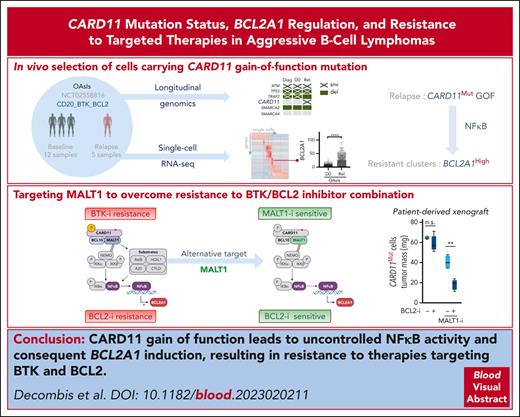
Key Points
BCR-independent NF-κB1 identified as a mechanism of MCL relapse in vivo.
CARD11/NF-κB1/BCL2A1 axis is counteracted by MALT1/BCL2 dual targeting.
Abstract
A strategy combining targeted therapies is effective in B-cell lymphomas (BCL), such as mantle cell lymphoma (MCL), but acquired resistances remain a recurrent issue. In this study, we performed integrative longitudinal genomic and single-cell RNA-sequencing analyses of patients with MCL who were treated with targeted therapies against CD20, BCL2, and Bruton tyrosine kinase (OAsIs trial). We revealed the emergence of subclones with a selective advantage against OAsIs combination in vivo and showed that resistant cells were characterized by B-cell receptor (BCR)–independent overexpression of NF-κB1 target genes, especially owing to CARD11 mutations. Functional studies demonstrated that CARD11 gain of function not only resulted in BCR independence but also directly increased the transcription of the antiapoptotic BCL2A1, leading to resistance against venetoclax and OAsIs combination. Based on the transcriptional profile of OAsIs-resistant subclones, we designed a 16-gene resistance signature that was also predictive for patients with MCL who were treated with conventional chemotherapy, underlying a common escape mechanism. Among druggable strategies to inhibit CARD11-dependent NF-κB1 transduction, we evaluated the selective inhibition of its essential partner MALT1. We demonstrated that MALT1 protease inhibition led to a reduction in the expression of genes involved in OAsIs resistance, including BCL2A1. Consequently, MALT1 inhibition induced synergistic cell death in combination with BCL2 inhibition, irrespective of CARD11 mutational status, both in vitro and in vivo. Taken together, our study identified mechanisms of resistance to targeted therapies and provided a novel strategy to overcome resistance in aggressive BCL. The OAsIs trial was registered at www.clinicaltrials.gov #NCT02558816.
Introduction
Over the past decade, efforts to characterize the molecular profiles of mantle cell lymphoma (MCL) cells and the multiple interactions that occur in their malignant ecosystems, have identified tumor vulnerabilities.1,2 Among them, elevated CD20 expression,3 imbalance in the Bcl2 family,4 and constitutive B-cell receptor (BCR) signaling5 have emerged as promising druggable pathways. Indeed, anti-CD20 antibodies (ie, rituximab), Bcl2 inhibitors (ie, venetoclax), and Bruton tyrosine kinase (BTK) inhibitors (ie, ibrutinib) have shown great promise as single agents6,7 or in combination with chemotherapy.8,9
Nevertheless, heterogeneity coupled with tumor-cell plasticity significantly limits the long-term efficacy of targeted monotherapies.10,11 As a result, multiple mechanisms of resistance arising from both mutations and microenvironmental cues have been described. For example, the selection of MCL cells with BCL2 amplicon loss has been observed during venetoclax treatment,12 and the microenvironment-dependent expression of alternative antiapoptotic proteins has been described as a major means of escape.13,14 Similarly, despite a strong dependence on BCR-dependent NF-κB activation, resistance to BTK inhibition rapidly emerges in MCL. Indeed, in addition to rare on-target mutations,15 mutational or microenvironment-dependent activation of the alternative NF-κB pathway, as well as metabolic reprogramming, have been reported in ibrutinib-resistant cells.16,17
To limit tumor adaptability, mechanism-based combinations of targeted therapies are being designed. We previously demonstrated that pretreatment with obinutuzumab, an anti-CD20 antibody13 and ibrutinib18 was able to counteract microenvironment-dependent resistance to venetoclax in MCL, mostly through the downregulation of NF-κB–dependent BCLXL expression. This rationale was involved in the design of the OAsIs trial, which consisted of a sequential combination of obinutuzumab and ibrutinib followed by venetoclax in patients with MCL. This triple “chemo-free” combination was well-tolerated and led to a complete response rate of 67% and 86.6% in patients who relapsed and those who remained untreated, respectively.19 Nevertheless, despite these high initial response rates, nearly one-third of the patients progressed or rapidly relapsed. In this study, by combining comprehensive genomics and single-cell transcriptomics, we established a novel signature of in vivo resistance and deciphered the molecular mechanisms underlying tumor escape. Based on the mechanisms we discovered, we performed functional studies to identify therapeutic vulnerabilities that could be exploited in order to overcome lymphoma resistance.
Methods
Patient samples cohort
Samples were collected from peripheral blood (PB), bone marrow (BM), or lymph nodes after obtaining informed consent from patients with MCL (Refract-Lyma cohort; ethical approval GNEGS-2015-09-1317) and in accordance with the Declaration of Helsinki. Characteristics of patients who participated in the OAsIs trial (NCT02558816) are summarized in supplemental Table 1 (available on the Blood website).
Deep DNA sequencing
All samples from patients enrolled in the OAsIs trial available at our institution and containing at least 5% of circulating MCL cells (CD19+ CD5+) were used for comprehensive genomics. DNA was extracted from CD19+ MCL cells (17 samples from 14 patients) previously enriched using anti-human CD19-conjugated magnetic beads (Miltenyi). A panel including 49 genes and 170 exons from the 9p21 region was designed to identify recurrent mutations and copy number variations, based on hits described in the literature for their role in lymphomagenesis (supplemental Table 2). Probes were produced by Integrated DNA Technologies.
Single-cell RNA-sequencing (scRNA-seq)
Seven BM samples from 6 patients included in the OAsIs trial were analyzed at the single-cell resolution. In brief, live MCL cells were first enriched by Ficoll-Paque Plus and anti-human CD19 magnetic beads. Depletion of human erythroid precursors and erythrocytes was then performed using CD235a (glycophorin-A) magnetic beads. A total of 15 000 cells from the resulting suspensions were loaded on a Chromium Controller to generate single-cell partitions following the manufacturer’s protocol (10× genomics).
scRNA-seq libraries were constructed using the Chromium Next GEM Single Cell 3′ GEM, Library & Gel Bead Kit v3.1 and sequenced using the NovaSeq 6000 system (Illumina). Overall, 88 034 cells were sequenced with a median of 14 805 per sample and an average of 50 686 reads per cell. Data processing and bioinformatics analysis are detailed in the supplemental Methods section.
Chick embryo chorioallantoic membrane (CAM) model
Fertilized white Leghorn chicken eggs were purchased from Granja Santa Isabel, S. L. (Córdoba, Spain), and incubated for 9 days at 37°C at 55% humidity. At day 9 of their embryonic development, a window of an ∼2 cm-diameter was drilled on top of the air chamber of the eggshell. Then, half million DFBL-44685v2 CARD11D230N patient-derived xenografts MCL cells were suspended in 25 μL RPMI medium containing 10% fetal bovine serum, 100 U/mL penicillin and streptomycin, and 25 μL Matrigel. The mix was incubated for 15 minutes at 37°C and subsequently implanted into the CAM of each egg. Two hundred fifty nM JNJ-67856633 (day 12 and 13), 2 nM of venetoclax (day 13), or both were administered topically on the tumor-bearing CAMs. Seven days after implantation (day 16), chick embryos were killed by decapitation. Tumors were excised and carefully weighed to determine their mass.
Additional methods are detailed in the supplemental Methods and supplemental Table 3 or have been previously described.20
Results
Selective advantage of CARD11GOF mutated cells under OAsIs combination
Fresh tumor tissues from patients with MCL enrolled in the OAsIs trial were collected and comprehensive genomic analysis (single nucleotide variants/copy number variations, n = 17) was performed on CD19+ MCL cells (Figure 1A; supplemental Tables 1 and 2). The most frequently altered genes at inclusion included ATM (6/12), TP53 (5/12), and KMT2D (5/12) (Figure 1B; supplemental Table 2). Only CARD11 mutations were enriched at relapse (n = 3/5, Fisher t = 0.01), suggesting a selective advantage of CARD11 mutated cells under OAsIs therapy (Figure 1C).
Comprehensive genomic analysis of MCL patients enrolled in the OAsIs trial. (A) Schematic representation of the experimental design to identify therapeutic resistance in MCL. Patients with MCL were included in the OAsIs clinical trial (obinutuzumab [anti-CD20], ibrutinib [BTK-i], and venetoclax [BCL2-i]). Blood or BM samples were collected at inclusion (incl.) or relapse (Rel) and targeted DNA-seq of 49 selected genes plus the 9p21 region was performed. (B-D) Landscape of recurrent mutations (SNV/indel) and copy number variations (gain/deletion) in the patients enrolled in OAsIs and detected by deep DNA-seq at incl. (n = 12) (B), Rel (n = 5) (C), or in longitudinal samples (D). Each column represents an individual sample/patient. Only genes with at least 2 anomalies detected in the cohort and with a variant frequency >5% are represented. Statistical significance was determined by a 2-tailed Fisher exact test. (E) The fish plot shows the clonal evolution pattern of patient 34 based on the variant frequency (VaF) measured at diagnosis (Diag), incl., and Rel (supplemental Table 2) and normalized to the TP53Y220C VaF. (F) Schematic representation of CARD11 domains and the mutations found in the Refract-Lyma cohort (n = 62 sequenced for CARD11), including patients enrolled in the OAsIs trial. CR, complete response; PD, progression disease; PR, partial response; SNV, single nucleotide variant; SD, stable disease.
Comprehensive genomic analysis of MCL patients enrolled in the OAsIs trial. (A) Schematic representation of the experimental design to identify therapeutic resistance in MCL. Patients with MCL were included in the OAsIs clinical trial (obinutuzumab [anti-CD20], ibrutinib [BTK-i], and venetoclax [BCL2-i]). Blood or BM samples were collected at inclusion (incl.) or relapse (Rel) and targeted DNA-seq of 49 selected genes plus the 9p21 region was performed. (B-D) Landscape of recurrent mutations (SNV/indel) and copy number variations (gain/deletion) in the patients enrolled in OAsIs and detected by deep DNA-seq at incl. (n = 12) (B), Rel (n = 5) (C), or in longitudinal samples (D). Each column represents an individual sample/patient. Only genes with at least 2 anomalies detected in the cohort and with a variant frequency >5% are represented. Statistical significance was determined by a 2-tailed Fisher exact test. (E) The fish plot shows the clonal evolution pattern of patient 34 based on the variant frequency (VaF) measured at diagnosis (Diag), incl., and Rel (supplemental Table 2) and normalized to the TP53Y220C VaF. (F) Schematic representation of CARD11 domains and the mutations found in the Refract-Lyma cohort (n = 62 sequenced for CARD11), including patients enrolled in the OAsIs trial. CR, complete response; PD, progression disease; PR, partial response; SNV, single nucleotide variant; SD, stable disease.
This was further confirmed by the longitudinal analysis of samples from patient MCL34 at the time of diagnosis, before inclusion in the OAsIs trial, and at relapse. In contrast to a poor response to first-line chemotherapy (R-CHOP [rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone], stable disease), the patient achieved a complete molecular response before relapsing after 12 months of OAsIs treatment.19 Although most hits were similar across the samples, we observed the emergence of a gain-of-function (GOF) CARD11 mutation (G123S, latch domain)21,22 at relapse and the downfall of a loss-of-function (LOF) TRAF2 variant (C49F, ring domain,23Figure 1D; supplemental Table 2). Indeed, although CARD11G123S MCL cells remained discrete after first-line treatment, they made up the majority of the tumor after OAsIs (Figure 1E). Notably, a deeper analysis of the G123 position revealed the presence of few c.367G>A reads before treatment (G123S frequency, 0.12% at diagnosis and 0.19% at inclusion), confirming the hypothesis of selection, under OAsIs, of a rare subclone already present at diagnosis (supplemental Table 4). Regarding CARD11 mutations found in other OAsIs relapse biopsies, one was at the same position (G123D in MCL24) and another was located in the coiled-coil (CC) domain (R235Q in MCL40), both of which led to GOF.21,24 Although we were unable to collect samples at OAsIs inclusion for these patients, paired analysis at diagnosis vs OAsIs relapse showed an increased frequency of CARD11 mutated cells at relapse (R235Q frequency, 31% and 48%, respectively) (supplemental Figure 1). Finally, we detected CARD11 mutations in 4 additional patients with MCL from our local cohort (Refract-Lyma, n = 48 sequenced), including 2 additional G123S mutants, suggesting that mutations at position G123 are not infrequent in MCL (Figure 1F; supplemental Table 4).
Both TRAF2 LOF and CARD11 GOF have been reported as being involved in ibrutinib resistance, inducing BCR-independent NF-κB2 and NF-κB1 constitutive activity, respectively.25,26 To understand the selective advantage of CARD11 GOF in MCL under OAsIs, we performed a transcriptional analysis at the single-cell resolution of the samples of patient MCL34, before and at OAsIs relapse.
Longitudinal single-cell analysis uncovered a minor OAsIs-resistant subpopulation
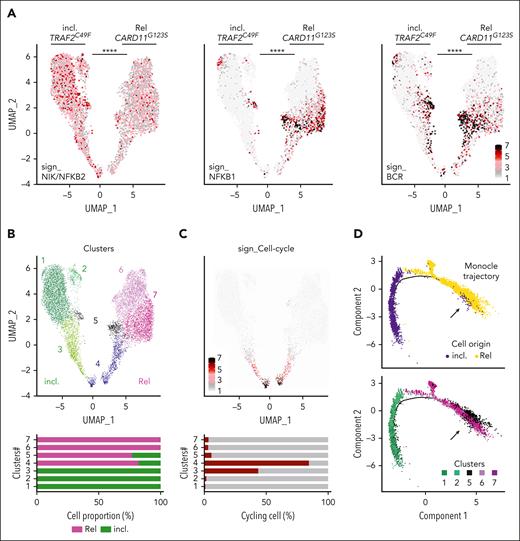
We were able to sequence 4860 BM MCL cells before OAsIs (incl.), and 5345 cells at relapse (Rel) (supplemental Figure 2A). We observed an elevated NIK_NF-κB2 transcriptional signature27 at inclusion (P < .0001), consistent with the functional consequences of the TRAF2 LOF mutation observed in this sample (Figure 2A). In contrast, cells from the sample collected at relapse displayed higher NF-κB1 and BCR signatures27 (P < .0001), consistent with the acquired CARD11 GOF mutation (Figure 2A). We did not observe transcriptional modulation of the OAsIs targets (supplemental Figure 2B).
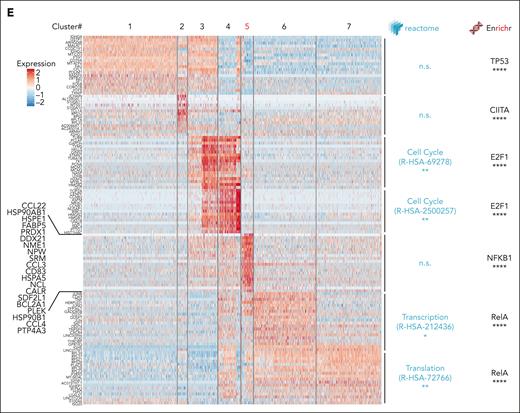
Longitudinal single-cell RNA-seq analysis of MCL during OAsIs. (A) UMAP plots of the cells from patient MCL34 at inclusion (incl.) and at relapse (Rel) after OAsIs treatment. Each dot represents a single cell. UMAP plots display the NIK/ NFκB2 (left panel), NFκB1 (middle panel) and BCR (right panel) signature scores at incl. and Rel. Wilcoxon sign-rank test; ∗∗∗∗Padj < .0001. (B) UMAP plot of the 7 clusters identified by Seurat at incl. and at Rel after OAsIs treatment for the patient MCL34. The cell proportion for each cluster at incl. (green) or Rel (pink) is indicated at the bottom. (C) UMAP plot displaying the cell-cycle signature scores at incl. and Rel for the patient MCL34. Cell proportion of cycling (red) or resting (gray) in each cluster is indicated at the bottom. (D) Trajectory of single cells at incl. and at Rel using Monocle2. Clusters defined in panel B are identified in the lower panel. (E) Heatmap showing the top 20 genes for each cluster. The color gradient indicates the intensity of the median gene expression as indicated. Functional enrichment pathway was calculated using Reactome and Enrichr. ∗∗∗∗Padj < .0001. UMAP, Uniform Manifold Approximation and Projection.
Longitudinal single-cell RNA-seq analysis of MCL during OAsIs. (A) UMAP plots of the cells from patient MCL34 at inclusion (incl.) and at relapse (Rel) after OAsIs treatment. Each dot represents a single cell. UMAP plots display the NIK/ NFκB2 (left panel), NFκB1 (middle panel) and BCR (right panel) signature scores at incl. and Rel. Wilcoxon sign-rank test; ∗∗∗∗Padj < .0001. (B) UMAP plot of the 7 clusters identified by Seurat at incl. and at Rel after OAsIs treatment for the patient MCL34. The cell proportion for each cluster at incl. (green) or Rel (pink) is indicated at the bottom. (C) UMAP plot displaying the cell-cycle signature scores at incl. and Rel for the patient MCL34. Cell proportion of cycling (red) or resting (gray) in each cluster is indicated at the bottom. (D) Trajectory of single cells at incl. and at Rel using Monocle2. Clusters defined in panel B are identified in the lower panel. (E) Heatmap showing the top 20 genes for each cluster. The color gradient indicates the intensity of the median gene expression as indicated. Functional enrichment pathway was calculated using Reactome and Enrichr. ∗∗∗∗Padj < .0001. UMAP, Uniform Manifold Approximation and Projection.
Unsupervised clustering of these longitudinal samples was performed to segregate the cells according to their transcriptional profile. Seven distinct clusters were identified, 2 of them being common at inclusion and relapse (clusters #4 and #5; Figure 2B). The cell-cycle signature analysis highlighted a proliferative heterogeneity among the clusters, clusters #3 and #4 brought together cycling MCL cells (44% and 85%, respectively, Figure 2C). Inference of the cell-cycle transcriptional program reassigned cycling cells to their quiescent counterpart clusters (supplemental Figure 2C), and hierarchical clustering highlighted a close transcriptional proximity between clusters #1 and #2 (incl.), as well as between clusters #6 and #7 (Rel.) (supplemental Figure 2D). Finally, single-cell trajectory analysis identified a small number of cells from the inclusion sample, which belonged to cluster #5, within the branches that contributed to the relapse, suggesting that cells belonging to this minor cluster were at the initiation of the relapse process (Figure 2D).
Functional annotation of each cluster’s transcriptional signature confirmed enrichment of the cell-cycle program (#3 and #4) and highlighted increased transcriptional and translational activity at relapse (#6 and #7). Enriched activity of the transcription factors NF-κB1 and RELA were predicted at relapse (#5 and #6/#7, P < 5 × 10–5), suggesting the central role of this transcriptional program in OAsIs resistance (Figure 2E). Altogether, we identified a minor subclone (cluster#5 187/4860 cells at inclusion) before OAsIs therapy that was resistant in vivo, which survived upon treatment (393/5345 cells at relapse) and was at the initiation of the clusters exclusively detected at relapse (#6 and #7). All relapse clusters were characterized by elevated NF-κB1/RELA activity, which was consistent with the CARD11 GOF mutation previously identified.
A 16-gene signature characteristic of OAsIs-resistant cells and associated with poorer outcome in MCL
We further focused our analysis on the NF-κB1–related transcriptional signature characteristic of the OAsIs-resistant cluster #5 (n = 742 genes, Figure 2E, supplemental Table 5) and assessed it at single-cell resolution in additional samples from 5 patients enrolled on OAsIs (supplemental Figure 3A). Unsupervised clustering highlighted intratumoral heterogeneity in all the samples, with 4 to 5 clusters identified in each case (supplemental Figure 3B), all of them displayed expression of the OAsIs targets, with the exception of CD20 loss in all the cells from the patient MCL10 with refractory disease (supplemental Figure 3C). Proliferating clusters (supplemental Figure 3D) were inferred to limit the impact of heterogeneous proliferation in further analysis (supplemental Figure 4A).
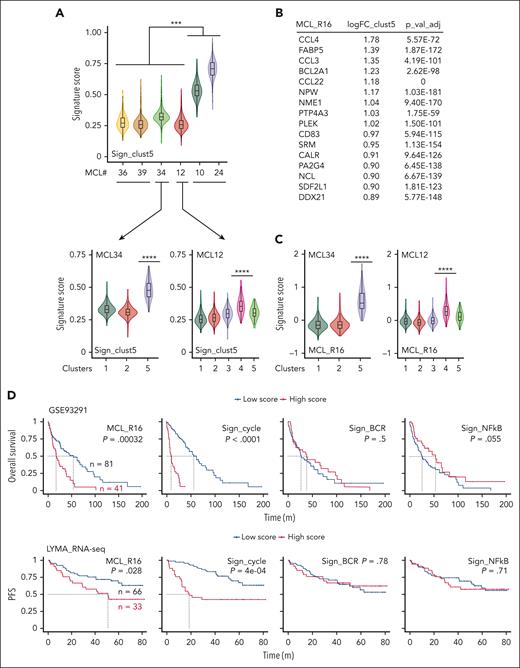
We then observed that the OAsIs-resistant cluster #5 signature was significantly more elevated in patients with refractory disease when compared with that in responders (P < .001; Figure 3A, upper panel). This signature score was homogeneously elevated across the cellular clusters in nonresponders and low for long-term responders (supplemental Figure 4B). In contrast, the score was significantly more elevated in 1 cluster from the patient who rapidly relapsed after initial PR (MCL12, cluster #4), as previously seen for MCL34 (cluster #5) (Figure 3A, lower panel).
OAsIs resistance score predicts a less favorable outcome in MCL. (A) The sign_clust5 score was computed based on the 742 genes characteristic of cluster #5 of patient MCL34, as represented in Figure 2E. The violin plot shows the sign_clust5 score for patients who achieved complete response (MCL36, MCL39), rapid relapse after the initial response (MCL34, MCL12), or progression (MCL10, MCL24). The analysis was performed at the whole sample level (upper panel) or at Seurat-computed cluster level (lower panel). Mann-Whitney test; ∗∗∗P < .001, ∗∗∗∗P < .0001. (B) Sixteen genes comprising a novel OAsIs resistance score (MCL_R16) based on the top genes of the sign_clust5 signature. (C) The MCL_R16 score was computed at Seurat-computed cluster level in MCL34 and MCL12 at the time of inclusion. Mann-Whitney test; ∗∗∗∗P < .0001. (D) Kaplan-Meier analyses of OS or PFS for the MCL_R16, cell-cycle, BCR, and NFκB signatures. Probabilities were calculated on 122 MCL patients treated with R-CHOP (GSE93291) or on 99 MCL patients included in the LYMA cohort (LYMA_RNA-seq) respectively. Blue curve: low score, red curve: high score. Log-rank test.
OAsIs resistance score predicts a less favorable outcome in MCL. (A) The sign_clust5 score was computed based on the 742 genes characteristic of cluster #5 of patient MCL34, as represented in Figure 2E. The violin plot shows the sign_clust5 score for patients who achieved complete response (MCL36, MCL39), rapid relapse after the initial response (MCL34, MCL12), or progression (MCL10, MCL24). The analysis was performed at the whole sample level (upper panel) or at Seurat-computed cluster level (lower panel). Mann-Whitney test; ∗∗∗P < .001, ∗∗∗∗P < .0001. (B) Sixteen genes comprising a novel OAsIs resistance score (MCL_R16) based on the top genes of the sign_clust5 signature. (C) The MCL_R16 score was computed at Seurat-computed cluster level in MCL34 and MCL12 at the time of inclusion. Mann-Whitney test; ∗∗∗∗P < .0001. (D) Kaplan-Meier analyses of OS or PFS for the MCL_R16, cell-cycle, BCR, and NFκB signatures. Probabilities were calculated on 122 MCL patients treated with R-CHOP (GSE93291) or on 99 MCL patients included in the LYMA cohort (LYMA_RNA-seq) respectively. Blue curve: low score, red curve: high score. Log-rank test.
We further designed the MCL_R16 signature composed of 16 top genes upregulated in OAsIs-resistant cluster #5 (MCL34, Figure 3B; supplemental Table 5). Similar to the full resistance signature, the MCL_R16 signature was significantly more elevated in clusters #5 of MCL34 and #4 of the MCL12 at inclusion, as well as after OAsIs relapse (Figure 3C; supplemental Figure 4C). The MCL_R16 signature partially overlapped and significantly correlated with previously described BCR and NF-κB1 signatures (supplemental Figure 4D and 4E). Akin to all these signatures,27 MCL_R16 was significantly higher in MCL lymph nodes when compared with PB (P < .0001) (supplemental Figure 4F), in line with the previously described increased resistance in MCL protective niches.13,28
To investigate whether this resistance profile could be extended to other regimen, we computed the MCL_R16 score in gene expression data from patients with MCL previously treated with chemotherapy (R-CHOP; n = 122). Patients having a high MCL_R16 score (upper tercile) had significantly lower overall survival (OS), the median survival time being 17.3 months for patients with a high MCL_16R score compared with 53.8 months for others (P < .001) (Figure 3D, upper panel). This observation was confirmed with an intensive regimen (Lyma-trial, n = 99), in which patients with a higher MCL_R16 score had significantly shorter progression-free survival (PFS; 51 months vs not reached; P = .028) (Figure 3D, lower panel) and OS (P = .042) (supplemental Figure 4G). Although the proliferation score was also a strong predictor of clinical response, previously described BCR and NF-κB signatures were not associated with OS or PFS (Figure 3D; supplemental Figure 4G).
BCL2A1 is frequently expressed in MCL and triggers resistance by buffering BH3-only proteins
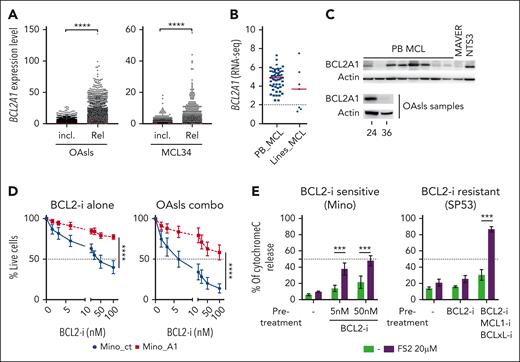
Among the top genes associated with OAsIs resistance (Figure 3B), BCL2A1, an antiapoptotic member of the Bcl2 family, has previously been described as a major venetoclax resistance factor.12,29,30 In accordance with the MCL_R16 signature pattern, the BCL2A1 RNA level was (1) significantly higher at relapse than at inclusion (P < .0001) (Figure 4A); (2) especially elevated in nonresponders (MCL10 and MCL24) (supplemental Figure 5A); and (3) enriched in a specific cluster of cells in the 2 patients who experienced rapid relapse after initial response (cluster #5 of MCL34 and #4 of MCL12) (supplemental Figure 5B). Frequent expression of BCL2A1 was observed in PB MCL samples (n = 63), whereas 3 out of 7 MCL cell lines displayed low levels (Figure 4B). This pattern of expression was further confirmed at the protein level in cell lines (supplemental Figure 5C) and primary cells, including OAsIs-resistant cells (BCL2A1high in MCL24) and OAsIs-sensitive cells (BCL2A1low in MCL36, Figure 4C).
Identification of BCL2A1 as a resistance factor to OAsIs combination. (A) scRNA-seq analysis of BCL2A1 gene expression at inclusion (incl.) or relapse (Rel.) in all OAsIs samples available (left panel) or longitudinal samples from patient MCL34. Each dot represents a cell. Wilcoxon-sign-rank test; ∗∗∗∗P < .0001. (B) Bulk RNA-seq analysis of BCL2A1 gene expression in PB MCL cells (PB, n = 63) or MCL cell lines (n = 7). (C) BCL2A1 protein expression in primary PB MCL cells (n = 8), MAVER, and NTS3 cell lines, and in OAsIs samples (MCL 24 and MCL36) was evaluated by immunoblot. (D) Survival of Mino_ct and Mino_A1 (supplemental Figure 5) treated with single-agent venetoclax BCL2-i at indicated doses or with OAsIs combination (ibrutinib, 500 nM; obinutuzumab 500 ng/mL) for 48 hours. Cell viability was measured by the lack of annexin-V staining. ANOVA test; ∗∗∗∗P < .0001. The mean and standard deviation of 3 independent experiments are represented. (E) BCL2A1 specific BH3-profiling was performed using the selective peptide FS2 (20 μM) in venetoclax-sensitive Mino cells (left panel) and venetoclax-resistant SP53 cells (right panel). Mino cells were pretreated or not with venetoclax (BCL2-i) at indicated doses. SP53 cells were pretreated or not with venetoclax (BCL2-i, 500 nM), S63845 (MCL1-i, 500 nM) and A1155463 (BCLxL-i, 500 nM) for 6 hours before cytochrome-C release measurement. ANOVA test; ∗∗∗P < .001. The mean and standard deviation of 3 independent experiments are represented. ANOVA, analysis of variance.
Identification of BCL2A1 as a resistance factor to OAsIs combination. (A) scRNA-seq analysis of BCL2A1 gene expression at inclusion (incl.) or relapse (Rel.) in all OAsIs samples available (left panel) or longitudinal samples from patient MCL34. Each dot represents a cell. Wilcoxon-sign-rank test; ∗∗∗∗P < .0001. (B) Bulk RNA-seq analysis of BCL2A1 gene expression in PB MCL cells (PB, n = 63) or MCL cell lines (n = 7). (C) BCL2A1 protein expression in primary PB MCL cells (n = 8), MAVER, and NTS3 cell lines, and in OAsIs samples (MCL 24 and MCL36) was evaluated by immunoblot. (D) Survival of Mino_ct and Mino_A1 (supplemental Figure 5) treated with single-agent venetoclax BCL2-i at indicated doses or with OAsIs combination (ibrutinib, 500 nM; obinutuzumab 500 ng/mL) for 48 hours. Cell viability was measured by the lack of annexin-V staining. ANOVA test; ∗∗∗∗P < .0001. The mean and standard deviation of 3 independent experiments are represented. (E) BCL2A1 specific BH3-profiling was performed using the selective peptide FS2 (20 μM) in venetoclax-sensitive Mino cells (left panel) and venetoclax-resistant SP53 cells (right panel). Mino cells were pretreated or not with venetoclax (BCL2-i) at indicated doses. SP53 cells were pretreated or not with venetoclax (BCL2-i, 500 nM), S63845 (MCL1-i, 500 nM) and A1155463 (BCLxL-i, 500 nM) for 6 hours before cytochrome-C release measurement. ANOVA test; ∗∗∗P < .001. The mean and standard deviation of 3 independent experiments are represented. ANOVA, analysis of variance.
To address the impact of BCL2A1 expression in the OAsIs response, we carried out further functional assays. BCL2A1 downregulation sensitized BCL2A1high SP53 and OCILY3 resistant cells to venetoclax (supplemental Figure 5D-E). Consistently, ectopic BCL2A1 expression resulted in a dramatic loss of sensitivity to selective BCL2 inhibition (LD50 venetoclax in Mino_ct = 25 nM vs Mino_A1 > 100 nM) and to the OAsIs combination (LD50 Mino_ct = 6.25 nM vs Mino_A1 > 100 nM) (Figure 4D; supplemental Figure 5F). As observed in Mino cells, the BCL2A1high MCL cells from the patient MCL10 with refractory disease were more resistant to the combination ex vivo than the MCL cells from responding patients with MCL7 and MCL12 (supplemental Figure 5G).
We then performed dynamic BH3-profiling using the BCL2A1-specific peptide FS2.31 We first observed that cytochrome-c release was not significantly increased in the presence of FS2 at baseline, suggesting that MCL cells were not primed on BCL2A1 (Figure 4E). In contrast, pretreatment with venetoclax in venetoclax-sensitive Mino cells (LD50 = 25 nM) or with a combination of BH3-mimetics in venetoclax-resistant SP53 cells (LD50 > 2500 nM), resulted in elevated priming to BCL2A1 (P < .001; 48% and 87% of cytochrome-c release, respectively; Figure 4E). These results suggest that BCL2A1 was able to capture back proapoptotic BH3-only, released upon the targeting of other antiapoptotic members of the Bcl2-family, resulting in diminished apoptosis induction (supplemental Figure 5H-I). Finally, we observed that BCL2A1 protein expression increased upon treatment with venetoclax in vitro (supplemental Figure 5J), consistent with its stabilization once it is in complex with BH3-only proteins.30 Overall, our results highlighted that elevated expression of BCL2A1 could be involved in resistance to selective BCL2-targeted monotherapy but also in combination, such as OAsIs.
BCR-dependent expression of BCL2A1 is disrupted by BTK inhibition, resulting in cell-death synergy in combination with BCL2 inhibition
We observed a strong correlation between BCL2A1 RNA level and the BCR signature in MCL cells (r = 0.5; P < .0001; Figure 5A; supplemental Figure 6A). In line with a BCR-dependent expression, BCL2A1 was induced 3 hours after IgM engagement (median increase of 5.25-fold, n = 5; P < .05; Figure 5B). This was further confirmed at the protein level (Figure 5C).
BCR-dependent expression of BCL2A1 in MCL. (A) Correlation between BCR signature and BCL2A1 gene expression in PB MCL cells (n = 63) and MCL cell lines (n = 7). Spearman r and P-value are indicated on the graph. (B) BCL2A1 expression in PB MCL cells (n = 3), and MCL cell lines (MAVER, UPN1) treated or not for 3 hours with anti-IgM (αIgM, 10 μg/mL) was measured by RT-qPCR. Paired t test; ∗P < .05. (C) BCL2A1 protein expression in MCL primary cells (n = 6) and NTS3 cell line treated or not for 3 hours with anti-IgM (αIgM, 10 μg/mL) was assessed by immunoblot. (D) CD83, BCL2A1, BCL2, MCL1 and BCLXL gene expression in BCL2A1 positive MCL cell lines (SP53, NTS3, REC-1, Mino) treated for 24 hours with ibrutinib (500 nM) was evaluated by bulk RNA-sequencing. ANOVA test; ∗∗P < .01, ∗P < .05. (E) Synergy score (highest single-agent [HSA] method) computed for 6 MCL cell lines treated with venetoclax and ibrutinib for 48 hours. Unpaired t test; ∗P < .05. Detailed results are represented in supplemental Figure 6C. The mean and standard deviation of 3 independent experiments are represented.
BCR-dependent expression of BCL2A1 in MCL. (A) Correlation between BCR signature and BCL2A1 gene expression in PB MCL cells (n = 63) and MCL cell lines (n = 7). Spearman r and P-value are indicated on the graph. (B) BCL2A1 expression in PB MCL cells (n = 3), and MCL cell lines (MAVER, UPN1) treated or not for 3 hours with anti-IgM (αIgM, 10 μg/mL) was measured by RT-qPCR. Paired t test; ∗P < .05. (C) BCL2A1 protein expression in MCL primary cells (n = 6) and NTS3 cell line treated or not for 3 hours with anti-IgM (αIgM, 10 μg/mL) was assessed by immunoblot. (D) CD83, BCL2A1, BCL2, MCL1 and BCLXL gene expression in BCL2A1 positive MCL cell lines (SP53, NTS3, REC-1, Mino) treated for 24 hours with ibrutinib (500 nM) was evaluated by bulk RNA-sequencing. ANOVA test; ∗∗P < .01, ∗P < .05. (E) Synergy score (highest single-agent [HSA] method) computed for 6 MCL cell lines treated with venetoclax and ibrutinib for 48 hours. Unpaired t test; ∗P < .05. Detailed results are represented in supplemental Figure 6C. The mean and standard deviation of 3 independent experiments are represented.
Accordingly, BCL2A1 was inhibited by ibrutinib in all BCL2A1+ cell lines (median reduction, 34%, n = 4; P < .05), in a manner similar to the sensitive BCR activation marker CD8332 but unlike other antiapoptotic Bcl2 family proteins (Figure 5D). Consequently, ibrutinib reduced the BCL2A1 protein level (supplemental Figure 6B), leading to synergistic apoptosis when associated with venetoclax in BCL2A1+ but not in BCL2A1− cells (mean synergy score, 17 vs 1) (Figure 5E; supplemental Figure 6C). Overall, we identified that (i) the antiapoptotic protein BCL2A1 is a BCR-dependent resistance factor to Bcl2 inhibition; (ii) BCL2A1 is selectively downregulated through BTK inhibition, leading to ibrutinib and venetoclax synergistic apoptosis.
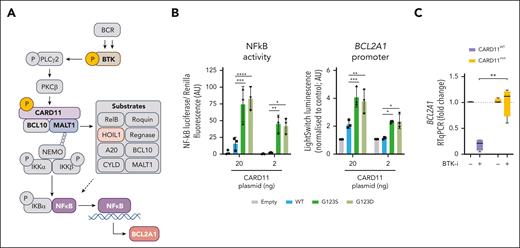
CARD11 GOF mutations result in BCR-independent BCL2A1 overexpression
The CARD11-BCL10-MALT1 (CBM) complex is involved in the transduction of BCR-specific NF-κB signaling by triggering MALT1 scaffold and protease activities (Figure 6A).33 We first observed constitutive CBM activity, through the cleavage of the MALT1 substrate HOIL1, in BCL2A1high MCL cells but not in BCL2A1low MCL cells (supplemental Figure 7A). This suggested that GOF mutations of CARD11, which are enriched in patients who have relapsed after treatment with OAsIs, could have a direct impact on BCL2A1 expression. To test this hypothesis, we overexpressed CARD11WT, CARD11G123S, and CARD11G123D in HEK293 cells, which lack CARD11 but express both BCL10 and MALT1. Both mutations resulted in increased activity of the CBM, measured by the cleavage of HOIL1 and CYLD (supplemental Figure 7B). Using reporter assay experiments, we showed that CARD11 GOF mutations led directly to increased activity of NF-κB (fold change in CARD11MUT = 4.2; P < .001) as well as BCL2A1 promoter activity (fold change in CARD11MUT = 2.7; P < .001; Figure 6B). These results suggest that GOF mutations in the latch domain of CARD11 lead directly to BCR-independent induction of NF-κB/BCL2A1. Similar findings were obtained with GOF mutations in the CC domain (fold change in CARD11MUT = 1.7; P < .05; supplemental Figure 7C). Accordingly, ibrutinib greatly reduced BCL2A1 level in CARD11WT (fold change = 0.21, n = 4) but not in CARD11MUT (fold change = 1.12, n = 4) lymphoma cells (Figure 6C).
BCR-independent expression of BCL2A1 in CARD11 mutated cells. (A) BCR signaling triggers the NF-κB1 pathway through the activation of several proteins such as BTK, which can be inhibited by ibrutinib, and complexes such as CBM. Several previously described CBM substrates are mentioned including HOIL1, which is used in the following experiments as a marker of CBM activity. (B) NF-κB luciferase assay (left panel) and BCL2A1 promoter LightSwitch luciferase assay (right panel) in HEK293 cells transfected with a pUNO1 (empty), pUNO1-hCARD11-WT, pUNO1-hCARD11-G123S or pUNO1-hCARD11-G123D vector (48 hours). Two-way ANOVA; ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, ∗P < .05. The mean and standard deviation of 3 independent experiments are represented. (C) BCL2A1 gene expression was measured by RT-qPCR in CARD11WT MCL (SP53, NTS3, and REC-1) and DLBCL (U2932) cell lines and CARD11MUT MCL cells (001-024, DFBL) and DLBCL (OCILY3, SUDHL16) cell lines treated with ibrutinib (500 nM) for 6 hours. Detailed CARD11 mutational status is described in supplemental Table 4. Mann-Whitney test; ∗P < .05.
BCR-independent expression of BCL2A1 in CARD11 mutated cells. (A) BCR signaling triggers the NF-κB1 pathway through the activation of several proteins such as BTK, which can be inhibited by ibrutinib, and complexes such as CBM. Several previously described CBM substrates are mentioned including HOIL1, which is used in the following experiments as a marker of CBM activity. (B) NF-κB luciferase assay (left panel) and BCL2A1 promoter LightSwitch luciferase assay (right panel) in HEK293 cells transfected with a pUNO1 (empty), pUNO1-hCARD11-WT, pUNO1-hCARD11-G123S or pUNO1-hCARD11-G123D vector (48 hours). Two-way ANOVA; ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, ∗P < .05. The mean and standard deviation of 3 independent experiments are represented. (C) BCL2A1 gene expression was measured by RT-qPCR in CARD11WT MCL (SP53, NTS3, and REC-1) and DLBCL (U2932) cell lines and CARD11MUT MCL cells (001-024, DFBL) and DLBCL (OCILY3, SUDHL16) cell lines treated with ibrutinib (500 nM) for 6 hours. Detailed CARD11 mutational status is described in supplemental Table 4. Mann-Whitney test; ∗P < .05.
MALT1 targeting results in BCL2A1 inhibition and consequent synergistic cell death with BCL2 targeting, irrespective of CARD11 mutational status
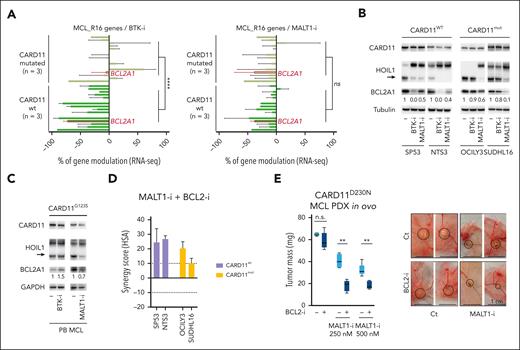
MALT1, in contrast to CARD11 and BCL10, is significantly upregulated in MCL cells when compared with normal B cells (supplemental Figure 7D), and selective inhibitors against its protease activity are currently being tested in early-phase trials. To address the potential efficacy of MALT1 inhibition for counteracting both BCR-dependent (CARD11WT) and independent (CARD11MUT) BCL2A1 expression, we tested 2 highly selective MALT1 allosteric inhibitors, MLT-74834 and JNJ-67856633.35 Using reporter assay experiments, we first observed that MALT1 inhibition resulted in the inhibition of CARD11MUT-dependent NF-κB activity (fold change = 0.59 for G123S, 0.56 for D230N; P < .05; supplemental Figure 7C). Likewise, we observed that both inhibitors partially counteracted CARD11G123S-dependent BCL2A1 promoter activity (fold inhibition MALT1-i vs untreated = 0.59 for G123S; supplemental Figure 7E). Based on these encouraging results, we further performed RNA-seq of CARD11WT and CARD11MUT lymphoma cells treated with the BTK inhibitor ibrutinib or the MALT1 inhibitor MLT-748. BTK targeting resulted in the inhibition of genes belonging to the MCL_R16 signature, including BCL2A1, in CARD11WT but not in CARD11MUT cells (P < .0001; Figure 7A left panel). In contrast, MALT1 inhibition similarly lowered gene expression in both groups including BCL2A1, which was downregulated irrespective of CARD11 status (Figure 7A, right panel). This was further confirmed by quantitative polymerase chain reaction in CARD11WT (fold change = 0.54, n = 3) and CARD11MUT cells (fold change = 0.51, n = 3) and similar results were obtained using MLT-748 and JNJ-67856633 (supplemental Figures 8A-B).
Identification of MALT1 as a target to overcome OAsIs resistance. (A) RNA-seq expression of the 16 genes of the MCL_R16 resistance signature in BCL2A1+CARD11WT cell lines (SP53, NTS3, REC-1) and BCL2A1+CARD11MUT cells (001-024, OCILY3, SUDHL16) treated 24 hours with the BTK inhibitor ibrutinib (500 nM) (left panel) or the MALT1 inhibitor MLT-748 (1 μM) (right panel). Mann-Whitney test; ∗∗∗∗P < .0001. BCL2A1 is shown in red. Genes from bottom to top: CCL4, FABP5, CCL3, BCL2A1, CCL22, NPW, NME1, PTP4A3, PLEK, CD83, SRM, CALR, PA2G4, NCL, SDF2L1, DDX21. (B-C) CARD11, HOIL1, and BCL2A1 protein expression were assessed by immunoblot in indicated cells treated or not with the BTK inhibitor ibrutinib (500 nM) or the MALT1 inhibitor MLT-748 (1 μM) for 24 hours. The arrow highlights cleaved form of HOIL1. (D) Synergy score (HSA method) computed for 4 cell lines treated with venetoclax and MLT-748 for 48 hours. Unpaired t-test; ∗P < .05. Detailed results are represented in supplemental Figure 8C. The mean and SD of 3 independent experiments are represented. (E) Tumor weights at day 16 for CARD11D230N MCL PDX (DFBL44685-v2) treated with the BCL2-i venetoclax (2 nM), the MALT1-i JNJ-67856633 (250-500 nM) or both. n = 5 per group. Mann-Whitney test; ∗∗P < .01. Representative pictures of engrafted tumors in CAM at day 16 are shown in the right panel. n.s., not significant.
Identification of MALT1 as a target to overcome OAsIs resistance. (A) RNA-seq expression of the 16 genes of the MCL_R16 resistance signature in BCL2A1+CARD11WT cell lines (SP53, NTS3, REC-1) and BCL2A1+CARD11MUT cells (001-024, OCILY3, SUDHL16) treated 24 hours with the BTK inhibitor ibrutinib (500 nM) (left panel) or the MALT1 inhibitor MLT-748 (1 μM) (right panel). Mann-Whitney test; ∗∗∗∗P < .0001. BCL2A1 is shown in red. Genes from bottom to top: CCL4, FABP5, CCL3, BCL2A1, CCL22, NPW, NME1, PTP4A3, PLEK, CD83, SRM, CALR, PA2G4, NCL, SDF2L1, DDX21. (B-C) CARD11, HOIL1, and BCL2A1 protein expression were assessed by immunoblot in indicated cells treated or not with the BTK inhibitor ibrutinib (500 nM) or the MALT1 inhibitor MLT-748 (1 μM) for 24 hours. The arrow highlights cleaved form of HOIL1. (D) Synergy score (HSA method) computed for 4 cell lines treated with venetoclax and MLT-748 for 48 hours. Unpaired t-test; ∗P < .05. Detailed results are represented in supplemental Figure 8C. The mean and SD of 3 independent experiments are represented. (E) Tumor weights at day 16 for CARD11D230N MCL PDX (DFBL44685-v2) treated with the BCL2-i venetoclax (2 nM), the MALT1-i JNJ-67856633 (250-500 nM) or both. n = 5 per group. Mann-Whitney test; ∗∗P < .01. Representative pictures of engrafted tumors in CAM at day 16 are shown in the right panel. n.s., not significant.
MALT1 targeting resulted partial BCL2A1 protein downregulation in all samples, including CARD11G123S MCL cells, in contrast to BTK targeting whose efficacy was restricted to CARD11wt cells (Figure 7B-C). Supporting the role of BCL2A1 in venetoclax resistance, we observed synergistic apoptosis after a MALT1/BCL2 targeted combination in vitro in both BCL2A1+CARD11WT and BCL2A1+CARD11MUT (synergy score>10) (Figure 7D). As expected, synergy was not observed in BCL2A1– Z138 cells (synergy score, –4.7) (supplemental Figure 8C).
To address MALT1/BCL2 dual targeting in CARD11MUT MCL cells in vivo, we used the chick embryo CAM. This vascularized and immunodeficient alternative in vivo model allows the growth of numerous cell types.36 In this study, we established a CAM model using BTK-resistant37CARD11D230NBCL2A1+ patient-derived xenografts cells, which were treated with the clinically available MALT1 inhibitor JNJ-67856633 alone or in combination with the BCL2 inhibitor venetoclax (Figure 7E; supplemental Figure 9A-C). Although low doses of venetoclax showed no significant inhibition of tumor growth, JNJ-67856633 reduced tumor growth to 40% of control. The combination highlighted a strong synergy leading to 70% of tumor growth inhibition after 4 days of treatment (Figure 7E). These results were confirmed in NSG mice, the CARD11D230NBCL2A1+ tumor burden (PB and spleen) being reduced using a combination of JNJ-67856633 and venetoclax, both in suboptimal concentrations as a single agent (supplemental Figure 9D-F).
Discussion
Based on single-cell resolution analysis of longitudinal samples from a patient enrolled on the OAsIs trial, we identified, at baseline, a minor cellular population that persisted upon treatment in vivo and ultimately gave rise to relapse. The transcriptomic signature of this minor population was enriched in genes that are controlled by NF-κB1, highlighting the fact that a BTK-independent NF-κB program was involved in the resistance to the OAsIs regimen in vivo. The resistance signature was the basis for a 16-gene score, which also retrospectively predicted lower OS/PFS in patients treated with chemotherapy. Overall, our results suggested overlapping resistance mechanisms for both chemotherapy and targeted therapies in MCL. A subset of patients with multiresistant MCL, whose disease progressed on chemotherapy, BTK/BCL2-based targeted therapy and chimeric antigen receptor T-cell therapy cells, is emerging, however little is known about the (poly)resistance mechanisms involved.38 It would now be interesting to examine whether tumoral cells from these patients with MCL who are at very high-risk display a similar NF-κB1–related resistance profile and whether alternative strategies, such as bispecific antibodies,39,40 can counteract this resistance.
BTK-independent NF-κB activation can be initiated in malignant B cells using multiple pathways which have been implicated in BTK-inhibition resistance.16,41 This signaling is triggered by genetic events,15,26 epigenetic regulation,42 and microenvironmental cues.13,20 In this study, we have observed that CARD11 GOF mutations were enriched at OAsIs relapse and resulted in increased NF-κB activity. CARD11 GOF mutations are observed in ∼10% of non-Hodgkin lymphomas at diagnosis,43 and allow cell survival upon BTK inhibition by restoring NF-κB signaling downstream. Accordingly, CARD11 GOF mutations have been associated with in vivo ibrutinib resistance in diffuse large B-cell lymphoma (DLBCL)25 and follicular lymphoma (FL).44 In this study, we have demonstrated that a similar pattern is found in MCL. Intriguingly, we have observed enrichment of CARD11 mutations within the latch domain (supplemental Table 4). This pattern was confirmed in a recent analysis of recurrently mutated genes in several B-cell non-Hodgkin lymphoma subtypes at diagnosis, in which CARD11 mutants were enriched in the G123 (3/11) and D230 (3/11) positions in MCL, but not in FL, DLBCL, or Burkitt lymphoma.43 In this study, we have demonstrated that both variants induced elevated NF-κB activity in vitro, and a recent multiplex functional assessment of genetic variants ranked the G123S as one of the most efficient in escaping ibrutinib treatment.45 In addition, elevated NF-κB activation associated with CARD11 germ line mutation at the G123 position has been described in the context of B-cell expansion with NF-κB and T-cell anergy (BENTA) disease,21 reinforcing the potent GOF associated with this hotspot.
By integrating multiomics and functional analysis, we have demonstrated that GOF mutations result in NF-κB–dependent induction of BCL2A1. Recent publications have highlighted its critical role in venetoclax resistance in hematological malignancies.12,29,46 Our BH3-profiling results suggest the dynamic sequestration of the BH3-only proteins by BCL2A1 at the mitochondrial level after targeting BCL2, MCL1, and BCLXL, as recently described in chronic lymphoid leukemia.30 Accordingly, overexpression of BCL2A1 led to increased resistance to the OAsIs combination, highlighting the need to target this antiapoptotic protein to counteract resistance. No selective small molecule has yet been developed to specifically target BCL2A1, but in this study we have demonstrated that BCL2A1 is tightly controlled by the BTK/BCR pathway in CARD11wt MCL. Altogether, our results position BCL2A1 as a selective bridge between the BCR pathway and mitochondrial apoptosis. Accordingly, BCL2A1 expression is greatly inhibited by ibrutinib in CARD11WT cells, but not in CARD11MUT cells, which results in ibrutinib/venetoclax synergy exclusively in CARD11WT BCL2A1+ lymphoma cells.
CARD11 forms the CBM complex with MALT1, which has emerged as an attractive therapeutic target, and several molecules that target its protease function are being tested in vitro and in vivo. 47,48,49 Moreover, recent studies have demonstrated that MCL cells are addicted to MALT1, at least partially inherited from their B1a B-cell origin.50,51 We have demonstrated that inhibition of the MALT1 protease function resulted in the inhibition of the MCL_R16 signature, including BCL2A1 and synergized with BCL2 inhibition to induce apoptosis, irrespective of the CARD11 mutational status. Overall, our results have highlighted that MALT1 targeting could be an interesting alternative to BTK inhibitors, especially in the context of CARD11MUT cells, although MALT1 inhibitors lead to a less profound inhibition of BCL2A1 compared with BTK inhibition in CARD11WT cells. Several strategies for inhibiting both MALT1 scaffold and protease functions, such as the PROTAC technology that leads to selective protein degradation,52 are under preclinical evaluation and may lead to increased efficacy of MALT1 targeting in the near future.
To summarize, our data have uncovered the control of mitochondrial apoptosis by CARD11 through BCL2A1 expression. This network is crucially involved in the resistance to BTK/BCL2-based targeted inhibition, because CARD11 GOF mutations lead not only to BCR independence and consequently ibrutinib resistance, but also to BCL2A1-mediated venetoclax resistance. Nevertheless, our data have shown that targeting the CBM through MALT1 inhibition reverts this resistance, and MALT1 targeting appears to be a promising therapeutic option for counteracting resistance, especially in the context of CARD11 mutation. Mutations in CARD11 are frequently found in DLBCL, for which there is a growing interest in BTK/BCL2-based targeted therapy.53 Our results, which describe the role of the CARD11-NF-κB-BCL2A1 axis in MCL resistance in vivo, could be applied to a broad spectrum of hematological malignancies.
Acknowledgments
The authors thank the patients who agreed to be part of the Refract-Lyma cohort. The authors thank la Ligue Contre le Cancer Grand-Ouest, i-Site NexT (ANR-16-IDEX-0007), the SIRIC ILIAD (INCa-DGOS-Inserm-ITMO Cancer_12558 and INCa-DGOS-Inserm-ITMO Cancer_18011) and Action Cancer 44. The authors thank L. Gangoda and M.J. Herold for kindly providing the Bfl1-specific monoclonal antibody and Andrew L. Snow for kindly providing the pUNO1-hCARD11 plasmids. The authors thank Magali Devic for expert technical assistance. The authors are most grateful to the Genomics Core Facility GenoA, member of Biogenouest and France Genomique and to the Bioinformatics Core Facility BiRD, member of Biogenouest and Institut Français de Bioinformatique (ANR-11-INBS-0013) for the use of their resources and their technical support. The authors acknowledge the Cytocell-Flow Cytometry and fluorescence-activated cell sorting core facility (SFR Bonamy, BioCore, INSERM UMS 016, CNRS UAR 3556, Nantes, France) for its technical expertise and help, member of the Scientific Interest Group Biogenouest and the Labex IGO program supported by the French National Research Agency (n°ANR-11-LABX-0016-01).
Authorship
Contribution: S.D. designed and performed the experiments and analyzed data; C.B. performed the experiments and bioinformatics analysis and analyzed data; Y.L.B., C.M., D.B., C.D., A.M., E.D., and A.M.-A. performed experiments and analyzed data; J.J. and N.B. performed NF-κB and BCL2A1 luciferase assays and analyzed data; J.C.S. and G.R. performed experiments on CAM model and analyzed data; C.K. performed experiments on NSG mice and analyzed data; P.M., S.M., B.T., and S.L.G. participated in the design of the study; C.P.-D. participated in the design of the study, in the data analysis, and in writing the article; and D.C. designed the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David Chiron, Nantes Université, INSERM, CNRS, Université d'Angers, CRCI2NA, 8 quai Moncousu, 44007 Nantes, France; e-mail: david.chiron@univ-nantes.fr.
References
Author notes
∗S.D. and C.B. contributed equally to this study.
RNA-sequencing datasets are publicly available in the Gene Expression Omnibus under accession numbers GSE239497 and GSE239353. All other datasets analyzed during the current study are available on reasonable request from the corresponding author, David Chiron (david.chiron@univ-nantes.fr).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![Comprehensive genomic analysis of MCL patients enrolled in the OAsIs trial. (A) Schematic representation of the experimental design to identify therapeutic resistance in MCL. Patients with MCL were included in the OAsIs clinical trial (obinutuzumab [anti-CD20], ibrutinib [BTK-i], and venetoclax [BCL2-i]). Blood or BM samples were collected at inclusion (incl.) or relapse (Rel) and targeted DNA-seq of 49 selected genes plus the 9p21 region was performed. (B-D) Landscape of recurrent mutations (SNV/indel) and copy number variations (gain/deletion) in the patients enrolled in OAsIs and detected by deep DNA-seq at incl. (n = 12) (B), Rel (n = 5) (C), or in longitudinal samples (D). Each column represents an individual sample/patient. Only genes with at least 2 anomalies detected in the cohort and with a variant frequency >5% are represented. Statistical significance was determined by a 2-tailed Fisher exact test. (E) The fish plot shows the clonal evolution pattern of patient 34 based on the variant frequency (VaF) measured at diagnosis (Diag), incl., and Rel (supplemental Table 2) and normalized to the TP53Y220C VaF. (F) Schematic representation of CARD11 domains and the mutations found in the Refract-Lyma cohort (n = 62 sequenced for CARD11), including patients enrolled in the OAsIs trial. CR, complete response; PD, progression disease; PR, partial response; SNV, single nucleotide variant; SD, stable disease.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/142/18/10.1182_blood.2023020211/2/m_blood_bld-2023-020211-gr1.jpeg?Expires=1771045547&Signature=D4LbdRVUUIw3Ntm4OYl5TdrWp0VH-2pvKZPDDZSGGtFtJFNLN-bVRmeG0MzDY9PA1cyr~V1dJR8ZNjPokGrul44V0jHqPNizDkn0TxazrsGDOcBpmfKxmvF9QbBg9jXVGcpygarsqFpX82JjXtbrKfoEDOFuIhjsWSy3T7L~dQWPkf3V64FlDBBTPFEMmMzLoq2y8E4sAy-NX4IVKtsp4FXgXC6TsoPODBE1W19dLKMOJMqji3061jJKB60tT6qtCqmQ7710uZfkWeQ0GZFFV5LilBNueMfYBkQvuktdYNNzbSmJ9cwM4Ig~Vl076sPiWxOWFAjxxk1wlFTAnw~3TA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![BCR-dependent expression of BCL2A1 in MCL. (A) Correlation between BCR signature and BCL2A1 gene expression in PB MCL cells (n = 63) and MCL cell lines (n = 7). Spearman r and P-value are indicated on the graph. (B) BCL2A1 expression in PB MCL cells (n = 3), and MCL cell lines (MAVER, UPN1) treated or not for 3 hours with anti-IgM (αIgM, 10 μg/mL) was measured by RT-qPCR. Paired t test; ∗P < .05. (C) BCL2A1 protein expression in MCL primary cells (n = 6) and NTS3 cell line treated or not for 3 hours with anti-IgM (αIgM, 10 μg/mL) was assessed by immunoblot. (D) CD83, BCL2A1, BCL2, MCL1 and BCLXL gene expression in BCL2A1 positive MCL cell lines (SP53, NTS3, REC-1, Mino) treated for 24 hours with ibrutinib (500 nM) was evaluated by bulk RNA-sequencing. ANOVA test; ∗∗P < .01, ∗P < .05. (E) Synergy score (highest single-agent [HSA] method) computed for 6 MCL cell lines treated with venetoclax and ibrutinib for 48 hours. Unpaired t test; ∗P < .05. Detailed results are represented in supplemental Figure 6C. The mean and standard deviation of 3 independent experiments are represented.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/142/18/10.1182_blood.2023020211/2/m_blood_bld-2023-020211-gr5.jpeg?Expires=1771045547&Signature=fZ9GtZRVSj0gD2rsNZENoqI1gvjrKj~LDSeaPaTf37oB7cg8dGX5QwD0Kv86Q7yg4FkCAMQfwwTjkOQU30yUCeQjeEdawthhgADNjjOdjMVP-Idpw1fjQWX8iubwIciQfrExLWPfWWkIoDoiw9Rliq5u~-BTeSDYyLpcsmQvBfqoi2iagEZA7NnGZ-thRWsdgvPHFL5RjdNCmI1EI-1vmIgSIlgxeA6haxMpNiyWhT7w7fsSr5hsbqxUtGmuPoMKZh-OdQhGkaFeiWN~71oh-ICzqbk3qqdV5a76xrGYTN9HESG3G3MuE6fE-AIc6PNrrTkR9OmdDKe-Lc-g9tPsVA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal