Key Points
PR can be predicted from scattergrams generated by hematology analyzers of a type that is in widespread clinical use.
Genetic analysis of predicted PR reveals associations of PR with the risk of thrombotic diseases, including stroke.
Abstract
Genetic studies of platelet reactivity (PR) phenotypes may identify novel antiplatelet drug targets. However, such studies have been limited by small sample sizes (n < 5000) because of the complexity of measuring PR. We trained a model to predict PR from complete blood count (CBC) scattergrams. A genome-wide association study of this phenotype in 29 806 blood donors identified 21 distinct associations implicating 20 genes, of which 6 have been identified previously. The effect size estimates were significantly correlated with estimates from a study of flow cytometry–measured PR and a study of a phenotype of in vitro thrombus formation. A genetic score of PR built from the 21 variants was associated with the incidence rates of myocardial infarction and pulmonary embolism. Mendelian randomization analyses showed that PR was causally associated with the risks of coronary artery disease, stroke, and venous thromboembolism. Our approach provides a blueprint for using phenotype imputation to study the determinants of hard-to-measure but biologically important hematological traits.
Introduction
Platelets are small, anucleate blood cells that contribute to physiological clot formation, ensuring hemostasis and healing after vascular injury. However, they also contribute to pathological thrombosis, which underlies venous thromboembolic disease, acute coronary syndrome, and ischemic stroke, including microinfarcts, a leading cause of dementia.1 Platelets in circulation activate in response to stimulation by agonists such as collagen (which is exposed by injury), adenosine diphosphate (ADP, which is released by other activated platelets), thromboxane A2 (which is also released by other activated platelets), and thrombin (which is generated by the coagulation cascade). Pathological platelet activation can be caused by atherosclerotic plaque rupture, which exposes collagen and releases tissue factor, which triggers the coagulation cascade. Antiplatelet therapies are the leading pharmaceutical strategy for the primary and secondary prevention of pathological thrombosis. They reduce platelet reactivity (PR) by blocking specific activation pathways. For example, clopidogrel inhibits the ADP receptors on the surface of platelets, whereas aspirin prevents the production of thromboxane A2. The sensitivity of platelets to stimulation is finely balanced. People with inherited platelet disorders that impair PR have a high risk of bleeding.2 Patients with atherosclerosis treated with percutaneous coronary intervention and antiplatelet drugs have a greater risk of bleeding if they have a lower PR and a greater risk of forming thrombi that occlude blood vessels, causing heart attacks or strokes, if they have a higher PR.3 An improved understanding of the mechanisms that govern activation pathways in platelets could unveil novel drug targets with improved pharmacological safety profiles and potentially offer a means to stratify patients to improve the safety and efficacy of existing therapies.4
PR is typically characterized using light transmission aggregometry (LTA) to measure the aggregation response of platelets to stimulation by agonists. Alternatively, PR to stimulation by an agonist can be measured by flow cytometry (FC), using surface markers of activation, such as the externalization of P-selectin, or the binding of fibrinogen to surface receptors that have undergone conformational change. Estimates of the heritabilities of PR phenotypes typically range from 30% to 60%.5-8 A comparison of 2 recent genetic association studies, one using FC9 and the other using LTA phenotypes,10 demonstrates that FC provides a more heritable measure of PR than LTA: evidence of similar strength was obtained for an association between a variant in GRK5 and PR to thrombin in both studies, but the study using FC relied on half the sample size of the study using LTA. Both the LTA and FC approaches are time consuming, difficult to standardize, and require the processing of fresh citrated blood samples soon after venipuncture. This has limited the sample sizes of genetic association meta-analyses9-15 (supplemental Table 1, available on the Blood website).
To address the limitations of LTA and FC-measured PR, we explored the possibility of imputing PR phenotypes from measurements made using an alternative technology in widespread clinical use. Sysmex XN hematology analyzers are sophisticated, high-throughput, clinically standardized instruments containing a miniaturized flow cytometer, a device to measure cellular impedance, and a spectrophotometer. The primary function of a Sysmex analyzer is to generate a complete blood count (CBC) from the data measured by these internal devices. A CBC is a standard clinical report that summarizes cellular and biochemical properties of the blood, including cell concentrations, cell volume distributions, and the concentration of hemoglobin. We hypothesized that the cell-level measurements of platelets generated by the internal flow cytometer of a Sysmex instrument carry information that could be exploited to study PR in large genotyped cohorts. To explore this, we designed a study encompassing 3 cohorts for which it was possible to access or generate data of different types (Figure 1A). First, we generated FC (Figure 1B) and Sysmex (Figure 1C) data on 533 participants in the Cambridge Platelet Function Cohort (PFC). This allowed us to train models to predict PR to 4 agonists, including ADP, from cell-level measurements of platelets untreated with agonist generated by the Sysmex instrument. Second, we applied the trained model to predict PR to ADP (only PR to ADP could be predicted with useful accuracy) from Sysmex data on 29 806 genotyped participants in the INTERVAL cohort of blood donors.16 Third, we performed a genome-wide association study (GWAS) of the predicted PR (PPR) phenotype in INTERVAL. Fourth, we estimated the effects of the variants identified by the GWAS on the means of (1) FC-derived PR phenotypes and (2) an in vitro thrombus formation phenotype. Fifth, we built a genetic score of PR using the variants identified by the PPR GWAS. Sixth, we computed the genetic score for 384 059 British-ancestry participants in the UK Biobank study and tested for associations between the score and 524 health outcomes. Finally, we conducted Mendelian randomization analyses using large GWAS summary statistics to test for causal associations between PR and the risks of coronary artery disease (CAD), stroke, and venous thromboembolism (VTE).
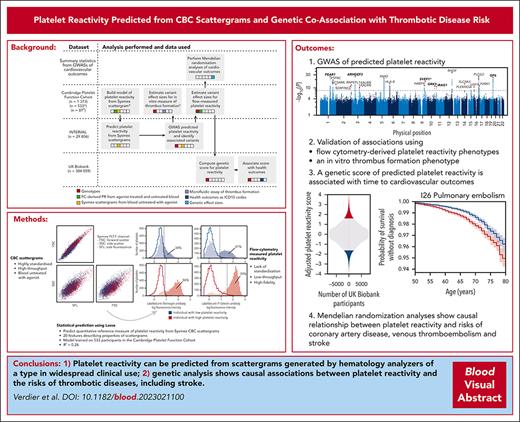
Overview of the study design and data types. (A) Schematic diagram of the analyses; gray blocks represent distinct analyses, which are grouped row-wise according to the data set to which they were applied; the arrows illustrate the flow of information between analyses; the colors in the embedded strips indicate the nature of the primary data used for each analysis. (B) Examples of histograms of log fluorescence intensity from the FC measurements used to quantify PR; the left and right hand panels show measurements of fluorescence generated by an anti-human fibrinogen polyclonal antibody that labels bound fibrinogen and an anti–P-selectin monoclonal antibody, respectively; the upper and lower panels correspond to an individual with a high (red) and an individual with a low (blue) PR, respectively; the filled histograms represent the distributions of log intensity measurements from activated platelets (using the agonist ADP, in this example) incubated with a labeled antifibrinogen antibody or a labeled anti-P-selectin antibody, whereas the open histograms represent negative controls (“Methods”); the dotted lines correspond to the positivity thresholds (ie, to the 98% quantiles of the distributions illustrated by the open histograms); the percentages of agonist-treated platelets that exceed the positivity thresholds are shown. (C) Composite of the 2 CBC scattergrams measured from blood taken from the 2 individuals in panel B, which are distinguished using colors, as in panel B.
Overview of the study design and data types. (A) Schematic diagram of the analyses; gray blocks represent distinct analyses, which are grouped row-wise according to the data set to which they were applied; the arrows illustrate the flow of information between analyses; the colors in the embedded strips indicate the nature of the primary data used for each analysis. (B) Examples of histograms of log fluorescence intensity from the FC measurements used to quantify PR; the left and right hand panels show measurements of fluorescence generated by an anti-human fibrinogen polyclonal antibody that labels bound fibrinogen and an anti–P-selectin monoclonal antibody, respectively; the upper and lower panels correspond to an individual with a high (red) and an individual with a low (blue) PR, respectively; the filled histograms represent the distributions of log intensity measurements from activated platelets (using the agonist ADP, in this example) incubated with a labeled antifibrinogen antibody or a labeled anti-P-selectin antibody, whereas the open histograms represent negative controls (“Methods”); the dotted lines correspond to the positivity thresholds (ie, to the 98% quantiles of the distributions illustrated by the open histograms); the percentages of agonist-treated platelets that exceed the positivity thresholds are shown. (C) Composite of the 2 CBC scattergrams measured from blood taken from the 2 individuals in panel B, which are distinguished using colors, as in panel B.
Methods
A comprehensive description of the methods is given in supplemental Methods, a summary of which is provided here.
Measuring FC-derived PR
Isolating platelets in Sysmex XN scattergrams
The PLT-F channel of the Sysmex XN instrument is designed to measure properties of platelets. The channel generates a 3-dimensional FC scattergram, each point of which corresponds to a cell. We developed a gating procedure to identify platelets (supplemental Table 2) and validated it by comparing the number of cells lying inside the gates with the corresponding platelet count value generated by the instrument for each sample (supplemental Figure 1).
Adjusting for technical variability in PLT-F scattergrams
We adjusted for between-instrument and time-dependent technical variation in the INTERVAL PLT-F scattergrams using a breakpoint detection algorithm to partition the time series. We adjusted the scattergrams within each class of the partition by applying an affine transformation chosen to match the mean and covariance structure of the scattergram data aggregated within the class to those of the aggregated scattergram data from the PFC.
Extracting features from PLT-F scattergrams
We developed a procedure based on principal components analysis to extract 15 features capturing interindividual variation (including measures of central tendency, variation, and covariation) from PLT-F scattergrams.
Building a predictive model of FC-derived PR from Sysmex PLT-F scattergrams
We fitted a Lasso regression18,19 model to predict PR in response to a given agonist from the 15 scattergram-derived features and 5 standard CBC platelet traits (20 features in total) using data from 533 participants in the PFC. We optimized the penalty parameter of the Lasso using cross-validation. In the case of ADP, the optimal value of the penalty parameter yielded a mean prediction R2 = 0.26 (supplemental Figure 2). Conditioning on the optimal penalty parameter value, 14 covariates were included in at least 50% of the Lasso model fitting iterations during cross-validation. We fitted a linear regression model of the ADP response on these 14 covariates using the entire data set.
GWAS of PPR in INTERVAL
We predicted PR in response to ADP in European-ancestry participants in the INTERVAL cohort using the model fitted to the PFC data and performed a GWAS of the predicted phenotype using a standard approach. We tested variants with a minor allele frequency (MAF) >1% and an INFO score >0.4 and adopted a significance threshold of P < 10-8.
Imputation of genotypes in the PFC
We imputed the genotypes in the PFC from a combined 1000 Genomes Phase 3 and UK10K whole genome sequencing panel, as described in Downes et al.14
Regression of FC-derived phenotypes on variant imputed allele counts
We regressed the 4 FC-derived PR phenotypes on the imputed allele counts of each of the 21 PPR-associated variants (identified in INTERVAL) using data from 1373 participants in the PFC.14 Two PPR-associated variants (rs3819288 and rs59001897) had no imputed PFC genotypes. For each of these, we used the LDproxy tool20 to identify the most strongly correlated alternative variant (rs17881225 and rs12905925, respectively) in the “British in England and Scotland” reference panel.
Replication in the LTA study by Keramati et al
We downloaded publicly available summary statistics from the LTA study by Keramati et al,15 which are restricted to the set of variants with association test P values <3 × 10–4. We identified the variant in the summary statistics exhibiting the strongest linkage disequilibrium (LD) (r2 in INTERVAL Europeans) with each of the 21 PPR-associated variants and we identified the phenotype exhibiting the smallest P value of association with each identified variant.
Regression of an in vitro thrombus formation phenotype on variant imputed allele counts
A 48-dimensional in vitro thrombus formation phenotype was measured on 87 genotyped participants in the PFC.21 After standardizing each dimension to have a mean of zero and a variance of 1, we performed a principal components analysis. We regressed the leading principal component on the imputed variant allele counts corresponding to the 21 PPR-associated variants identified in INTERVAL. We compared the P values and effect sizes computed for the genetic analysis of PPR in INTERVAL with those computed for the genetic analysis of the in vitro formation phenotype in the PFC. We used proxies for 2 PPR-associated variants (rs3819288 and rs59001897), as described earlier.
Building a genetic score of general PR
For each of the variants identified by the INTERVAL GWAS of PPR (or their corresponding proxies, see above), we obtained the vector of previously published PFC effect sizes with respect to FC-measured PR in response to each of the 4 agonists.14 We sought to calibrate the effect sizes of the genetic variants across agonists, to place them on a scale measuring a general propensity of platelets to activate (a latent, agonist-independent form of PR). Assuming that each causal variant was involved in only 1 of the 4 activation pathways (corresponding to the 4 agonists), we linked each variant to the agonist yielding the smallest P value of association in the PFC and assigned it to that pathway. We then applied a standardization procedure to calibrate the effect sizes corresponding to each agonist. Finally, we computed a polygenic score of general PR as the sum of the variants’ imputed alternative allele counts weighted by the calibrated effect sizes corresponding to the assigned pathways.
Survival analysis in UK Biobank
We performed Cox regression analyses using the UK Biobank to test for associations between the genetic score of PR and 524 health outcomes (ICD10 codes) derived from electronic health records, adjusting for several covariates known to play a role in cardiovascular diseases and for 5 standard CBC traits.
Mendelian randomization analyses
We performed 2-sample Mendelian randomization analyses to estimate the causal effect of general PR (variant effect sizes calibrated as earlier) on the log odds of disease events for CAD,22 stroke,23 and VTE.24 We excluded rs61751937 because evidence in the literature suggests that variation in SVEP1 expression may be a PR-independent risk factor for atherosclerosis.25 We selected the 10 remaining variants in Table 1 with a P value of association (with a PR phenotype) <.05 in the PFC as primary instruments. We meta-analyzed the instrument-specific ratio estimates using the standard inverse variance weighted (IVW) fixed-effects estimator. We then performed a series of concordancy analyses using the robust MR Egger, IVW random effects, weighted median, and weighted mode estimators.51-53
GWAS of PPR and genetic score of general PR
| Chr . | Position . | rsID . | Ref . | Alt . | MAF . | PINTERVAL . | βINTERVAL . | SEINTERVAL . | Gene . | Comments . | CBC associations . | βscore . | Pscore . | Phenotype for βscore . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 156869047 | rs12566888 | G | T | 0.10 | 5.0 × 10–24 | −0.12 | 0.012 | PEAR1 | Variant is associated with ADP- and epinephrine-induced platelet aggregation.11,12 Platelet receptor that signals upon platelet-platelet contact in response to and independently of the activation response.26 | PLT, MPV, and PDW | −0.99 | 3.2 × 10–8 | ADP |
| 1 | 199010721 | rs1434282 | C | T | 0.27 | 1.9 × 10–17 | −0.07 | 0.008 | PTPRC | Protein regulates GP6-mediated signaling during platelet activation via Src family kinase.27 | PLT, MPV, and PCT | −0.12 | 4.5 × 10–3 | CRP-XL |
| 1 | 247712303 | rs41315846 | T | C | 0.47 | 1.4 × 10–11 | −0.05 | 0.007 | GCSAML | Protein identified as a mediator of platelet activation downstream of cAMP/protein kinase A signaling using protein-protein interaction analysis.28 | PLT, MPV, PDW, and PCT | −0.17 | 1.1 × 10–3 | PAR1 |
| 2 | 224874874 | rs13412535 | G | A | 0.24 | 2.2 × 10–10 | −0.06 | 0.009 | SERPINE2 | Protein is a serine protease inhibitor with activity against the potent platelet activator thrombin.29 | MPV | 0.07 | 1.3 × 10–1 | CRP-XL |
| 2 | 241510903 | rs78909033 | G | A | 0.13 | 2.5 × 10–11 | −0.07 | 0.010 | RNPEPL1 | PLT, MPV, PDW, and PCT | 0.10 | 6.6 × 10–2 | CRP-XL | |
| 3 | 56849749 | rs1354034 | T | C | 0.40 | 1.2 × 10–24 | −0.07 | 0.007 | ARHGEF3 | Variant is associated with ADP-induced aggregation.14 Guanine nucleotide exchanger for ρA, implicated in platelet production and ADP-mediated platelet activation.30 | PLT, MPV, PDW, and PCT | −0.64 | 4.2 × 10–10 | ADP |
| 3 | 124340093 | rs13067286 | G | A | 0.48 | 2.2 × 10–11 | 0.05 | 0.007 | KALRN | Protein is a ρ-GTPase activator, activated by ADP receptor signaling in platelets.31 Another variant in KALRN (rs3772800) is associated with the risk of myocardial infarction.32 | PLT, MPV, and PDW | −0.14 | 4.9 × 10–3 | PAR1 |
| 3 | 124366890 | rs76445378 | C | T | 0.02 | 5.8 × 10–9 | −0.17 | 0.029 | KALRN | PLT, MPV, and PDW | −0.41 | 8.4 × 10–2 | PAR1 | |
| 5 | 122088890 | rs922140 | A | G | 0.40 | 9.4 × 10–21 | 0.07 | 0.007 | SNX2 | MPV and PCT | −0.08 | 1.3 × 10–1 | PAR1 | |
| 6 | 31322694 | rs3819288† | T | C | 0.10 | 1.2 × 10–12 | −0.08 | 0.012 | HLA-B | Antigen-presenting major histocompatibility complex class I molecule that mediates alloimmune clearance of circulating platelets.33 Other variants in this region are associated with unstable angina pectoris,34 myocardial infarction, use of antithrombotic agents,35 and rheumatoid arthritis,36 among many other phenotypes. | PLT and MPV | 0.19 | 1.8 × 10–3 | PAR4 |
| 9 | 99234329 | rs55665228 | C | T | 0.19 | 4.0 × 10–11 | −0.06 | 0.009 | HABP4 | PLT, MPV, PDW, and PCT | −0.35 | 1.2 × 10–2 | ADP | |
| 9 | 113312231 | rs61751937 | G | C | 0.03 | 9.2 × 10–16 | 0.17 | 0.021 | SVEP1∗ | Gene recently associated with ADP-induced platelet aggregation using a gene-based approach.15 Extracellular matrix protein that interacts with PEAR1 to activate platelets.37 | PLT, MPV, and PDW | 1.45 | 8.2 × 10–6 | ADP |
| 10 | 121010256 | rs10886430 | A | G | 0.14 | 2.3 × 10–10 | 0.07 | 0.011 | GRK5 | Variant is associated with thrombin-induced platelet aggregation and VTE.10,14 The protein is a serine/threonine kinase GPCR regulator that modulates thrombin-mediated platelet activation.14 | PLT, MPV, and PDW | 0.90 | 2.8 × 10–40 | PAR1 |
| 11 | 10711817 | rs7123827 | A | C | 0.50 | 7.2 × 10–10 | 0.04 | 0.007 | IRAG1 | Other variants in this region are associated with ADP- and epinephrine-induced platelet aggregation.11,12 Inositol 1,45-triphosphate receptor regulator during nitric oxide/cyclic GMP modulation of PR.38 | PLT and MPV | 0.05 | 1.7 × 10–1 | CRP-XL |
| 12 | 122216910 | rs11553699 | A | G | 0.15 | 4.2 × 10–35 | 0.13 | 0.011 | RHOF | Variant is associated with serum levels of heparin-binding EGF.39 Small GTPase actin regulator that mediates platelet filopodia formation.40 | PLT, MPV, PDW, and PCT | 0.06 | 3.2 × 10–1 | CRP-XL |
| 14 | 70653758 | rs61978213 | G | A | 0.05 | 2.4 × 10–12 | 0.12 | 0.018 | SLC8A3 | Protein is a sodium/calcium exchanger involved in platelet calcium homeostasis.41 Another variant near gene (rs55784307) is associated with peripheral arterial disease.42,43 | 0.20 | 5.9 × 10–2 | PAR4 | |
| 15 | 65160392 | rs59001897✝ | T | A | 0.18 | 6.4 × 10–9 | 0.05 | 0.010 | PLEKHO2 | Another variant near PLEKHO2 (rs832890) is associated with pulse pressure.44 | PLT, MPV, PDW, and PCT | 0.15 | 2.8 × 10–1 | ADP |
| 16 | 9052989 | rs8057254 | T | A | 0.19 | 2.8 × 10–9 | 0.05 | 0.009 | USP7 | Inhibition of USP7 blocks collagen-stimulated aggregation.45 | PLT, MPV, and PDW | 0.10 | 3.4 × 10–2 | CRP-XL |
| 16 | 81870969 | rs12445050 | C | T | 0.14 | 5.2 × 10–25 | 0.11 | 0.010 | PLCG2 | Variant associated with VTE46 and CAD.34 Protein is a member of the phospholipase C family and mediates GP6- and αIIbβ3-mediated platelet activation.47 | PLT, MPV, and PDW | 0.09 | 1.0 × 10–1 | CRP-XL |
| 17 | 3819002 | rs11078475 | T | C | 0.47 | 2.5 × 10–10 | 0.05 | 0.007 | P2RX1 | Gene is overexpressed in reticulated platelets.48 Protein is a platelet ATP receptor contributing to platelet granule release.49 | PLT and MPV | 0.16 | 1.1 × 10–1 | ADP |
| 19 | 55538980 | rs1654425 | T | C | 0.17 | 3.7 × 10–22 | 0.09 | 0.009 | GP6 | A variant in strong LD (rs1671152) is associated with collagen-induced platelet aggregation mediated by the surface receptor encoded by GP6.12 The protein is a platelet receptor for the potent activating agonist collagen. | PLT, MPV, and PDW | 0.89 | 5.4 × 10–100 | CRP-XL |
| Chr . | Position . | rsID . | Ref . | Alt . | MAF . | PINTERVAL . | βINTERVAL . | SEINTERVAL . | Gene . | Comments . | CBC associations . | βscore . | Pscore . | Phenotype for βscore . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 156869047 | rs12566888 | G | T | 0.10 | 5.0 × 10–24 | −0.12 | 0.012 | PEAR1 | Variant is associated with ADP- and epinephrine-induced platelet aggregation.11,12 Platelet receptor that signals upon platelet-platelet contact in response to and independently of the activation response.26 | PLT, MPV, and PDW | −0.99 | 3.2 × 10–8 | ADP |
| 1 | 199010721 | rs1434282 | C | T | 0.27 | 1.9 × 10–17 | −0.07 | 0.008 | PTPRC | Protein regulates GP6-mediated signaling during platelet activation via Src family kinase.27 | PLT, MPV, and PCT | −0.12 | 4.5 × 10–3 | CRP-XL |
| 1 | 247712303 | rs41315846 | T | C | 0.47 | 1.4 × 10–11 | −0.05 | 0.007 | GCSAML | Protein identified as a mediator of platelet activation downstream of cAMP/protein kinase A signaling using protein-protein interaction analysis.28 | PLT, MPV, PDW, and PCT | −0.17 | 1.1 × 10–3 | PAR1 |
| 2 | 224874874 | rs13412535 | G | A | 0.24 | 2.2 × 10–10 | −0.06 | 0.009 | SERPINE2 | Protein is a serine protease inhibitor with activity against the potent platelet activator thrombin.29 | MPV | 0.07 | 1.3 × 10–1 | CRP-XL |
| 2 | 241510903 | rs78909033 | G | A | 0.13 | 2.5 × 10–11 | −0.07 | 0.010 | RNPEPL1 | PLT, MPV, PDW, and PCT | 0.10 | 6.6 × 10–2 | CRP-XL | |
| 3 | 56849749 | rs1354034 | T | C | 0.40 | 1.2 × 10–24 | −0.07 | 0.007 | ARHGEF3 | Variant is associated with ADP-induced aggregation.14 Guanine nucleotide exchanger for ρA, implicated in platelet production and ADP-mediated platelet activation.30 | PLT, MPV, PDW, and PCT | −0.64 | 4.2 × 10–10 | ADP |
| 3 | 124340093 | rs13067286 | G | A | 0.48 | 2.2 × 10–11 | 0.05 | 0.007 | KALRN | Protein is a ρ-GTPase activator, activated by ADP receptor signaling in platelets.31 Another variant in KALRN (rs3772800) is associated with the risk of myocardial infarction.32 | PLT, MPV, and PDW | −0.14 | 4.9 × 10–3 | PAR1 |
| 3 | 124366890 | rs76445378 | C | T | 0.02 | 5.8 × 10–9 | −0.17 | 0.029 | KALRN | PLT, MPV, and PDW | −0.41 | 8.4 × 10–2 | PAR1 | |
| 5 | 122088890 | rs922140 | A | G | 0.40 | 9.4 × 10–21 | 0.07 | 0.007 | SNX2 | MPV and PCT | −0.08 | 1.3 × 10–1 | PAR1 | |
| 6 | 31322694 | rs3819288† | T | C | 0.10 | 1.2 × 10–12 | −0.08 | 0.012 | HLA-B | Antigen-presenting major histocompatibility complex class I molecule that mediates alloimmune clearance of circulating platelets.33 Other variants in this region are associated with unstable angina pectoris,34 myocardial infarction, use of antithrombotic agents,35 and rheumatoid arthritis,36 among many other phenotypes. | PLT and MPV | 0.19 | 1.8 × 10–3 | PAR4 |
| 9 | 99234329 | rs55665228 | C | T | 0.19 | 4.0 × 10–11 | −0.06 | 0.009 | HABP4 | PLT, MPV, PDW, and PCT | −0.35 | 1.2 × 10–2 | ADP | |
| 9 | 113312231 | rs61751937 | G | C | 0.03 | 9.2 × 10–16 | 0.17 | 0.021 | SVEP1∗ | Gene recently associated with ADP-induced platelet aggregation using a gene-based approach.15 Extracellular matrix protein that interacts with PEAR1 to activate platelets.37 | PLT, MPV, and PDW | 1.45 | 8.2 × 10–6 | ADP |
| 10 | 121010256 | rs10886430 | A | G | 0.14 | 2.3 × 10–10 | 0.07 | 0.011 | GRK5 | Variant is associated with thrombin-induced platelet aggregation and VTE.10,14 The protein is a serine/threonine kinase GPCR regulator that modulates thrombin-mediated platelet activation.14 | PLT, MPV, and PDW | 0.90 | 2.8 × 10–40 | PAR1 |
| 11 | 10711817 | rs7123827 | A | C | 0.50 | 7.2 × 10–10 | 0.04 | 0.007 | IRAG1 | Other variants in this region are associated with ADP- and epinephrine-induced platelet aggregation.11,12 Inositol 1,45-triphosphate receptor regulator during nitric oxide/cyclic GMP modulation of PR.38 | PLT and MPV | 0.05 | 1.7 × 10–1 | CRP-XL |
| 12 | 122216910 | rs11553699 | A | G | 0.15 | 4.2 × 10–35 | 0.13 | 0.011 | RHOF | Variant is associated with serum levels of heparin-binding EGF.39 Small GTPase actin regulator that mediates platelet filopodia formation.40 | PLT, MPV, PDW, and PCT | 0.06 | 3.2 × 10–1 | CRP-XL |
| 14 | 70653758 | rs61978213 | G | A | 0.05 | 2.4 × 10–12 | 0.12 | 0.018 | SLC8A3 | Protein is a sodium/calcium exchanger involved in platelet calcium homeostasis.41 Another variant near gene (rs55784307) is associated with peripheral arterial disease.42,43 | 0.20 | 5.9 × 10–2 | PAR4 | |
| 15 | 65160392 | rs59001897✝ | T | A | 0.18 | 6.4 × 10–9 | 0.05 | 0.010 | PLEKHO2 | Another variant near PLEKHO2 (rs832890) is associated with pulse pressure.44 | PLT, MPV, PDW, and PCT | 0.15 | 2.8 × 10–1 | ADP |
| 16 | 9052989 | rs8057254 | T | A | 0.19 | 2.8 × 10–9 | 0.05 | 0.009 | USP7 | Inhibition of USP7 blocks collagen-stimulated aggregation.45 | PLT, MPV, and PDW | 0.10 | 3.4 × 10–2 | CRP-XL |
| 16 | 81870969 | rs12445050 | C | T | 0.14 | 5.2 × 10–25 | 0.11 | 0.010 | PLCG2 | Variant associated with VTE46 and CAD.34 Protein is a member of the phospholipase C family and mediates GP6- and αIIbβ3-mediated platelet activation.47 | PLT, MPV, and PDW | 0.09 | 1.0 × 10–1 | CRP-XL |
| 17 | 3819002 | rs11078475 | T | C | 0.47 | 2.5 × 10–10 | 0.05 | 0.007 | P2RX1 | Gene is overexpressed in reticulated platelets.48 Protein is a platelet ATP receptor contributing to platelet granule release.49 | PLT and MPV | 0.16 | 1.1 × 10–1 | ADP |
| 19 | 55538980 | rs1654425 | T | C | 0.17 | 3.7 × 10–22 | 0.09 | 0.009 | GP6 | A variant in strong LD (rs1671152) is associated with collagen-induced platelet aggregation mediated by the surface receptor encoded by GP6.12 The protein is a platelet receptor for the potent activating agonist collagen. | PLT, MPV, and PDW | 0.89 | 5.4 × 10–100 | CRP-XL |
The first 10 columns show the results of the GWAS of a PR phenotype predicted from Sysmex scattergrams in INTERVAL. The coordinates are given with respect to genome build GRCh37. The subsequent columns include (1) a comment on each gene mapped to the associated single nucleotide polymorphism, (2) a list of CBC trait associations identified previously by GWAS,50 and (3) the effect size and P value corresponding to the PR trait with the smallest P value for association in the PFC and the agonist corresponding to that trait. The bold gene names indicate loci previously associated with a PR phenotype by GWAS analysis (P < 5 × 10–8) or, in the case of SVEP1, by gene-based analysis (P < 10–5). The underlined gene names indicate associations with evidence for replication in partial summary statistics from LTA studies (P < 3 × 10–4).10,15
Alt, alternate allele; ATP, adenosine triphosphate; cAMP, cyclic adenosine monophosphate; Chr, chromosome; EGF, epidermal growth factor; GMP, guanosine monophosphate; GPCR, G protein-coupled receptor; GTP, guanosine triphosphate; MAF, minor allele frequency; MPV, mean platelet volume; PCT, platelet crit; PLT, platelet count; PDW, platelet distribution width; Ref, reference allele; SE, standard error.
Locus previously associated with a PR phenotype by gene-based analysis using the significance threshold P < 10−5.
Because statistics summarizing the associations in the PFC between PR phenotypes and these variants were not available, we identified suitable proxies in the PFC to build a genetic score (supplemental Table 3).
Studies involving participants in the Cambridge PFC received approval from the National Research Ethics Service Committee for East of England, Cambridge East, with the following Research Ethics Committee (REC) reference numbers: REC 05/Q0104/27; REC 05/Q0104/27; BLUEPRINT REC 12/EE/0040; HipSci REC 09/H0304/77, V2 04/01/2013, and V3 15/03/2013; and Genes and Platelets REC 10/H0304/65. The INTERVAL study received approval from the National Research Ethics Service Committee for East of England, Cambridge East, with reference number 11/EE/0538. The UK Biobank study received approval from the North West Multicentre REC as a research tissue bank. As such, researchers do not require separate ethical clearance and can operate under the research tissue bank approval.
Results
We attempted to predict from hematology analyzer data 4 FC-derived phenotypes measuring PR in response to each of (1) ADP, (2) a synthetic cross-linked collagen-related peptide (CRP-XL54), (3) a peptide targeting the thrombin receptor PAR1, and (4) a peptide targeting the thrombin receptor PAR4. Linear regression of each of the 4 phenotypes measuring PR to agonists on the 5 standard platelet CBC traits (platelet count, mean platelet volume, platelet crit, platelet distribution width, and immature platelet fraction) showed poor predictive performance (all R2 < 0.10). Consequently, we applied a gating procedure to select platelets from the PLT-F channel scattergrams and extracted 15 quantitative summary features from each of the resulting subscattergrams (“Methods”; supplemental Table 2). We fitted Lasso regressions to predict each of the 4 FC-derived phenotypes measuring PR to agonists from the 15 features and 5 platelet CBC traits using data from 533 individuals in the PFC. We used 100 repetitions of fivefold cross-validation to tune the Lasso penalty parameters. Although PR in response to each of CRP-XL, PAR1-targeting peptide, and PAR4-targeting peptide could not be predicted usefully (mean R2 < 0.05 in held-out data, see “Discussion”), PR in response to ADP could be predicted with mean R2 = 0.26. Consequently, we investigated whether a GWAS of this phenotype using tens of thousands of individuals could identify genetic variants related to PR.
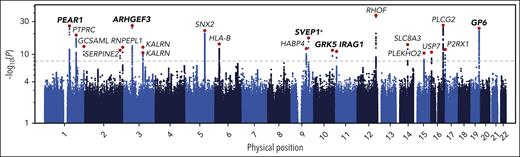
We fitted a linear regression to predict PR to ADP using the covariates identified by the Lasso regression without holding data out. Using this fitted model, we predicted PR to ADP from Sysmex PLT-F scattergram data generated on 29 806 participants in the INTERVAL cohort. We performed univariable tests for additive allelic association between the PPR phenotype and genotypes at 10 013 294 imputed genetic variants. Using stepwise regression, we identified a parsimonious subset of 21 variants explaining the significant (at P < 10-8) genetic associations with PPR and determined the protein-coding gene nearest to each variant (Figure 2; Table 1).
Genome-wide associations with PPR. A Manhattan plot showing the P values of linear mixed model tests for association between genetic variants and PR predicted from CBC scattergrams in INTERVAL. Each dot corresponds to a genetic variant in the Haplotype Reference Consortium r1.1 reference panel. The position on the x-axis indicates the physical position of the variant; the position on the y-axis indicates the log of –log10 of the P value corresponding to the χ2BOLT-LMM statistic. Only variants with an imputation information score >0.4 and a P value <.1 are shown. The horizontal dashed line corresponds to the genome-wide significance threshold (α = 10–8). The red dots correspond to the variants showing the strongest evidence for association in loci containing significantly associated variants. The gene names indicate the protein-coding gene that is nearest to each of these associated variants. The genes in bold mark loci previously associated with a PR phenotype by GWAS analysis (P < 5 × 10–8) or by gene-based analysis (P < 10–5; marked with an asterisk).
Genome-wide associations with PPR. A Manhattan plot showing the P values of linear mixed model tests for association between genetic variants and PR predicted from CBC scattergrams in INTERVAL. Each dot corresponds to a genetic variant in the Haplotype Reference Consortium r1.1 reference panel. The position on the x-axis indicates the physical position of the variant; the position on the y-axis indicates the log of –log10 of the P value corresponding to the χ2BOLT-LMM statistic. Only variants with an imputation information score >0.4 and a P value <.1 are shown. The horizontal dashed line corresponds to the genome-wide significance threshold (α = 10–8). The red dots correspond to the variants showing the strongest evidence for association in loci containing significantly associated variants. The gene names indicate the protein-coding gene that is nearest to each of these associated variants. The genes in bold mark loci previously associated with a PR phenotype by GWAS analysis (P < 5 × 10–8) or by gene-based analysis (P < 10–5; marked with an asterisk).
Of the 20 genes identified, ARHGEF3, GP6, GRK5, IRAG1, and PEAR1 have been implicated as mediators of variation in PR previously by GWAS (Table 1). Although no genome-wide significant single-variant association implicating SVEP1 in the mediation of PR had been reported before the present study (rs61751937; P = 9.2 × 10–16), a gene-based genetic association between SVEP1 and PR has recently been reported with a border-line P value (P = 2.6 × 10−6; α = 2.82 × 10–6). However, that gene-based test relied principally on evidence from a single variant (rs61751937, the same variant identified in this study), which had a univariable P = 5.84 × 10−6.15
ARHGEF3 encodes a megakaryocyte-expressed Rho-guanine nucleotide exchange factor that has previously been associated with PR.14,30,55GP6 encodes 1 of the 2 major collagen receptors on the surface of platelets. GRK5 is a G protein-coupled receptor kinase that regulates thrombin signaling, possibly by phosphorylating the receptors PAR1 and PAR4, leading to their internalization and destruction.10,14IRAG1 plays a role in the inhibition of platelet aggregation and in vivo thrombosis in mice.56PEAR1 encodes a platelet aggregation receptor that signals secondarily to αIIbβ3-mediated contact between platelets.26SVEP1 encodes a protein that may mediate variation in PR through cell-cell adhesion, cell differentiation, or mechanisms in bone marrow niches.15,57 All but 3 of the remaining 14 genes tagged by variants associated with PPR in INTERVAL have plausible roles in biological processes underlying platelet activation (Table 1).
Of the 6 genes previously implicated in the variation of PR phenotypes by genetic association analyses, PEAR1, ARHGEF3, SVEP1, and IRAG1 mediated associations with PR to ADP; PEAR1 also mediated PR to epinephrine; GRK5 mediated PR to thrombin; and GP6 mediated PR to collagen. Therefore, the GWAS of PPR (to ADP) had power to identify genes that play a role in multiple PR pathways, suggesting that the predictive signature in the Sysmex scattergrams captures biological variation downstream of the convergence point of the activation pathways initiated by these different agonists.
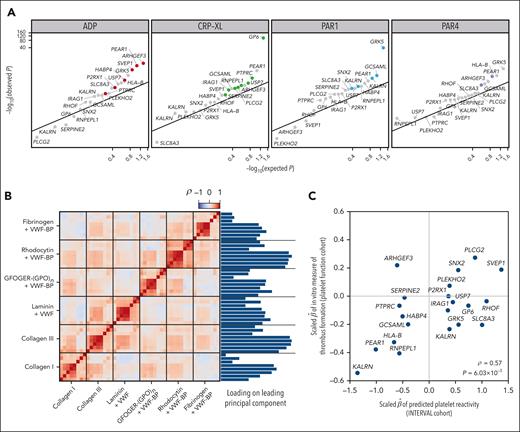
To strengthen the evidence that the PPR phenotype derived from Sysmex scattergrams can be a useful proxy for identifying associations with PR in general, we tested the 21 variants associated with PPR for association with each of the phenotypes measuring an agonist-induced PR response in the PFC. We regressed the 4 FC-measured PR phenotypes (measuring responses to ADP, CRP-XL, PAR1-targeting peptide, and PAR4-targeting peptide) on the imputed allele count of each variant (supplemental Table 3). The P values for each agonist were skewed toward zero relative to the uniform distribution on the interval [0,1] (Figure 3A). When controlling the false discovery rate (Benjamini-Hochberg procedure) at 0.05, 5 variants were significantly associated with PR to ADP, 3 variants with PR to CRP-XL, 3 variants with PR to PAR1-targeting peptide, and 3 variants with PR to PAR4-targeting peptide. The variants exhibiting the strongest evidence for association with the PR phenotypes (with minimum P values across the agonists ranging from 5.37 × 10–100 to 8.24 × 10–6), tagged 5 genes previously implicated in the variation of PR: GP6, GRK5, ARHGEF3, PEAR1, and SVEP1. Variants tagging 4 genes that had not been previously implicated in the variation of PR (GCSAML, HLA-B, PTPRC, and KALRN) exhibited minimum P values ranging from 1.05 × 10–3 to 4.91 × 10–3, strongly suggesting that the analysis of PPR can reveal novel mediators of PR. To demonstrate that the PPR associations were enriched for associations with PR phenotypes relative to standard CBC platelet traits, we compared the distribution of minimum P values (over PR phenotypes) between PPR-associated variants and variants associated with standard CBC traits in INTERVAL.58 The distribution of P values for the PPR-associated variants was the lowest (supplemental Figure 3).
Regression of PR-related phenotypes on PPR-associated variants. (A) log10 scale Q-Q plot comparing the P values obtained by regressing FC-derived PR to ADP, CRP-XL, PAR1-targeting peptide, and PAR4-targeting peptide on the imputed allele counts at 21 PPR-associated variants in 1373 PFC participants with the P values obtained under the null hypothesis of no association. The smallest P value obtained across agonists for each variant is highlighted using an agonist-specific color. (B) Heatmap showing the correlation structure among 48 in vitro phenotypes of thrombus formation and their relative loadings on the leading principal component of a principal components analysis. (C) Scatterplot of the scaled effect sizes of the 21 PPR-associated variants with respect to PPR in INTERVAL and the first principal component of the in vitro thrombus formation phenotypes in 87 PFC participants. The correlation (ρ) and the P value under the null hypothesis, ρ = 0, are embedded.
Regression of PR-related phenotypes on PPR-associated variants. (A) log10 scale Q-Q plot comparing the P values obtained by regressing FC-derived PR to ADP, CRP-XL, PAR1-targeting peptide, and PAR4-targeting peptide on the imputed allele counts at 21 PPR-associated variants in 1373 PFC participants with the P values obtained under the null hypothesis of no association. The smallest P value obtained across agonists for each variant is highlighted using an agonist-specific color. (B) Heatmap showing the correlation structure among 48 in vitro phenotypes of thrombus formation and their relative loadings on the leading principal component of a principal components analysis. (C) Scatterplot of the scaled effect sizes of the 21 PPR-associated variants with respect to PPR in INTERVAL and the first principal component of the in vitro thrombus formation phenotypes in 87 PFC participants. The correlation (ρ) and the P value under the null hypothesis, ρ = 0, are embedded.
We sought to replicate the PPR associations using publicly available GWAS summary statistics of LTA-measured PR phenotypes from Keramati et al.15 PPR-associated variants in 5 loci (PEAR1, ARHGEF3, GP6, SVEP1, and PTPRC) were in strong LD (r2 > 0.8), with a variant exhibiting an LTA association (P < 3 × 10–4). PPR-associated variants in 3 loci (HLA-B [r2 = 0.47], IRAG1 [r2 = 0.35], and USP7 [r2 = 0.07]) were in moderate LD, with a variant exhibiting an LTA association (P < 3 × 10–4). Finally, 2 PPR-associated variants (tagging GCSAML and PLCG2) were within 10 kb of, but not in LD with, a variant exhibiting an LTA association (P < 3 × 10–4), providing supporting evidence that these 2 genes are mediators of variation in PR. The GRK5 variant has been associated with LTA phenotypes in a separate study.10 Therefore, of the 20 genes identified by the PPR GWAS, we found replicative evidence from GWAS of LTA phenotypes for 11 genes.
To assess whether variants identified by the genetic analysis of PPR are associated with the tendency of blood to form thrombi, we analyzed previously published measurements made with an in vitro assay of thrombus formation performed on fresh blood samples from 87 PFC participants.21 Briefly, glass coverslips were coated with 6 microspots, each containing a different platelet agonist. Whole blood was perfused onto the microspots using a parallel-plate flow chamber. Eight variables representing phenotypes related to platelet adhesion, aggregation, or activation were measured on each microspot using a fluorescence microscope. To account for the correlation structure between the 48 parameters of thrombus formation, we performed dimensionality reduction by principal components analysis and regressed the leading principal component on the imputed allele count of each variant (Figure 3B). Although the sample size was insufficient for any individual variant to exhibit a statistically significant association (supplemental Table 3), the effect size estimates were significantly correlated with the effect size estimates for PPR from INTERVAL (Figure 3C; ρ = 0.57; P = 6.03 × 10−3), providing good evidence that some variants associated with PPR play a role in the formation of thrombi.
To explore whether variation in PR might be a predictor of health outcomes, we built a genetic score of PR using the 21 variants identified by the GWAS of PPR. Although the functions of the genes proximal to the variants associated with PPR implied that most were also associated with PR, we were cautious about relying on effect sizes estimated by the GWAS of PPR to weight the genetic score. PPR was only weakly predictive of the FC-measured PR to ADP (R2 = 0.26); therefore, in principle, the phenotype could have a component of variation that depends on biological mechanisms extraneous to PR. Consequently, we sought to identify an estimate of the effect of each variant on a general propensity of platelets to activate (general PR), unbiased by extraneous variation. We assumed that the effect of each variant on PR is mediated by one of the pathways activated by ADP, CRP-XL, PAR1-targeting peptide, and PAR4-targeting peptide. We assigned each variant ad hoc to the pathway corresponding to the agonist yielding the smallest P value of association in the PFC. We standardized the estimated effect sizes in the PFC to render them commensurable across PR phenotypes (ie, FC-measured PR to ADP, CRP-XL, PAR1-targeting peptide, and PAR4-targeting peptide) and used the standardized estimates to weight the imputed allele counts in the polygenic score (“Methods”). The weights assigned to the 21 variants were only moderately correlated with the effect sizes for association with PPR (Figure 4A; R2 = 0.47), so the genetic score of general PR differs substantially from the score that would be derived from the effect size estimates of the PPR GWAS.
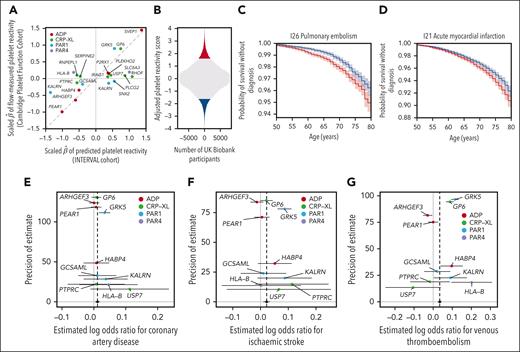
Genetic score of PR and its association with health outcomes. (A) A scatterplot, each dot of which corresponds to a variant identified by stepwise multiple regression analysis of genetic associations with PPR in INTERVAL. The x-axis shows the estimated additive effect size of each variant on mean PPR in INTERVAL. The y-axis shows the estimated additive effect size of each variant on the mean of the agonist-specific PR phenotype with the smallest P value in the PFC. The 2 sets of effect sizes were scaled in order to render them commensurable, as described in “Methods.” (B) Histogram of the genetic score for PR after adjusting for known risk factors and platelet traits computed in UK Biobank participants, with participants in the upper and lower vigintiles of the distribution marked in red and blue, respectively. (C-D) Kaplan-Meier plots corresponding to the 2 vigintile subgroups for the 2 health outcomes significantly associated with the genetic score. Plain lines indicate the estimated survival functions and the shaded areas represent the 90% confidence intervals. (E-G) Funnel plots showing estimates of the log odds ratios for a disease event per standard deviation increase in PPR. The horizontal line segments represent the corresponding 95% confidence intervals. The black circle and line segment lying on the line y = 0 show the estimate and 95% confidence interval for the log odds ratio derived from an IVW fixed-effects meta-analysis. The confidence intervals were derived using second-order weights. Each point is colored according to the PR phenotype used to calculate the general PR effect size of the corresponding variant.
Genetic score of PR and its association with health outcomes. (A) A scatterplot, each dot of which corresponds to a variant identified by stepwise multiple regression analysis of genetic associations with PPR in INTERVAL. The x-axis shows the estimated additive effect size of each variant on mean PPR in INTERVAL. The y-axis shows the estimated additive effect size of each variant on the mean of the agonist-specific PR phenotype with the smallest P value in the PFC. The 2 sets of effect sizes were scaled in order to render them commensurable, as described in “Methods.” (B) Histogram of the genetic score for PR after adjusting for known risk factors and platelet traits computed in UK Biobank participants, with participants in the upper and lower vigintiles of the distribution marked in red and blue, respectively. (C-D) Kaplan-Meier plots corresponding to the 2 vigintile subgroups for the 2 health outcomes significantly associated with the genetic score. Plain lines indicate the estimated survival functions and the shaded areas represent the 90% confidence intervals. (E-G) Funnel plots showing estimates of the log odds ratios for a disease event per standard deviation increase in PPR. The horizontal line segments represent the corresponding 95% confidence intervals. The black circle and line segment lying on the line y = 0 show the estimate and 95% confidence interval for the log odds ratio derived from an IVW fixed-effects meta-analysis. The confidence intervals were derived using second-order weights. Each point is colored according to the PR phenotype used to calculate the general PR effect size of the corresponding variant.
We computed the genetic score of general PR for 384 059 British-ancestry participants in UK Biobank. For each of 524 ICD10 codes recording diagnostic events in at least 1000 participants, we applied Cox proportional hazards regression to estimate the association between the survival time from birth to the event and the genetic score of PR (“Methods”). To adjust the estimates for variation mediated through known risk factors for cardiovascular disease, we included the following variables as covariates in each regression: sex, tobacco use, total cholesterol level, HDL cholesterol level, systolic blood pressure, C-reactive protein concentration, and history of diabetes.59 To ensure that any identified associations were mediated independently of standard platelet parameters, we also included the 4 platelet traits measured in UK Biobank (platelet count, mean platelet volume, platelet crit, and platelet distribution width) as covariates. The score was significantly (family-wise error rate controlled at 0.05 by the Bonferroni method) associated with 2 ICD10 codes, both of which record cardiovascular events with an etiological link to PR: pulmonary embolism (I26) (P = 5.14 × 10-8) and acute myocardial infarction (I21) (P = 5.50 × 10-6; Table 2). We compared the survival distributions of the individuals in the upper and lower 5% tails of the score distribution. The time required to achieve a 2% cumulative probability of a pulmonary embolism diagnosis was ∼3 years longer for the lower tail than for the upper tail, whereas the time required to achieve a 5% cumulative probability of an acute myocardial infarction diagnosis was ∼2 years longer for the lower tail than for the upper tail (Figure 4B–D).
Cox regression association statistics
| P . | ICD10 code . | Description . | No. of participants with an event . | log hazard ratio . |
|---|---|---|---|---|
| 5.14 × 10–8 | I26 | Pulmonary embolism | 6 942 | 0.07 |
| 5.50 × 10–6 | I21 | Acute myocardial infarction | 14 198 | 0.04 |
| P . | ICD10 code . | Description . | No. of participants with an event . | log hazard ratio . |
|---|---|---|---|---|
| 5.14 × 10–8 | I26 | Pulmonary embolism | 6 942 | 0.07 |
| 5.50 × 10–6 | I21 | Acute myocardial infarction | 14 198 | 0.04 |
Significant associations (at a family-wise error rate <0.05, equivalent to P < 9.54 × 10–5) between the genetic score of PR and ICD10-coded health outcomes in 384 059 unrelated, British-ancestry participants in UK Biobank. ICD10 subterms (ie, containing a “.”) were collapsed into the parent term. Only collapsed terms assigned to at least 1000 participants were analyzed (524 terms in total).
Next, we sought to validate the association between the genetic component of variability in general PR and cardiovascular outcomes using recently published large case-control GWASs of CAD, stroke, and VTE.22-24 Without access to individual-level data, we were unable to compute the genetic score in the study participants; instead, we performed 2-sample Mendelian randomization analyses using the 2-sample IVW estimator and complementary robust methods.51 After removing weak instruments (minimum P value >.05 in the PFC) and the variant in SVEP1, which may affect vascular risk through a horizontal pathway,25 10 variants remained. Because at least 1 of these was assigned to each of the 4 agonists in the construction of the score, variation in PR mediated by all 4 pathways contributed to the analyses. Variation in PR was significantly and positively associated with the risks of CAD (P = .019), stroke (P = 2.58 × 10–4), and VTE (P = 6.55 × 10-11; Figure 4E-G). None of the estimates of the intercepts of Egger regression models differed significantly (P > .05) from zero, implying an absence of evidence for directional pleiotropy. The point estimates and confidence intervals derived from the complementary robust estimators were broadly consistent with the IVW estimates (supplemental Table 4; supplemental Figure 4).
Discussion
Despite its clinical importance, PR is challenging to measure, which has limited GWASs of PR phenotypes to sample sizes of a few thousand participants (supplemental Table 2). Our GWAS of PPR in 29 806 blood donors was able to identify variants known to be associated with PR to various agonists, without the need for technically challenging platelet stimulation experiments, by exploiting previously unrecognized information on PR contained in Sysmex XN scattergrams derived from EDTA-treated whole blood. We hypothesize that the variants can be identified in this way because the blood contains small quantities of ADP, collagen, and thrombin, and interindividual variation in PR to these agonists generates variation downstream of the convergence of the activation pathways, which is reflected in the Sysmex scattergrams. It may be that the scattergram signature reflects variation in the stimulation of platelet surface receptors in EDTA-treated blood, where they are stimulated in sufficient numbers to cause morphological changes but in insufficient numbers to cause activation. When training from the Sysmex data, we were able to predict FC-measured PR to ADP better (R2 = 0.26) than PR to CRP-XL, PAR1-targeting peptide, and PAR4-targeting peptide (all R2 < 0.05). Furthermore, our GWAS of PPR (Figure 2) produced much smaller P values for genetic variants known to cause variation in PR to ADP (PEAR1 and ARHGEF3) than for genetic variants known to cause variation in PR to collagen (GP6) or thrombin (GRK5) despite the fact that the power to detect associations by FC was lower for PR to ADP than for PR to collagen or thrombin.14 We speculate that this was because ADP is present in EDTA-treated blood in more potent quantities than collagen or thrombin.
We identified 6 genes previously found by GWAS or by gene-based association analysis of PR phenotypes, including all 3 genes previously identified in at least 2 nonoverlapping study cohorts: GP6, GRK5, and PEAR1 (supplemental Tables 1 and 3). In addition, we identified 14 highly credible candidate genes. For example, one of the candidates, SERPINE2, encodes Serpin Family E Member 2, a natural inhibitor of thrombin, a strong platelet activator that binds to protease-activated receptors on the surface of platelets. The 21 SNPs identified by our GWAS were collectively associated with an in vitro measure of thrombus formation, supporting the hypothesis that the identified genes mediate biological mechanisms involved in thrombosis. Detailed laboratory follow-up of these mechanisms, beyond the scope of the present study, will be required to determine whether they present viable drug targets.
We sought to identify causal associations between PR phenotypes and health outcomes using genetics. However, because PPR is not a direct measure of PR, the effect sizes computed from the INTERVAL GWAS of PPR were potentially biased as estimates of PR. Consequently, we used estimates of effect sizes for association with PR phenotypes in the PFC to quantify the variation in general PR explained by each of the genetic variants associated with PPR, decoupling detection from estimation. We demonstrated the effectiveness of this approach by showing that a genetic score of general PR predicts health outcomes that are closely linked to platelet function, namely, survival without pulmonary embolism and survival without acute myocardial infarction. In addition, 2-sample Mendelian randomization analyses demonstrated an association between variation in PR and the risks of CAD, stroke, and VTE. These results represent the first time the causality of PR as a risk factor for cardiovascular events has been demonstrated using genome-wide instrumental analyses. Other difficult-to-measure risk factors with correlates that are easy to measure may benefit from a similar approach.
Acknowledgments
The authors are grateful to Stephen Garner for generating part of the Cambridge PFC FC data set and for his invaluable advice on the operation of Sysmex hematology analyzers. The authors thank Jarob Saker and Joachim Linssen of Sysmex Europe for the invaluable technical assistance and advice. The authors gratefully acknowledge the participation of all UK Biobank, NIHR Cambridge BioResource, and INTERVAL volunteers. Participants in the INTERVAL randomized controlled trial were recruited through the active collaboration of NHS Blood and Transplant England, which supported field work and other elements of the trial. A complete list of the investigators of and contributors to the INTERVAL trial is provided in a previous publication.16
This research has been conducted using the UK Biobank resource under application number 13745. The authors are grateful to the Lowy Foundation USA for supporting this work. The authors thank the members of the Cambridge BioResource Scientific Advisory Board and Management Committee for supporting our study and the NIHR Cambridge Biomedical Research Centre for funding (RG64219). DNA extraction and genotyping were cofunded by the National Institute for Health and Care Research (NIHR), the NIHR BioResource, and the NIHR Cambridge Biomedical Research Centre (BRC-1215-20014). The academic coordinating centre for INTERVAL was supported by core funding from the NIHR Blood and Transplant Research Unit in Donor Health and Genomics (NIHR BTRU-2014-10024), the NIHR Blood and Transplant Research Unit in Donor Health and Behaviour (NIHR203337), the UK Medical Research Council (MR/L003120/1), the British Heart Foundation (SP/09/002, RG/13/13/30194, and RG/18/13/33946) and the NIHR Cambridge BRC (BRC-1215-20014 and NIHR203312). W.J.A. is supported financially by NHS Blood and Transplant.
The views expressed are those of the authors and not necessarily those of the NIHR, National Health Service Blood and Transplant, or the Department of Health and Social Care.
Authorship
Contribution: H.V. conducted all analyses and wrote the manuscript; P.T., J.B., C.K., and H.M. generated FC and Sysmex data for the PFC; N.G. conducted the imputation of the PFC genotyping data; J.D. established the INTERVAL and provided critical comments on the manuscript; A.M. provided clinical and biological interpretations; J.W.M.H. provided in vitro data on thrombus formation phenotypes; W.H.O. established the PFC and INTERVAL; K.D. supervised the experiments and oversaw analyses; and W.J.A. and E.T. supervised the project and wrote the manuscript jointly.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ernest Turro, Department of Genetics and Genomic Sciences, Hess Center for Science and Medicine at Mount Sinai, 8–119, 1470 Madison Ave, New York City, NY 10029; e-mail: ernest.turro@mssm.edu.
References
Author notes
∗W.J.A. and E.T. jointly supervised this work and contributed equally.
The genotype data for the genotyped participants in UK Biobank are available by application at https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access.
The GWAS summary statistics for CAD, stroke, and VTE are available from the NHGRI-EBI Catalog and deCODE genetics at https://www.ebi.ac.uk/gwas/studies/GCST90132314, https://www.ebi.ac.uk/gwas/studies/GCST90104539, and https://download.decode.is/form/2022/vte_meta.txt.gz, respectively.
The code for the analysis is available at https://github.com/hippover/sysmex2pf.
Deidentified INTERVAL participant data are available on request from the INTERVAL Data Access Committee at helpdesk@intervalstudy.org.uk.
Summary statistics for our PPR GWAS are available from the NHGRI-EBI Catalog at https://www.ebi.ac.uk/gwas/.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal