Oncogenic NF-κB gene signatures induction was partially diminished by the individual mutations in MALT1 PM and TBM and nearly abrogated by the combined destruction of TRAF6-binding and protease activity in MALT1 PM/TBM. Optimal expression of most CARD11 L232LI–induced genes relied on MALT1 protease activity and TRAF6 binding (Figure 3F-G; supplemental Figure 3D). However, clustering and crosscomparisons revealed that induction of specific subsets of genes were more strongly affected by either the individual PM or TBM variants of MALT1. Again, genes that strongly relied on MALT1 protease encompassed NFKBID, ZC3H12A, NFKBIZ, IL10, ZC3H12D, and BCL2A1, which have been identified either as genes that are largely independent of TRAF6 and/or downregulated by MALT1 inhibition in the absence of TRAF6 (compare Figure 2C; supplemental Figure 2B). qRT-PCR validated the strong dependency of NFKBIZ, NFKBID, and ZC3H12A induction on MALT1 catalytic activity downstream of oncogenic CARD11 L232LI (Figure 3H). In line, only MALT1 WT but not PM was able to completely (NFKBID and ZC3H12A) or partially (NFKBIZ) rescue induction of these genes in MALT1/TRAF6 double-KO BJAB cells. NFKBID/IκBNS protein was induced in a MALT1 protease–dependent and TRAF6-independent manner (Figure 2C).
To confirm MALT1 protease–driven regulation of selected target genes in an independent setting, we used MALT1 KO Jurkat T cells reconstituted with MALT1B WT and mutants.4 Acute NF-κB reporter gene activation by P/I stimulation was strongly impaired by the MALT1B TBM, which retained the ability to catalyze cleavage of RBPs Regnase-1 and Roquin-1/2 (supplemental Figure 4A-B). In contrast, protease-defective MALT1 PM, which was unable to promote substrate cleavage, still retained NF-κB responsiveness but slightly below that of MALT1 WT. Although P/I induction of NFKBIZ and ZC3H12A mRNAs as well as NFKBIZ/IκBζ protein was codependent on MALT1 scaffolding and protease function, expression of NFKBID transcripts and NFKBID/IκBNS protein was strongly relied on MALT1 protease function (supplemental Figure 4B-C). Furthermore, MALT1 protease activation but not TRAF6 binding was required to induce NFKBID/IκBNS downstream of CARD11 L232LI in Jurkat cells (supplemental Figure 4D-E).
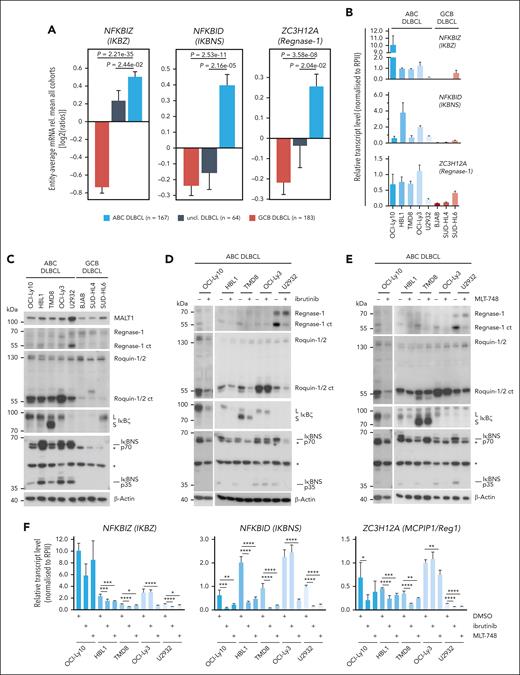
Upregulation of MALT1 protease target genes in ABC DLBCL
We assessed mRNA expression levels of TRAF6- and/or MALT1 protease–controlled genes in a gene expression data set comprising tumors from 414 well-characterized samples from patients with primary DLBCL and a panel of 5 ABC- and 3 GCB–derived DLBCL cells lines (Figure 4A-B; supplemental Figure 5A-B). As previously observed, NFKBIZ expression, which is controlled by both MALT1-TRAF6 interaction and MALT1 protease activation, is higher in primary ABC compared with GCB or unclassified DLBCL.42 Furthermore, induction of other genes that we had identified as highly MALT1-protease dependent (refer to Figure 3F-G), such as NFKBID, ZC3H12A, BCL2A1, IL10, CCL22, FCRL5, TM2D3, FCRL2, and TNFRSF13B, are enriched in ABC DLBCL samples, suggesting that the MALT1 protease directly engages in the regulation of these transcripts independent of TRAF6 and NF-κB (Figure 4A-B; supplemental Figure 5A-B). Accordingly, ABC DLBCL cell lines show higher constitutive mRNA expression of the MALT1 protease–dependent genes when compared with GCB DLBCL cell lines. Among the GCB DLBCL cell lines, only SUD-HL6, which has retained a functional BCR, displayed high expression of some of these genes.49 Notably, despite evidence that MALT1 protease enhances expression of ZC3H12D and TLR3, we could not detect enriched expression of these gene in ABC DLBCL.
Expression and regulation of MALT1 protease–dependent genes in ABC DLBCL. (A) Differential gene expression for n = 414 cases (167 ABC-like DLBCL, 64 unclassified, and 183 GCB-like DLBCL; Gene Expression Omnibus accession GSE10846) for the genes ZC3H12A, NFKBIZ, and NFKBID in ABC, unclassified, and GCB DLBCLs. (B) Relative transcript expression of ZC3H12A, NFKBID, and NFKBIZ by qRT-PCR in a panel of ABC and GCB DLBCLs; n ≥ 3, all error bars depict the mean ± SD. (C) Western blot analysis of MALT1 substrates Regnase-1 and Roquin-1/2 and their targets IκΒζ and IκBNS in a panel of ABC and GCB DLBCL cells. Unspecific bands are marked with an asterisk. (D-E) Western blot analysis of (D) ibrutinib- or (E) MLT-748–treated ABC DLBCL. Activity of the MALT1 protease was determined by cleavage of Regnase-1 and Roquin-1/2 and their targets IκBζ and IκBNS. “L” and “S” depict long and short IκBζ isoforms, respectively. Unspecific bands are marked with an asterisk. (F) Transcript analysis of ZC3H12A, NFKBIZ, and NFKBID after inhibition with ibrutinib or MLT-748 in ABC DLBCL. n = 4, all error bars depict the mean ± SD; ordinary 1-way ANOVA with Dunnett multiple comparisons; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001.
Expression and regulation of MALT1 protease–dependent genes in ABC DLBCL. (A) Differential gene expression for n = 414 cases (167 ABC-like DLBCL, 64 unclassified, and 183 GCB-like DLBCL; Gene Expression Omnibus accession GSE10846) for the genes ZC3H12A, NFKBIZ, and NFKBID in ABC, unclassified, and GCB DLBCLs. (B) Relative transcript expression of ZC3H12A, NFKBID, and NFKBIZ by qRT-PCR in a panel of ABC and GCB DLBCLs; n ≥ 3, all error bars depict the mean ± SD. (C) Western blot analysis of MALT1 substrates Regnase-1 and Roquin-1/2 and their targets IκΒζ and IκBNS in a panel of ABC and GCB DLBCL cells. Unspecific bands are marked with an asterisk. (D-E) Western blot analysis of (D) ibrutinib- or (E) MLT-748–treated ABC DLBCL. Activity of the MALT1 protease was determined by cleavage of Regnase-1 and Roquin-1/2 and their targets IκBζ and IκBNS. “L” and “S” depict long and short IκBζ isoforms, respectively. Unspecific bands are marked with an asterisk. (F) Transcript analysis of ZC3H12A, NFKBIZ, and NFKBID after inhibition with ibrutinib or MLT-748 in ABC DLBCL. n = 4, all error bars depict the mean ± SD; ordinary 1-way ANOVA with Dunnett multiple comparisons; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001.
In agreement with transcript expression, IκBζ and IκBNS (p70 and p35), the protein products of NFKBIZ and NFKBID genes, respectively, are more highly expressed in ABC DLBCL compared with GCB DLBCL cell lines (Figure 4C; supplemental Figure 5C). In contrast, Regnase-1, the product of the ZC3H12A gene, was barely detectable in ABC DLBCL cells but visible in GCB DLBCL cells. However, despite the low expression of Regnase-1 full length, a C-terminal cleavage fragment of Regnase-1 appeared in ABC DLBCL cells, indicating that MALT1-catalyzed cleavage inactivates this RBP on a posttranslational level, selectively in ABC DLBCL. Similarly, MALT1 substrates Roquin-1 and Roquin-2 are constitutively cleaved in ABC DLBCL cells (Figure 4C; supplemental Figure 5C). Regnase-1 and Roquin-1/2 directly bind to, and antagonize NFKBIZ, NFKBID, and ZC3H12A transcripts,34,50 suggesting that expression of these genes is directly linked to MALT1-dependent cleavage of these RBPs.
To determine whether cleavage of RBPs and expression of their putative targets relies on chronic BCR signaling and MALT1 protease activity, we treated ABC DLBCL cells with the BTK inhibitor ibrutinib acting upstream of the CBM complex or the MALT1 inhibitors MLT-748 and S-mepazine (Figure 4D-E; supplemental Figure 5D). In cell lines carrying mutations in the BCR adaptors CD79A/B (HBL1, TMD8, and OCI-Ly10) or TAK1 (U2932) BTK, or MALT1 inhibition led to a substantial reduction of Regnase-1 and Roquin-1/2 cleavage bands and, concomitantly, IκBζ and IκBNS expression was decreased. OCI-Ly-3 cells, which carry the oncogenic CARD11 mutation, were resistant to BTK inhibition but RBP cleavage and IκBζ and IκBNS expression were still sensitive to MALT1 inhibitor treatment. Interestingly, despite a severe reduction in the RBP cleavage products upon inhibitor treatment, there was hardly an enrichment in full length proteins, suggesting a counter selection against high expression of these active RBPs in ABC DLBCL cells. We performed qRT-PCR to confirm that chronic BCR signaling via BTK and MALT1 protease activity regulates the expression of NFKBIZ, NFKBID, and ZC3H12A on transcript levels (Figure 4F). BTK inhibitor ibrutinib significantly decreased NFKBIZ, ZC3H12A, and NFKBID in all ABC DLBCL cells lines except OCI-Ly3. In all ABC DLBCL cell lines and independent of the CARD11 mutational status, MALT1 inhibitor MLT-748 caused a decrease in the level of all 3 genes. Especially high expression of ZC3H12A and NFKBID, which are not controlled by TRAF6-dependent NF-κB activation, strictly relies on MALT1 protease activity. Thus, our data suggest that by cleaving Regnase-1 and Roquin-1/2, MALT1 protease restricts the expression of the RBPs at the posttranslational level in ABC DLBCL cells. In turn, inactivation of the RBPs in ABC DLBCL causes an increase in NFKBIZ, NFKBID, and ZC3H12A mRNA and protein expression, which is impeded by inhibition of chronic BCR signaling by BTK or MALT1 inhibition.
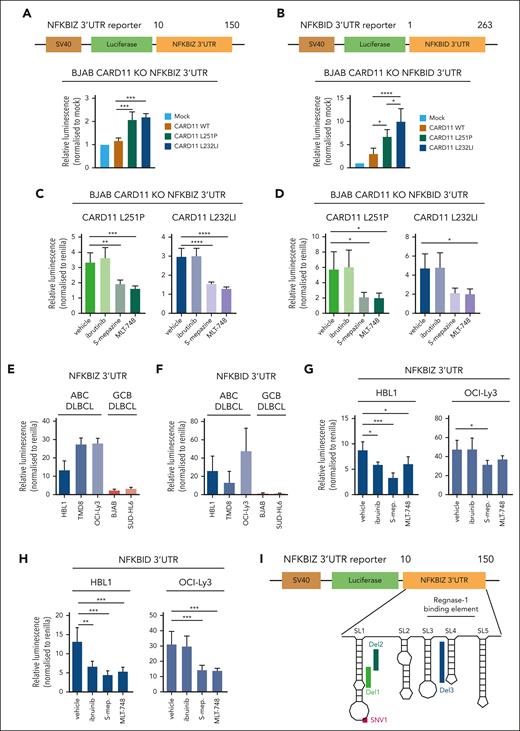
Control of posttranscriptional NFKBIZ and NFKBID expression by MALT1 in DLBCL
RBPs Regnase-1 and Roquin-1/2 decrease NFKBIZ and NFKBID mRNA stability by binding to secondary stem-loop (SL) structures in the 3′ UTR of both genes.51,52 To determine the effects of the RBP-controlled binding to UTR sequences on oncogene–induced gene regulation, we used CARD11 KO BJAB cells, which were transduced with CARD11 WT or mutants L251P or L232LI. As shown previously in parental BJAB cells, and also in CARD11 KO cells, expression of oncogenic CARD11 triggered robust cleavage of MALT1 substrate and induction of ZC3H12A, NFKBIZ, and NFKBID relied on MALT1 protease but not BTK kinase activity (supplemental Figure 6A-D). To uncouple posttranscriptional and transcriptional gene induction by oncogenic CARD11, we generated luciferase reporter constructs containing the 3′ UTRs of NFKBIZ (1-150) or NFKBID (1-263), which confer control by Regnase-1 and Roquin-1/2 (Figure 5A-B). Although the reporter lacking the UTRs was not differentially expressed, both oncogenic CARD11 mutants induced expression of the NFKBIZ- and NFKBID-containing 3′ UTR reporter constructs (Figure 5A-B; supplemental Figure 6E). Induction of the reporter via the 3′ UTRs of both genes was impaired by MALT1 but not BTK inhibitor treatment, demonstrating the strict dependency on MALT1 protease activation for posttranscriptional gene regulation downstream of oncogenic CARD11 (Figure 5C-D).
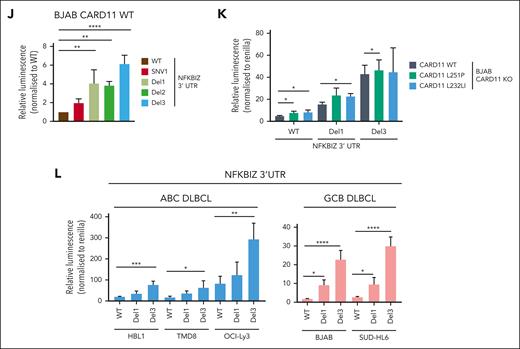
Regulation of the 3′ UTR of NFKBIZ and NFKBID in DLBCL. (A-B) Schematic depiction of the 3′ UTR constructs of the (A) NFKBIZ and (B) NFKBID luciferase reporter. Relative luminescence was measured 48 hours after cotransfection of the reporter with a Renilla construct in BJAB CARD11 KOs expressing CARD11 WT, L251P, or L232LI. Values were normalized to mock vector-infected cells; n ≥ 4, all error bars depict the mean ± SD, ordinary 1-way ANOVA with Tukey multiple comparisons; ∗P < .05; ∗∗∗P < .001; and ∗∗∗∗P < .0001. (C-D) Luciferase reporter assay of the 3′ UTR of (C) NFKBIZ and (D) NFKBID in BJAB CARD11 KOs reconstituted with CARD11 L251P and L232LI, respectively, after ibrutinib, S-mepazine, or MLT-748 treatment for 18 hours. n ≥ 3, all error bars depict the mean ± SD, ordinary 1-way ANOVA with Dunnett multiple comparisons; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001. (E-F) Luciferase reporter assay of the 3′ UTR reporter of (E) NFKBIZ and (F) NFKBID in ABC and GCB DLBCL cell lines. n = 3, all error bars depict the mean ± SD. (G-H) Luciferase reporter assay of the 3′ UTR of (G) NFKBIZ and (H) NFKBID in HBL1 and OCI-Ly3 after treatment with ibrutinib, S-mepazine, or MLT-748. n ≥ 4, all error bars depict the mean ± SD, ordinary 1-way ANOVA with Dunnett multiple comparisons; ∗P < .05; ∗∗P < .01; and ∗∗∗P < .001. (I) Schematic representation of the SL structure of the UTR of NFKBIZ, and location of patient-derived mutations in the UTR. (J) Luciferase reporter assay of WT and mutation variants SNV1, Del1, Del2, and Del3 UTR of NFKBIZ in BJAB CARD11 KO cells expressing CARD11 WT. The relative luminescence was normalized to the stability of the WT UTR of NFKBIZ. N = 3, all error bars depict the mean ± SD, ordinary 1-way ANOVA with Dunnett multiple comparisons; ∗∗P < .01 and ∗∗∗P < .001. (K) Luciferase reporter assay of the WT, Del1, and Del3 UTR reporter in BJAB CARD11 KO cells expressing CARD11 WT, L251P, or L232LI. n = 3, all error bars depict the mean ± SD, 2-way ANOVA with Dunnett multiple comparisons test; ∗P < .05. (L) Luciferase reporter assay of WT, Del1, and Del3 NFKBIZ UTR in a panel of ABC and GCB DLBCLs. n = 4, all error bars depict the mean ± SD, ordinary 1-way ANOVA with Dunnett multiple comparisons test; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001.
Regulation of the 3′ UTR of NFKBIZ and NFKBID in DLBCL. (A-B) Schematic depiction of the 3′ UTR constructs of the (A) NFKBIZ and (B) NFKBID luciferase reporter. Relative luminescence was measured 48 hours after cotransfection of the reporter with a Renilla construct in BJAB CARD11 KOs expressing CARD11 WT, L251P, or L232LI. Values were normalized to mock vector-infected cells; n ≥ 4, all error bars depict the mean ± SD, ordinary 1-way ANOVA with Tukey multiple comparisons; ∗P < .05; ∗∗∗P < .001; and ∗∗∗∗P < .0001. (C-D) Luciferase reporter assay of the 3′ UTR of (C) NFKBIZ and (D) NFKBID in BJAB CARD11 KOs reconstituted with CARD11 L251P and L232LI, respectively, after ibrutinib, S-mepazine, or MLT-748 treatment for 18 hours. n ≥ 3, all error bars depict the mean ± SD, ordinary 1-way ANOVA with Dunnett multiple comparisons; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001. (E-F) Luciferase reporter assay of the 3′ UTR reporter of (E) NFKBIZ and (F) NFKBID in ABC and GCB DLBCL cell lines. n = 3, all error bars depict the mean ± SD. (G-H) Luciferase reporter assay of the 3′ UTR of (G) NFKBIZ and (H) NFKBID in HBL1 and OCI-Ly3 after treatment with ibrutinib, S-mepazine, or MLT-748. n ≥ 4, all error bars depict the mean ± SD, ordinary 1-way ANOVA with Dunnett multiple comparisons; ∗P < .05; ∗∗P < .01; and ∗∗∗P < .001. (I) Schematic representation of the SL structure of the UTR of NFKBIZ, and location of patient-derived mutations in the UTR. (J) Luciferase reporter assay of WT and mutation variants SNV1, Del1, Del2, and Del3 UTR of NFKBIZ in BJAB CARD11 KO cells expressing CARD11 WT. The relative luminescence was normalized to the stability of the WT UTR of NFKBIZ. N = 3, all error bars depict the mean ± SD, ordinary 1-way ANOVA with Dunnett multiple comparisons; ∗∗P < .01 and ∗∗∗P < .001. (K) Luciferase reporter assay of the WT, Del1, and Del3 UTR reporter in BJAB CARD11 KO cells expressing CARD11 WT, L251P, or L232LI. n = 3, all error bars depict the mean ± SD, 2-way ANOVA with Dunnett multiple comparisons test; ∗P < .05. (L) Luciferase reporter assay of WT, Del1, and Del3 NFKBIZ UTR in a panel of ABC and GCB DLBCLs. n = 4, all error bars depict the mean ± SD, ordinary 1-way ANOVA with Dunnett multiple comparisons test; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001.
Next, we tested whether the NFKBIZ and NFKBID 3′ UTRs contribute to the higher expression of these genes in MALT1 protease–active ABC vs –inactive GCB DLBCL cells (refer to Figure 4B). Whereas the reporter construct without an UTR shows equivalent expression in ABC and GCB DLBCL cells, there was a strong increase in the expression of both NFKBIZ and NFKBID 3′ UTR reporters in ABC DLBCL (Figure 5E-F; supplemental Figure 6F). BTK and MALT1 inhibition did not affect reporter gene expression in GCB DLBCL cells (supplemental Figure 6G-H). However, in CD79B mutant HBL1 cells, both BTK and MALT1 activities and in CARD11-mutant OCI-Ly3 cells, only MALT1 activity maintain the high expression levels of the NFKBIZ and NFKBID 3′ UTR reporters (Figure 5G-H).
In ABC DLBCL, recurrent mutations in the Regnase-1 binding region in the 3′ UTR of NFKBIZ have been shown to stabilize the mRNA and enhance IκBζ protein expression.53 We introduced various patient-derived mutations in SL1 (SNV1, Del1, or Del2) or SL4 (Del3), which all augmented expression of the NFKBIZ 3′ UTR reporter construct in BJAB cells, even in the absence of oncogenic CARD11 (Figure 5I-J). Moreover, although the WT 3′ UTR of NFKBIZ was induced by oncogenic CARD11, the activating mutations (Del1 or Del3) conferred high expression of the NFKBIZ reporter, which was barely further increased by oncogenic CARD11 (Figure 5K). We determined the effect of the patient–derived Del1 and Del3 mutations in the NFKBIZ 3′ UTR on the reporter in different ABC DLBCL and GCB DLBCL cells. The deletion mutants augmented expression of the NFKBIZ 3′ UTR reporter in MALT1 protease–active ABC DLBCL cells as well as in GCB DLBCL cells devoid of constitutive MALT1 activity (Figure 5L). Thus, NFKBIZ 3′ UTR mutations or MALT1 activation represent 2 independent mechanisms to release NFKBIZ mRNA from posttranscriptional repression by the RBPs Regnase-1 and Roquin-1/2. Taken together, chronic BCR signaling and oncogenic CARD11 are controlling NFKBIZ and NFKBID mRNA stability on the posttranscriptional level.
Oncogenic API2-MALT1 paracaspase activity controls posttranscriptional gene regulation
The API2-MALT1 fusion oncoprotein created by the recurrent t(11;18)(q21;q21) translocation triggers chronic NF-κB activation in MALT lymphoma.54 TRAF6 recruitment to MALT1 and MALT1 protease activity contribute to NF-κB activation by the API2-MALT1 fusion.28,29,55 To investigate whether oncogenic API2-MALT1 controls gene expression also on the posttranscriptional level via MALT1 protease activity, we transduced constructs expressing API2-MALT1B WT, PM (C689A), or TBM (E1031A) in BJAB cells (Figure 6A). Equivalent expression of API2-MALT1B fusion was achieved at levels below that of endogenous MALT1 (Figure 6B). Although expression of API2-MALT1B WT activated the NF-κB reporter gene, mutations rendering the MALT1 moiety either catalytically inactive (PM) or TRAF6-binding defective (TBM) were impaired in promoting NF-κB activation (Figure 6C; supplemental Figure 7A). API2-MALT1 PM failed to induce processing of p100 to p52 (Figure 6B), revealing that both canonical (via TRAF6 recruitment) and noncanonical (via NIK [NF-κB inducing kinase] cleavage) NF-κB pathways are required for optimal NF-κB activation by the API2-MALT1 fusion.28,29 The API2-MALT1 fusion catalyzed cleavage of CYLD and the RBPs Regnase-1 and Roquin-1/2, which was abolished in the API2-MALT1 PM but not the TBM mutant.
API2-MALT1 protease activity controls posttranscriptional gene regulation. (A) Schematic depiction of API2-MALT1 protein and domains. The C689A mutation is the MALT1 PM and the E1031A is the TBM of MALT1. (B) Western blot analysis of BJAB cells overexpressing API2-MALT1B WT, PM, or TBM. Activation of the noncanonical NF-κB pathways was determined by p100 and p52 expression, and the activation of the MALT1 protease was shown by substrate cleavage and IκBNS expression. Nonspecific bands are marked with an asterisk. (C) Quantification of ΔMFI of the 6 × NF-κB–EGFP reporter in BJAB cells expressing API2-MALT1B WT, PM, or TBM. Data represent MFI of n = 3 replicates, all error bars depict the mean ± SD, ordinary 1-way ANOVA with Tukey multiple comparisons; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001. (D) GSVA quantifying the effects of API2-MALT1 WT and mutant expression on a published ABC DLBCL–derived NF-κB gene signature. (E) Heat map of differentially expressed genes in API2-MALT1 WT-, PM-, or TBM–expressing BJAB cells compared with parental cells. On the color scale, red denotes higher and blue denotes lower expression; n = 5 for each sample. Genes highlighted in green are MALT1-protease dependent and genes highlighted in blue are TRAF6-binding dependent. (F) Signed Padj plot comparing gene induction by API2-MALT1 PM– or TBM– vs API2-MALT1 WT–expressing BJAB cells. Genes in the top-left and bottom-right corner are strongly TRAF6-binding and MALT1-protease dependent, respectively. Annotated are the genes from the heatmap in panel E. (G) Transcriptional activation of NFKBIZ, NFKBID, and ZC3H12A was shown by qRT-PCR in BJAB cells expressing API2-MALT1B WT, PM, or TBM. Values were normalized to the control (mock) infected cells, n = 8, all error bars depict the mean ± SD, ordinary 1-way ANOVA with Tukey multiple comparisons; ∗P < .05 and ∗∗P < .01. (H) Analysis of the inhibition effects of the MALT1 protease after MLT-748 (2 μM, 18 hours) on NFKBIZ, ZC3H12A, and NFKBID in BJAB cells expressing API2-MALT1B WT by qRT-PCR. n = 4, all error bars depict the mean ± SD, unpaired Student t test; ∗∗P < .01. (I-J) Luciferase reporter assay of the 3′ UTR of (I) NFKBIZ and (J) NFKBID in BJAB cells overexpressing API2-MALT1B WT, PM, and TBM (left). Stability of the reporter was determined after inhibition of the MALT1 protease with MLT-748 (right) in BJAB cells expressing API2-MALT1B WT. n = 4, all error bars depict the mean ± SD; ∗P < .05; ∗∗P < .01; and ∗∗∗∗P < .0001. (K) Scheme of posttranscriptional gene regulation by the MALT1 protease in BCR-addicted ABC DLBCL (upper panel) and API2-MALT1-driven MALT lymphomas (lower panel).
API2-MALT1 protease activity controls posttranscriptional gene regulation. (A) Schematic depiction of API2-MALT1 protein and domains. The C689A mutation is the MALT1 PM and the E1031A is the TBM of MALT1. (B) Western blot analysis of BJAB cells overexpressing API2-MALT1B WT, PM, or TBM. Activation of the noncanonical NF-κB pathways was determined by p100 and p52 expression, and the activation of the MALT1 protease was shown by substrate cleavage and IκBNS expression. Nonspecific bands are marked with an asterisk. (C) Quantification of ΔMFI of the 6 × NF-κB–EGFP reporter in BJAB cells expressing API2-MALT1B WT, PM, or TBM. Data represent MFI of n = 3 replicates, all error bars depict the mean ± SD, ordinary 1-way ANOVA with Tukey multiple comparisons; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001. (D) GSVA quantifying the effects of API2-MALT1 WT and mutant expression on a published ABC DLBCL–derived NF-κB gene signature. (E) Heat map of differentially expressed genes in API2-MALT1 WT-, PM-, or TBM–expressing BJAB cells compared with parental cells. On the color scale, red denotes higher and blue denotes lower expression; n = 5 for each sample. Genes highlighted in green are MALT1-protease dependent and genes highlighted in blue are TRAF6-binding dependent. (F) Signed Padj plot comparing gene induction by API2-MALT1 PM– or TBM– vs API2-MALT1 WT–expressing BJAB cells. Genes in the top-left and bottom-right corner are strongly TRAF6-binding and MALT1-protease dependent, respectively. Annotated are the genes from the heatmap in panel E. (G) Transcriptional activation of NFKBIZ, NFKBID, and ZC3H12A was shown by qRT-PCR in BJAB cells expressing API2-MALT1B WT, PM, or TBM. Values were normalized to the control (mock) infected cells, n = 8, all error bars depict the mean ± SD, ordinary 1-way ANOVA with Tukey multiple comparisons; ∗P < .05 and ∗∗P < .01. (H) Analysis of the inhibition effects of the MALT1 protease after MLT-748 (2 μM, 18 hours) on NFKBIZ, ZC3H12A, and NFKBID in BJAB cells expressing API2-MALT1B WT by qRT-PCR. n = 4, all error bars depict the mean ± SD, unpaired Student t test; ∗∗P < .01. (I-J) Luciferase reporter assay of the 3′ UTR of (I) NFKBIZ and (J) NFKBID in BJAB cells overexpressing API2-MALT1B WT, PM, and TBM (left). Stability of the reporter was determined after inhibition of the MALT1 protease with MLT-748 (right) in BJAB cells expressing API2-MALT1B WT. n = 4, all error bars depict the mean ± SD; ∗P < .05; ∗∗P < .01; and ∗∗∗∗P < .0001. (K) Scheme of posttranscriptional gene regulation by the MALT1 protease in BCR-addicted ABC DLBCL (upper panel) and API2-MALT1-driven MALT lymphomas (lower panel).
We performed transcriptomic analyses to determine which genes induced by oncogenic API2-MALT1 rely on MALT1 scaffolding and protease. Compared with PCA in oncogenic CARD11, individual samples cluster closely together, which may, at least partially, be explained by the low API2-MALT1 protein abundance (supplementary Figure 7B). API2-MALT1 was able to induced ABC DLBCL–derived target gene signatures, however, to a much lower degree compared with CARD11 L232LI, and TRAF6 binding and MALT1 protease activity cooperate in the induction of NF-κB signature genes (Figure 6D-F; supplemental Figure 7C). Strikingly, API2-MALT1 also induced previously identified genes NFKBID, ZC3H12D, and ZC3H12A in a highly MALT1 protease–dependent manner (Figure 6F; supplemental Figure 7D). Similarly, NFKBIZ induction by API2-MALT1 was MALT1-protease dependent but expression was below the threshold to be included in the heat map. We confirmed by qRT-PCR that expression of NFKBIZ, NFKBID, and ZC3H12A was significantly decreased in protease-inactive API2-MALT1 PM fusion, when compared with the WT or TRAF6 binding–defective API2-MALT1 fusions (Figure 6G). Accordingly, IκBNS p70 expression by API2-MALT1 relies on protease activity (Figure 6B). MALT1 inhibitor MLT-748 impaired cleavage of API2-MALT1 substrates CYLD, Regnase-1, and Roquin-1/2, and caused a significant suppression of MALT1 protease targets NFKBID and ZC3H12A (Figure 6H; supplemental Figure 7E). The decrease in NFKBIZ mRNA expression was not significant, which may be because of the weak induction by API2-MALT1. To confirm that API2-MALT1 fusion can control posttranscriptional gene expression of both NFKBID and NFKBIZ, we transfected 3′ UTR constructs into BJAB cells expressing the oncogenic fusions (Figure 6I-J). The 3′ UTR of NFKBIZ and even more of NFKBID were induced by the API2-MALT1 WT or TBM fusion constructs, whereas MALT1 protease–defective PM fusion failed to enhance expression of NFKBIZ and NFKBID 3′ UTRs. Induction of both UTR reporters by API2-MALT1 was decreased by MALT1 inhibition. Thus, expression of the API2-MALT1 oncogenic fusion protein catalyzes cleavage of Regnase-1 and Roquin-1/2 and thereby induces gene expression on the posttranscriptional level.
Discussion
We demonstrate that MALT1 constitutes a bifurcation point that orchestrates transcriptional and posttranscriptional gene regulation in ABC DLBCL addicted to chronic BCR signaling or oncogenic CARD11 mutations (Figure 6K; upper panel). Although TRAF6 recruitment links MALT1 scaffolding to canonical NF-κB signaling, MALT1 protease activation catalyzes the cleavage and inactivation of the mRNA restriction factors Regnase-1 and Roquin-1/2, which enhance posttranscriptional gene expression. In parallel, cleavage of A20, CYLD, and RelB augments NF-κB signaling and transcriptional responses.17,27,31 We show that the MALT1 protease is involved in driving posttranscriptional expression of genes that are dependent (eg, NFKBIZ) or independent (eg, NFKBID and ZC3H12A) of NF-κB transcriptional activation, revealing that MALT1 mediates oncogenic functions beyond NF-κB. Similarly, a posttranscriptional release of these MALT1 protease–dependent genes is seen in the context of the oncogenic API2-MALT1 fusion protein, which also catalyzes the cleavage of the RBPs Regnase-1 and Roquin-1/2 (Figure 6K; lower panel). Of note, NFKBIZ is more strongly induced by oncogenic CARD11 compared with API2-MALT1. Differences in expression of the oncogenic drivers but also alterations in MALT1 substrates selectivity between oncogenic CARD11 and API2-MALT1 may be involved in such variations.28 Thus, the exact interplay between the noncatalytic and catalytic functions of MALT1 is responsible for driving addiction to chronic BCR or API2-MALT1 signaling in ABC DLBCL or MALT lymphomas, respectively.
Although NF-κB activation induced by antigenic stimulation or oncogenic CARD11 was severely impaired in the absence of TRAF6, HOIL-1 KO and the concomitant destruction of LUBAC was largely dispensable for NF-κB signaling. Accordingly, BCR-induced NF-κB activation relies on TRAF6 in chicken DT40 B cells.56 Previous results found reduced NF-κB activation upon depletion of LUBAC components in ABC DLBCL cells or in cells expressing oncogenic CARD11.32,33,57 However, in most cases the decrease in NF-κB activation was rather mild, suggesting that TRAF6 possesses an essential function, and LUBAC an auxiliary function, downstream of oncogenic CARD11. This could also explain why HOIL-1 cleavage by MALT1 is not detrimental to NF-κB survival signaling in lymphoma cells. Importantly, LUBAC associates with the CBM complex and is critical for survival of ABC DLBCL cells.32,33 In the tumor necrosis factor receptor pathway, LUBAC and inhibitor of apoptosis proteins counteract cell death signaling.58 Because LUBAC cooperates with inihibitor of apoptosis proteins in ABC DLBCL survival,44 similar mechanisms to prevent cell death may be involved in the context of the oncogenic CBM complex.
The pathological relevance of deregulated posttranscriptional gene expression is exemplified in the case of NFKBIZ/IκBζ, which is highly expressed and maintains expression of NF-κB survival genes in ABC DLBCL.42 Induction of NFKBIZ is under transcriptional control of NF-κB,42 but destruction of Regnase-1 binding sites by recurrent mutations in the noncoding region of the NFKBIZ gene lead to upregulation in DLBCL, revealing the need to inactivate the posttranscriptional control that limits IκBζ expression.53 As an alternative mechanism, we demonstrate that MALT1-catalyzed Regnase-1 cleavage contributes to NFKBIZ induction in BCR- or CARD11-addicted DLBCL that do not carry oncogenic NFKBIZ 3′ UTR mutations. Of note, ZC3H12A itself is recurrently mutated in DLBCL,53 and Regnase-1 decreases the stability of its own mRNA, ZC3H12A,50 indicating that a vicious cycle of posttranscriptional self-suppression and posttranslational MALT1 processing tightly balances Regnase-1 expression in ABC DLBCL cells. We show that RBPs Roquin-1/2 are also cleaved by MALT1 in ABC DLBCL, and other RNA regulators such as N4BP1 and Regnase-4/ZC3H12D have been identified as MALT1 substrates.59,60 Putative functions for DLBCL pathogenesis need to be determined, but the data indicate that MALT1 protease modulates the activity of multiple factors involved in posttranscriptional gene regulation. Posttranscriptional events are also governing gene induction by oncogenic API2-MALT1. Beyond this, the transcriptomic analyses revealed a striking interplay between MALT1-TRAF6 interaction and MALT1 protease activity in orchestrating API2-MALT1–triggered canonical and noncanonical NF-κB activation, which both contribute to optimal target gene expression.
MALT1 inhibition is a promising approach for the treatment of BTK-resistant DLBCL.15,20 However, our data reveal distinct modes of actions for MALT1 protease and BTK inhibitors in ABC DLBCL addicted to chronic BCR upstream signaling. Although BTK inhibitors impair the catalytic and noncatalytic functions of MALT1, MALT1 inhibitors selectively target the protease function and thus only indirectly affect NF-κB activation through the cleavage of signaling and transcriptional regulators (eg, A20, CYLD, and RelB).17,30,31 We demonstrate that maintenance of high NFKBID and ZC3H12A mRNA expression is exclusively connected to constitutive MALT1 protease function in ABC DLBCL, suggesting that these genes can serve as specific biomarkers in clinical trials evaluating the efficacy of MALT1 inhibitors either in monotherapy or in combination with BTK inhibitors.
Acknowledgments
The authors thank K. Demski for technical assistance and I. A. De la Rosa-Velázquez (Helmholtz Core Facility Genomics) for RNA sequencing.
This work was supported by the Deutsche Krebshilfe award grant 70112622; the Deutsche Forschungsgemeinschaft, project ID 210592381 SFB 1054 A04 and ID 360372040 SFB 1335 P07 (D.K.); the European Union’s Horizon 2020 Research and Innovation Program (grant agreement number 950293 COMBAT-RES [M.P.M.]); and by the Federal Ministry of Education and Research–funded German Network for Bioinformatics Infrastructure (de.NBI).
Authorship
Contribution: N.W. designed the research, performed experiments, analyzed data, and wrote the manuscript; F.O. and W.X. performed and analyzed experiments; G.A. and M.P.M. performed bioinformatic analysis of RNA-sequencing data; M.G. and G.L. performed and analyzed primary expression data; D.K. designed the research, supervised the studies, analyzed data, and wrote the manuscript; and all authors have read, reviewed, and approved the manuscript.
Conflict-of-interest disclosure: D.K. is a scientific adviser of Monopteros Therapeutics Inc. M.P.M. collaborates and receives funding from GlaxoSmithKline, Roche, and AstraZeneca. The remaining authors declare no competing financial interests.
Correspondence: Daniel Krappmann, Helmholtz Zentrum München-German Research Center for Environmental Health, Research Unit Signaling and Translation, Ingolstaedter Landstr 1, 85764 Neuherberg, Germany; e-mail: daniel.krappmann@helmholtz-munich.de.
References
Author notes
∗F.O. and G.A. contributed equally to this study.
RNA-sequencing data are available at Gene Expression Omnibus under accession number GSE215792. RNA-sequencing analysis code is available on https://github.com/MendenLab/MALT1_RNAseq. The published article includes all other data generated or analyzed during this study.
Material and original source data will be available upon reasonable request from the corresponding author, Daniel Krappmann (daniel.krappmann@helmholtz-munich.de).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![TRAF6-dependent and -independent gene expression profiling (GEP) of oncogenic CARD11. (A) Comparative gene set enrichment analysis of 3 ABC DLBCL–derived NF-κB gene signatures of CARD11 L232LI in parental vs TRAF6 KO BJAB cells. Three different NF-κB signatures were compared, with q values for the individual signatures shown in the figure. (B) Heat map of genes differentially expressed in parental and TRAF6 KO BJAB cells expressing CARD11 WT or L232LI (log fold change [LFC] > 1.3; adjusted P value [Padj] < .01). Genes are ordered based on the Padj values of parental CARD11 L232LI vs CARD11 WT. Red denotes higher, blue denotes lower expression on the color scale; n = 5 for each sample. Genes highlighted in blue are induced in the absence of TRAF6. (C) Signed Padj plot comparing gene induction by CARD11 L232LI in parental vs TRAF6 KO BJAB cells. Transcripts enriched in TRAF6 KO cells are depicted in blue. (D) Evaluation of the expression of selected transcripts by qRT-PCR in parental and TRAF6 KO BJAB cells transduced with CARD11 WT or CARD11 L232LI. Changes in transcript expression after MALT1 protease inhibition by MLT-748 (2 μM; 18 hours) in CARD11 L232LI–expressing TRAF6 KO BJAB cells. n = 5, all error bars depict the mean ± SD; left panel: ANOVA with Tukey multiple comparisons; right panel: unpaired Student t test; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001. (E) Western blot analysis of BJAB and TRAF6 KO BJAB cells expressing CARD11 WT and L232LI to determine cleavage of Regnase-1 and Roquin-1/2 and expression of IκBNS. Unspecific bands are marked with an asterisk.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/142/23/10.1182_blood.2023021299/1/m_blood_bld-2023-021299-gr2de.jpeg?Expires=1770130790&Signature=Y0J8HUUCUxOOT7fP09nfdpibdrTpqXbKdjUCQPP6Lq~e1zYqMapPZL6807CQdH4xKoAiNPPc71t6iK2V6y99FmYKkZTpC0wuJQbUv1rWAoxKUHZKOcWz2swsw3E0ZT5fNKo1YllDjBaZ8SnTp49uxV5MLYOOMKw6hEZQ0Dtfy2qBUnNYb2e726HB0b4F~iDFj6rYO2Rq4eSWvSviGUXR02TE0H9CGTdDFmCMmzQY~QevRRzupGie~4YUsfYJ5TZ3XWhvwau1ILgCvPF8~jXvyP4RogMImXPbgFqxS8nc0GrV759neByfnSIIU~SsG4Kmina91iVOgrxnFI-p5l8vqw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal