Key Points
STAT3 mutations and Vδ2 status are needed to properly stratify Tγδ LGLL patients.
Independently from STAT3 mutations, Tγδ LGLL represents a subset of T-LGLL characterized by dismal outcome as compared with Tαβ LGLL.
Abstract
Tγδ large granular lymphocyte leukemia (LGLL) is a rare variant of T-cell LGLL (T-LGLL) that has been less investigated as compared with the more frequent Tαβ LGLL, particularly in terms of frequency of STAT3 and STAT5b mutations. In this study, we characterized the clinical and biological features of 137 patients affected by Tγδ LGLL; data were retrospectively collected from 1997 to 2020 at 8 referral centers. Neutropenia and anemia were the most relevant clinical features, being present in 54.2% and 49.6% of cases, respectively, including severe neutropenia and anemia in ∼20% of cases each. Among the various treatments, cyclosporine A was shown to provide the best response rates. DNA samples of 97 and 94 cases were available for STAT3 and STAT5b mutation analysis, with 38.1% and 4.2% of cases being mutated, respectively. Clinical and biological features of our series of Tγδ cases were also compared with a recently published Tαβ cohort including 129 cases. Though no differences in STAT3 and STAT5b mutational frequency were found, Tγδ cases more frequently presented with neutropenia (P = .0161), anemia (P < .0001), severe anemia (P = .0065), and thrombocytopenia (P = .0187). Moreover, Vδ2− cases displayed higher frequency of symptomatic disease. Overall, Tγδ cases displayed reduced survival with respect to Tαβ cases (P = .0017). Although there was no difference in STAT3 mutation frequency, our results showed that Tγδ LGLL represents a subset of T-LGLL characterized by more frequent symptoms and reduced survival as compared with Tαβ LGLL.
Introduction
Large granular lymphocyte leukemia (LGLL) is a rare and heterogenous chronic lymphoproliferative disorder characterized by the clonal expansion of large granular lymphocytes (LGLs).1,2 The etiology of LGLL is unknown, but a constitutive activation of JAK/STAT pathway is involved in the pathogenesis of LGL proliferation,3 further supported by the discovery of somatic STAT3 and STAT5b mutations in ∼40% of patients.4-9 Among LGLLs, the latest World Health Organization classification recognizes a CD3+ T-cell LGLL (T-LGLL) and CD3– natural killer (NK)-LGLL, accounting for 85% and 15% of cases, respectively. Moreover, based on surface T-cell receptor expression, Tαβ and Tγδ subsets of LGLL can be identified.10
Although LGLL incidence ranges between 0.2 and 0.72 cases per 1 million individuals per year,1 the frequency of Tγδ proliferation is still not well defined, and most information has been collected through small retrospective studies. As compared with the more frequent Tαβ LGLL, Tγδ LGLL has been less investigated. First reported by Oshimi et al in 1988 in a 60-year-old woman exposed to the radiation in Nagasaki in 1945,11 Tγδ LGLL has been described in a sizable number of patients in 2006 by Sandberg et al, who reported an immunophenotypical analysis of 44 cases.12 Up to now, only 4 retrospective studies including more than 200 LGLL patients are available7,13-15; however, few cases of Tγδ LGLL were included and only in the Italian cohort.7 Consequently, the clinical features of Tγδ LGLL and information on the efficacy of treatments in this LGLL variant are still missing. Furthermore, data on the frequencies of STAT3 and STAT5b mutations are nowadays available for Tαβ LGLL, but still limited and controversial for Tγδ LGLL. The Italian group recently reported 25% and 19% of Tγδ cases mutated in STAT3 and STAT5b genes, respectively,7 and STAT3 mutations were found in all patients included in a small Japanese Tγδ LGLL cohort.16
With this as a background and lacking large cohorts of Tγδ patients, major referral groups dealing with LGLL were invited to join this collaborative study aimed at better characterizing Tγδ LGLL patients, pointing to the evaluation of putative correlations among mutations, phenotype, and clinical presentation, and the comparison of the clinical behavior of Tγδ LGLL with respect to the more common Tαβ variant. This large series of cases for the first time shows the dismal outcome of Tγδ LGLL with respect to Tαβ LGLL.
Methods
Study patients
The study cohort included 137 patients affected by Tγδ LGLL who were followed from 1997 to 2020 at 8 referral centers across the world (France, Italy, Japan, Spain, United States). All patients met the currently approved World Health Organization diagnostic criteria for T-LGLL.2,17 T-LGL clonality was assessed by TCRγ gene rearrangement.
Demographic and clinical features, including presence of cytopenias, concomitant autoimmune/inflammatory diseases, secondary primary malignancies (SPMs), treatment requirement, and response, were collected. Response to treatment was evaluated based on periodical clinical and laboratory examinations after at least 4 to 6 months of therapy, using the currently accepted response criteria for LGLL.18 The frequency of LGLs positive for the characteristic antigens was assessed by flow cytometry using direct immunofluorescence assays combining up to 6 markers per tube, according to standard operating procedures of individual centers. The investigation for LGL surface markers was performed on whole peripheral blood anticoagulated with EDTA or anticoagulant citrate dextrose and on purified peripheral blood mononuclear cells. The commercially available fluorescein isothiocyanate-conjugated; phycoerythrin (PE)-, PE-Cy5-, and PE-Cy7-conjugated; and allophycocyanin- and allophycocyanin-Cy7–conjugated mouse monoclonal antibodies used included anti-CD3, anti-CD4, anti-CD8, anti-CD16, anti-CD56 and anti-CD57, anti-TCRγδ, anti-KIRs (killer immunoglobulin-like receptors: CD158a, CD158b, CD158e), anti-NKG2A, anti-NKG2C, anti-Vγ9, anti-Vδ1, and anti-Vδ2 from Becton Dickinson (Sunnyvale, CA).
This international Tγδ LGL leukemia cohort was compared with a recently reported equal-size Italian Tαβ LGL leukemia cohort.7
This study was performed according to the Helsinki Declaration, and patients gave their written informed consent prior to inclusion in the study. The protocol and informed consent form were approved by the Padua ethics committee (approval number 4213/AO/17).
Screening for STAT3 and STAT5b mutations
STAT3 and STAT5b sequencing was performed by Sanger Sequencing or Next Generation Sequencing according to local practice. For the screening of STAT3 and STAT5b mutations by Sanger Sequencing, we used the set of primers reported by Koskela et al4 and by Rajala et al,8 respectively, to amplify the hot spot regions for mutations (exons 19-21 for STAT3 and exons 16-18 for STAT5b).
Statistical analysis
Patients’ demographic, clinical, and biological features expressed as categorical variables were compared using the Fisher exact test. Patient overall survival (OS) was calculated from the date of diagnosis to death by any cause or the last-known follow-up visit for censored patients. Survival curves were estimated using the Kaplan-Meier method and compared with respect to the patients’ demographic and clinical characteristics using the log-rank test. Schoenfeld residual testing was applied to assess the proportional hazards assumption. A univariate Cox proportional hazards regression analysis was employed to evaluate the prognostic relevance of each variable. Results for significant variables were presented as hazard ratios (HRs) and 95% confidence intervals (CIs).
To determine the effect of response to first-line treatment on progression-free survival (PFS) and OS, we performed a 6-month landmark analysis in treated patients categorized by their response status (at least partial response vs stable disease or progressive disease) at 6 months after the start of therapy. The 6-month landmark time was selected a priori, before the beginning of data analysis, since at least 4 to 6 months of treatment are recommended before correctly assessing the response. For landmark analyses, PFS and OS were recalculated by shifting the time origin to 6 months after the start of therapy, and patients who experienced the event of progression or death before this time were excluded from the PFS or OS landmark analyses, respectively.
A restricted mean survival time (RMST) analysis was also performed to compare the Tγδ and Tαβ LGLL cohorts. RMST is a robust and clinically interpretable summary measure of the survival time distribution, estimable even under heavy censoring and when the proportional hazards assumption is not satisfied, as an alternative to the HR approach.19,20 This analysis depends on the truncation time point fixed for the RMST calculation. Four different truncation time points (100, 120, 140, and 160 months) were evaluated for the comparison of Tγδ and Tαβ LGLL cohorts. P values < .05 were considered significant. Statistical analysis was conducted using R version 3.6.2.
Results
Clinical and immunophenotypic features of Tγδ LGLL patients
Clinical and biological features of cases under study are summarized in supplemental Table 1, available on the Blood website. Median age at diagnosis was 58.5 years (range, 18-92), with 29.4% of subjects being >65 years old. No relevant gender prevalence was clearly demonstrated (male 55.9%, female 44.1%). By immunophenotype, all cases showed an expansion of CD3+ TCRγδ+ T cells, demonstrated to be clonal on molecular grounds. Tγδ LGLs usually displayed CD8 positivity (64/105, 61.0%), with 23 of 105 (21.9%) cases showing partial CD8 expression; otherwise, CD4 was mostly absent, with only 3 cases showing partial expression. CD16 and CD57 were typical LGL markers, and they were expressed on the expanded Tγδ cells at the highest frequency (72.3% and 78.4%, respectively); CD56 was present in 31.1% of cases. A dominant KIR expression was demonstrated in 23 of 56 cases (41.1%), with CD158b being the most frequently expressed marker (13/56, 23.2%), followed by CD158a (8/56, 14.3%) and CD158e (5/56, 8.9%). CD94/NKG2 receptor expression was found in 32 of 75 cases (42.7%), with 12 cases displaying NKG2A (12/54, 22.2%) and 3 cases showing NKG2C positivity (3/30, 10%).
Not being part of the workup for the diagnosis of LGLL, bone marrow evaluation, either by flow cytometry or immunohistochemistry, was available for only 40 of 137 (29.2%) cases, showing variable degree of infiltration with a range from less than 1% to 60% of bone marrow cellularity.
Neutropenia (absolute neutrophil count [ANC] < 1500/mm3) and mild anemia (hemoglobin [Hb] < 120g/L) were the main relevant clinical features of the entire cohort, being present in 54.2% (65/120) and 49.6% (59/119) of cases, respectively. Severe neutropenia (ANC < 500/mm3) and severe anemia (Hb < 90 g/L) were observed in 25 of 120 cases (20.8%) and in 25 of 119 cases (21%), respectively. Thrombocytopenia (platelets [PLTs] < 100 000/mm3) and splenomegaly were detected in 18 of 119 (15.1%) and in 31 of 122 (21.4%) cases, respectively. Forty-nine cases (41.5%) were affected by concurrent autoimmune/inflammatory diseases, mostly rheumatoid arthritis (16/49), autoimmune hemolytic anemia (5/49), and pure red cell aplasia (PRCA) (5/49). Finally, SPMs were detected in 17 of 84 cases (20.2%), either at the time of diagnosis or during the follow-up. Seven SPMs were hematological (3 marginal zone lymphoma, 1 chronic lymphocytic leukemia, 1 myelodysplastic syndrome, 1 plasma cell dyscrasia, and 1 systemic mastocytosis) and 10 were nonhematological neoplasms, including 3 cases of thymoma, 3 cases of thyroid neoplasms, 1 lung cancer, 1 prostatic cancer, 1 cervical cancer, and 1 skin cancer.
Treatment of Tγδ patients
Overall, more than half (53.7%) of patients required therapy during the natural history of the disease. All these patients were treated according to currently accepted indications.2,21 In detail, 8 of 58 (13.8%) patients started therapy due to severe neutropenia, 4 of 58 (6.9%) due to symptomatic neutropenia, 14 of 58 (24.1%) for transfusion-dependent anemia, 13 of 58 (22.4%) for symptomatic anemia, 6 of 58 (10.4%) due to combined severe neutropenia and symptomatic anemia, and the remaining 5 of 58 (8.6%) for symptomatic concomitant autoimmune diseases. In 8 patients (13.8%) the primary diagnosis was settled by hematology centers without experience in LGLL, and subsequently the patients were moved to the referral centers. Consequently, a clear treatment indication was not available.
Considering first-line treatment, most patients (34/57, 59.6%) received methotrexate (MTX), 26.3% (15/57) were treated with cyclosporine A (CyA), and only 10.5% (6/57) received cyclophosphamide (CTX). The remaining 2 patients received cladribine and splenectomy as first-line treatment. Response rates and the absolute numbers of cases are reported in supplemental Figure 1 and Table 1. Overall response (ORR) and complete response (CR) rates were lower in MTX-treated patients (26.9% and 7.7%, respectively) compared with patients who received CyA and CTX (ORR: 53.9% and 40%, respectively; CR: 23.1% and 40%, respectively), although the latter therapies were used in lower numbers of cases, particularly CTX. Four patients treated with MTX discontinued the treatment due to toxicity.
Response to first-line treatment
| Treatment . | n . | Response rate to first-line therapy, n/N (%) . | Treatment interruption . | ||
|---|---|---|---|---|---|
| ORR . | CR . | N/A . | Toxicity . | ||
| MTX | 34 | 7/26 (26.9) | 2/26 (7.7) | 4 | 4 |
| CyA | 15 | 7/13 (53.9) | 3/13 (23.1) | 2 | — |
| CTX | 6 | 2/5 (40) | 2/5 (40) | 1 | — |
| Cladribine | 1 | 1/1 (100) | 0/1 (0) | — | — |
| Splenectomy | 1 | 0/1 (0) | 0/1 (0) | — | — |
| Treatment . | n . | Response rate to first-line therapy, n/N (%) . | Treatment interruption . | ||
|---|---|---|---|---|---|
| ORR . | CR . | N/A . | Toxicity . | ||
| MTX | 34 | 7/26 (26.9) | 2/26 (7.7) | 4 | 4 |
| CyA | 15 | 7/13 (53.9) | 3/13 (23.1) | 2 | — |
| CTX | 6 | 2/5 (40) | 2/5 (40) | 1 | — |
| Cladribine | 1 | 1/1 (100) | 0/1 (0) | — | — |
| Splenectomy | 1 | 0/1 (0) | 0/1 (0) | — | — |
N/A, not available.
Among patients requiring treatment (n = 57), landmark analyses for PFS and OS were performed according to response status at 6 months since therapy initiation, only in the subsets of patients for whom precise timing of response was available (n = 20 for PFS and n = 29 for OS). Irrespective from the type of first-line treatment, responders (patients reaching at least partial response) after 6 months from the start of therapy were characterized by an increase in PFS with respect to nonresponders (HR = 6.16, 95% CI: 0.77-50.00; log-rank test P = .05) (Figure 1A). Notably, although with a P value not statistically significant, responders at 6 months showed also longer OS as compared with nonresponders (log-rank test P = .13) (Figure 1B). These results suggest a possible prognostic role of early response to first-line therapy that should be further addressed in future prospective studies by systematically collecting response times.
PFS and OS landmark analysis of patients treated for Tγδ LGLL. Kaplan-Meier curves showing 6-month landmark analysis for PFS (A) and OS (B) of Tγδ LGLL patients achieving at least a partial response to first-line therapy (Responders) compared with nonresponding patients (Non-responders) at 6 months from the start of therapy. Curves were compared by log-rank test.
PFS and OS landmark analysis of patients treated for Tγδ LGLL. Kaplan-Meier curves showing 6-month landmark analysis for PFS (A) and OS (B) of Tγδ LGLL patients achieving at least a partial response to first-line therapy (Responders) compared with nonresponding patients (Non-responders) at 6 months from the start of therapy. Curves were compared by log-rank test.
STAT3 and STAT5b mutation analysis
DNA samples of 97 and 94 cases were available for STAT3 and STAT5b mutational analyses, respectively. STAT3 mutations were detected in 37 cases (38.1%), with a prevalence of variants as follows: Y640F was detected in 16 cases (43.2%), D661Y in 9 cases (24.4%), D661V and S614R in 2 cases each (5.4%), and the H410R, Q448E, G618R, E638Q, K658F, and N647I variants were found in 1 case each (2.7%). In the Italian cohort, 2 cases showed the A662_N663delinsH deletion and insertion and an in-frame insertion, G656_Y657ins, as previously reported.7 In contrast, STAT5b mutations were found in only 4 cases (4.2%), of whom 3 carried the N642H variant and 1 had the Y665F mutation. Of note, STAT3 and STAT5b mutations were mutually exclusive in Tγδ LGLL cases, never being detected concurrently in the same patient.
From the phenotypic point of view, cases with STAT3 mutations were characterized by lower frequency of expression of CD56 (3.8% vs 56.1%, P < .0001), Vδ2 (0% vs 50%, P = .0003), and Vγ9 (25% vs 57.1%, P = .04). In addition, they showed a higher frequency of neutropenia (65.7% vs 40.8%, P = .0288), severe neutropenia (31.4% vs 12.2%, P = .0519), anemia (55.9% vs 34.7%, P = .0726), and autoimmune/autoinflammatory disorders (59.4% vs 31.5%, P = .0139). They more frequently required therapy (67.9% vs 37.5%, P = .0169) (Table 2). In contrast, no significant differences were found between STAT3-mutated and wild-type Tγδ LGLL patients regarding the frequency of cases with LGL counts > 2000/mm3 (25% vs 15.2%, P = .3824), expression of KIRs (20% vs 50%, P = .1413) and CD94 (38.9% vs 57.1%, P = .2542), thrombocytopenia (17.6% vs 14.3%, P = .7628), splenomegaly (22.9% vs 20.8%, P > .9999), and SPM (21.4% vs 22.7%, P > .9999) (Table 2). Unlike cases with STAT3 mutations, cases with STAT5b mutations were mostly asymptomatic, with only 1 case experiencing mild neutropenia and splenomegaly.
Biological and clinical features of STAT3-mutated and STAT3 wild-type Tγδ LGLL patients
| . | STAT3 mutated, n/N (%) . | STAT3 wild-type, n/N (%) . | P value . |
|---|---|---|---|
| Age > 65 y | 15/37 (40.5) | 11/59 (18.6) | .0324 |
| LGL > 2000/mm3 | 8/32 (25.0) | 7/46 (15.2) | .3824 |
| CD16 expression | 18/27 (66.7) | 29/40 (72.5) | .7860 |
| CD56 expression | 1/26 (3.8) | 23/41 (56.1) | <.0001 |
| CD57 expression | 21/27 (77.8) | 34/41 (82.9) | .7541 |
| KIR expression | 2/10 (20.0) | 12/24 (50.0) | .1413 |
| CD158a expression | 0/10 (0) | 3/24 (12.5) | .5388 |
| CD158b expression | 1/10 (10.0) | 8/24 (33.3) | .2250 |
| CD158e expression | 1/10 (10.0) | 3/24 (12.5) | >.9999 |
| CD94 expression | 7/18 (38.9) | 20/35 (57.1) | .2542 |
| NKG2A expression | 1/10 (10.0) | 10/24 (41.7) | .1133 |
| NKG2C expression | 1/8 (12.5) | 2/22 (9.1) | >.9999 |
| Vδ1+ | 7/12 (58.3) | 9/24 (37.5) | .2983 |
| Vδ2+ | 0/17 (0) | 17/34 (50.0) | .0003 |
| Vγ9+ | 4/16 (25.0) | 20/35 (57.1) | .0400 |
| ANC < 1500/mm3 | 23/35 (65.7) | 20/49 (40.8) | .0288 |
| ANC < 500/mm3 | 11/35 (31.4) | 6/49 (12.2) | .0519 |
| Hb < 120 g/L | 19/34 (55.9) | 17/49 (34.7) | .0726 |
| Hb < 90 g/L | 10/34 (29.4) | 6/49 (12.2) | .0874 |
| PLTs < 100 000/mm3 | 6/34 (17.6) | 7/49 (14.3) | .7628 |
| Splenomegaly | 8/35 (22.9) | 11/53 (20.8) | >.9999 |
| Autoimmune/inflammatory diseases | 19/32 (59.4) | 17/54 (31.5) | .0139 |
| SPMs | 6/28 (21.4) | 10/44 (22.7) | >.9999 |
| Need for treatment | 19/28 (67.9) | 18/48 (37.5) | .0169 |
| . | STAT3 mutated, n/N (%) . | STAT3 wild-type, n/N (%) . | P value . |
|---|---|---|---|
| Age > 65 y | 15/37 (40.5) | 11/59 (18.6) | .0324 |
| LGL > 2000/mm3 | 8/32 (25.0) | 7/46 (15.2) | .3824 |
| CD16 expression | 18/27 (66.7) | 29/40 (72.5) | .7860 |
| CD56 expression | 1/26 (3.8) | 23/41 (56.1) | <.0001 |
| CD57 expression | 21/27 (77.8) | 34/41 (82.9) | .7541 |
| KIR expression | 2/10 (20.0) | 12/24 (50.0) | .1413 |
| CD158a expression | 0/10 (0) | 3/24 (12.5) | .5388 |
| CD158b expression | 1/10 (10.0) | 8/24 (33.3) | .2250 |
| CD158e expression | 1/10 (10.0) | 3/24 (12.5) | >.9999 |
| CD94 expression | 7/18 (38.9) | 20/35 (57.1) | .2542 |
| NKG2A expression | 1/10 (10.0) | 10/24 (41.7) | .1133 |
| NKG2C expression | 1/8 (12.5) | 2/22 (9.1) | >.9999 |
| Vδ1+ | 7/12 (58.3) | 9/24 (37.5) | .2983 |
| Vδ2+ | 0/17 (0) | 17/34 (50.0) | .0003 |
| Vγ9+ | 4/16 (25.0) | 20/35 (57.1) | .0400 |
| ANC < 1500/mm3 | 23/35 (65.7) | 20/49 (40.8) | .0288 |
| ANC < 500/mm3 | 11/35 (31.4) | 6/49 (12.2) | .0519 |
| Hb < 120 g/L | 19/34 (55.9) | 17/49 (34.7) | .0726 |
| Hb < 90 g/L | 10/34 (29.4) | 6/49 (12.2) | .0874 |
| PLTs < 100 000/mm3 | 6/34 (17.6) | 7/49 (14.3) | .7628 |
| Splenomegaly | 8/35 (22.9) | 11/53 (20.8) | >.9999 |
| Autoimmune/inflammatory diseases | 19/32 (59.4) | 17/54 (31.5) | .0139 |
| SPMs | 6/28 (21.4) | 10/44 (22.7) | >.9999 |
| Need for treatment | 19/28 (67.9) | 18/48 (37.5) | .0169 |
P values are calculated using Fisher exact test. Significant P values are reported in bold.
Vδ pattern of expression analysis
Tγδ cells usually express 5 different Vδ receptor families (from Vδ1 to Vδ5), Vδ2 being generally expressed in blood circulating Tγδ cells, and the other subsets are typically enriched in epithelia, liver, and spleen.22 In our cohort, flow cytometric Vδ analysis was available in 51 cases; 17 cases (33.3%) were Vδ2+ and the remaining 34 (66.7%) were Vδ2−. Within this latter subset of cases, 16 of 34 (47.1%) were Vδ1+ and 18 cases were neither Vδ1+ nor Vδ2+ (Table 3).
Biological and clinical features of Tγδ LGLL patients according to Vδ2 status
| . | Vδ2+, n/N (%) . | Vδ2−, n/N (%) . | P value . |
|---|---|---|---|
| LGL > 2000/mm3 | 2/17 (11.8) | 9/29 (31.0) | .1723 |
| CD16 expression | 14/14 (100) | 18/22 (81.8) | .1412 |
| CD56 expression | 14/14 (100) | 2/22 (9.1) | <.0001 |
| CD57 expression | 14/14 (100) | 18/22 (81.8) | .1412 |
| KIR expression | 9/14 (64.3) | 3/16 (18.8) | .0236 |
| CD158a expression | 1/14 (7.1) | 1/16 (6.2) | >.9999 |
| CD158b expression | 6/14 (42.9) | 2/16 (12.5) | .1010 |
| CD158e expression | 3/14 (21.4) | 1/16 (6.2) | .3155 |
| CD94 expression | 13/17 (76.5) | 12/28 (42.9) | .0351 |
| NKG2A expression | 10/14 (71.4) | 1/16 (6.2) | .0004 |
| NKG2C expression | 0/14 (0) | 3/16 (18.8) | .2276 |
| Vγ9+ | 17/17 (100) | 6/33 (18.2) | <.0001 |
| STAT3 mutated | 0/17 (0) | 17/34 (50) | .0003 |
| STAT5b mutated | 3/17 (17.6) | 0/34 (0) | .0327 |
| ANC < 1500/mm3 | 1/17 (5.9) | 21/32 (65.6) | <.0001 |
| ANC < 500/mm3 | 0/17 (0) | 10/32 (31.2) | .0094 |
| Hb < 120 g/L | 0/17 (0) | 18/32 (56.2) | <.0001 |
| Hb < 90 g/L | 0/17 (0) | 11/32 (34.4) | .0090 |
| PLTs < 100 000/mm3 | 1/17 (5.9) | 4/32 (12.5) | .6463 |
| Splenomegaly | 0/16 (0) | 8/30 (26.7) | .0371 |
| Autoimmune/inflammatory diseases | 1/16 (6.2) | 15/31 (48.4) | .0039 |
| SPMs | 4/14 (28.6) | 6/24 (25) | >.9999 |
| Need for treatment | 0/14 (0) | 12/22 (54.5) | .0007 |
| . | Vδ2+, n/N (%) . | Vδ2−, n/N (%) . | P value . |
|---|---|---|---|
| LGL > 2000/mm3 | 2/17 (11.8) | 9/29 (31.0) | .1723 |
| CD16 expression | 14/14 (100) | 18/22 (81.8) | .1412 |
| CD56 expression | 14/14 (100) | 2/22 (9.1) | <.0001 |
| CD57 expression | 14/14 (100) | 18/22 (81.8) | .1412 |
| KIR expression | 9/14 (64.3) | 3/16 (18.8) | .0236 |
| CD158a expression | 1/14 (7.1) | 1/16 (6.2) | >.9999 |
| CD158b expression | 6/14 (42.9) | 2/16 (12.5) | .1010 |
| CD158e expression | 3/14 (21.4) | 1/16 (6.2) | .3155 |
| CD94 expression | 13/17 (76.5) | 12/28 (42.9) | .0351 |
| NKG2A expression | 10/14 (71.4) | 1/16 (6.2) | .0004 |
| NKG2C expression | 0/14 (0) | 3/16 (18.8) | .2276 |
| Vγ9+ | 17/17 (100) | 6/33 (18.2) | <.0001 |
| STAT3 mutated | 0/17 (0) | 17/34 (50) | .0003 |
| STAT5b mutated | 3/17 (17.6) | 0/34 (0) | .0327 |
| ANC < 1500/mm3 | 1/17 (5.9) | 21/32 (65.6) | <.0001 |
| ANC < 500/mm3 | 0/17 (0) | 10/32 (31.2) | .0094 |
| Hb < 120 g/L | 0/17 (0) | 18/32 (56.2) | <.0001 |
| Hb < 90 g/L | 0/17 (0) | 11/32 (34.4) | .0090 |
| PLTs < 100 000/mm3 | 1/17 (5.9) | 4/32 (12.5) | .6463 |
| Splenomegaly | 0/16 (0) | 8/30 (26.7) | .0371 |
| Autoimmune/inflammatory diseases | 1/16 (6.2) | 15/31 (48.4) | .0039 |
| SPMs | 4/14 (28.6) | 6/24 (25) | >.9999 |
| Need for treatment | 0/14 (0) | 12/22 (54.5) | .0007 |
P values are calculated using Fisher exact test. Significant P values are reported in bold.
Vδ2+ cases displayed a higher frequency of expression of CD56 (100% vs 9.1%, P < .0001), KIR (64.3% vs 18.8%, P = .0236), CD94 (76.5% vs 42.9%, P = .0351), and NKG2A (71.4% vs 6.2%, P = .0004), and no significant differences were found (vs Vδ2− cases) regarding CD16 and CD57 expression (100% vs 81.8%, P = .1412, and 100% vs 81.8%, P = .1412, respectively). Interestingly, all Vδ2+ cases showed concomitant Vγ9 expression (100%), and only a small fraction of Vδ2− cases was also Vγ9+ (18.2%, P < .0001).
From the clinical point of view, Vδ2+ cases displayed a more indolent LGLL. They rarely presented with symptomatic disease including neutropenia (5.9% vs 65.6%, P < .0001), severe neutropenia (0% vs 31.2%, P = .0094), anemia (0% vs 56.2%, P < .0001), severe anemia (0% vs 34.4%, P = .0090), splenomegaly (0% vs 26.7%, P = .0371), and concurrent autoimmune/inflammatory disease (6.2% vs 48.4%, P = .0039), in the absence of treatment requirement (0% vs 54.5%, P = .0007). Interestingly, STAT mutations were mutually exclusive in Vδ2− and Vδ2+ cases, all cases with STAT5b mutation being Vδ2+ (P = .0327), whereas all cases with STAT3 mutations were Vδ2− (P = .0003) (Table 3).
Clinical and biological features of Tγδ vs Tαβ LGLL
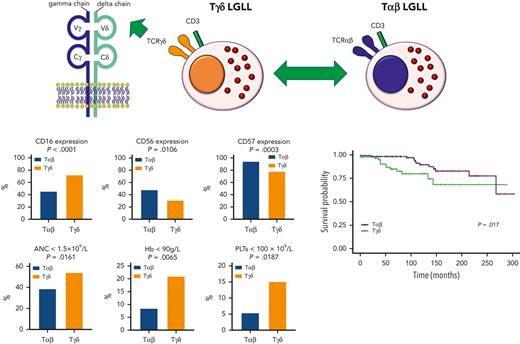
To get further insight into the unique clinical and biological features of Tγδ LGLL, we compared our cohort of patients with a recently published Tαβ LGLL cohort of comparable size7 (Table 4). No significant differences in gender and age were found between the 2 disease subtypes (P = .3906 and P = .2408, respectively), while Tαβ LGLL cases generally showed higher LGL counts than Tγδ LGLL cases (LGL count > 2000/mm3 in 54.3% vs 22% cases, respectively; P < .0001). By immunophenotype, Tγδ LGLL displayed a significantly higher frequency of expression of CD16 (72.3% vs 45.7%, P < .0001), CD94 (42.7% vs 14%, P < .0001), NKG2A (22.2% vs 10.1%, P = .0355), and CD158a (14.3% vs 4.7%, P = .0330) together with an increased KIR expression (41.1% vs 27.9%, P = .0876), and they showed a lower frequency of CD56 (31.1% vs 48.1%, P = .0106) and CD57 expression (78.4% vs 94.6%, P = .0003). Regarding STAT mutations, no significant differences were found between Tγδ and Tαβ LGLL cases in the frequency of STAT3 (38.1% vs 37.9%, respectively; P > .9999) and STAT5b mutations (4.8% vs 12.5%, respectively; P = .1130).
Biological and clinical features of the Tαβ and the Tγδ LGLL cohorts
| . | Tαβ LGLL,∗ n/N (%) . | Tγδ LGLL, n/N (%) . | P value . |
|---|---|---|---|
| Gender male | 65/129 (50.4) | 76/136 (55.9) | .3906 |
| Age > 65 y | 47/129 (36.4) | 40/136 (29.4) | .2408 |
| LGL > 2000/mm3 | 70/129 (54.3) | 24/109 (22.0) | <.0001 |
| CD16 expression | 59/129 (45.7) | 73/101 (72.3) | <.0001 |
| CD56 expression | 62/129 (48.1) | 32/103 (31.1) | .0106 |
| CD57 expression | 122/129 (94.6) | 80/102 (78.4) | .0003 |
| KIR expression | 36/129 (27.9) | 23/56 (41.1) | .0876 |
| CD158a expression | 6/129 (4.7) | 8/56 (14.3) | .0330 |
| CD158b expression | 29/129 (22.5) | 13/56 (23.2) | >.9999 |
| CD158e expression | 6/129 (4.7) | 5/56 (8.9) | .3124 |
| CD94 expression | 18/129 (14.0) | 32/75 (42.7) | <.0001 |
| NKG2A expression | 13/129 (10.1) | 12/54 (22.2) | .0355 |
| NKG2C expression | 5/129 (3.9) | 3/30 (10.0) | .1740 |
| STAT3 mutated | 39/103 (37.9) | 37/97 (38.1) | >.9999 |
| STAT5b mutated | 12/96 (12.5) | 4/94 (4.8) | .1130 |
| ANC < 1500/mm3 | 50/129 (38.8) | 65/120 (54.2) | .0161 |
| ANC < 500/mm3 | 29/129 (22.5) | 25/120 (20.8) | .7611 |
| Hb < 120 g/L | 15/129 (11.6) | 59/119 (49.6) | <.0001 |
| Hb < 90 g/L | 11/129 (8.5) | 25/119 (21.0) | .0065 |
| PLTs < 100 000/mm3 | 7/129 (5.4) | 18/119 (15.1) | .0187 |
| Splenomegaly | 22/129 (17.5) | 31/122 (21.4) | .1225 |
| Autoimmune/inflammatory diseases | 28/129 (21.7) | 49/118 (41.5) | .0009 |
| . | Tαβ LGLL,∗ n/N (%) . | Tγδ LGLL, n/N (%) . | P value . |
|---|---|---|---|
| Gender male | 65/129 (50.4) | 76/136 (55.9) | .3906 |
| Age > 65 y | 47/129 (36.4) | 40/136 (29.4) | .2408 |
| LGL > 2000/mm3 | 70/129 (54.3) | 24/109 (22.0) | <.0001 |
| CD16 expression | 59/129 (45.7) | 73/101 (72.3) | <.0001 |
| CD56 expression | 62/129 (48.1) | 32/103 (31.1) | .0106 |
| CD57 expression | 122/129 (94.6) | 80/102 (78.4) | .0003 |
| KIR expression | 36/129 (27.9) | 23/56 (41.1) | .0876 |
| CD158a expression | 6/129 (4.7) | 8/56 (14.3) | .0330 |
| CD158b expression | 29/129 (22.5) | 13/56 (23.2) | >.9999 |
| CD158e expression | 6/129 (4.7) | 5/56 (8.9) | .3124 |
| CD94 expression | 18/129 (14.0) | 32/75 (42.7) | <.0001 |
| NKG2A expression | 13/129 (10.1) | 12/54 (22.2) | .0355 |
| NKG2C expression | 5/129 (3.9) | 3/30 (10.0) | .1740 |
| STAT3 mutated | 39/103 (37.9) | 37/97 (38.1) | >.9999 |
| STAT5b mutated | 12/96 (12.5) | 4/94 (4.8) | .1130 |
| ANC < 1500/mm3 | 50/129 (38.8) | 65/120 (54.2) | .0161 |
| ANC < 500/mm3 | 29/129 (22.5) | 25/120 (20.8) | .7611 |
| Hb < 120 g/L | 15/129 (11.6) | 59/119 (49.6) | <.0001 |
| Hb < 90 g/L | 11/129 (8.5) | 25/119 (21.0) | .0065 |
| PLTs < 100 000/mm3 | 7/129 (5.4) | 18/119 (15.1) | .0187 |
| Splenomegaly | 22/129 (17.5) | 31/122 (21.4) | .1225 |
| Autoimmune/inflammatory diseases | 28/129 (21.7) | 49/118 (41.5) | .0009 |
P values are calculated using Fisher exact test. Significant P values are reported in bold.
The markedly different observation times of Tγδ LGLL and Tαβ LGLL patients prevented use of Fisher exact test for the comparison of time-dependent factors since this could lead to major bias due to lack of consideration of the time variable. Consequently, for SPMs and need for treatment, the data and the related P value were not available.
Tαβ LGLL cohort of comparable size.7
From the clinical point of view, Tγδ LGLL cases more frequently showed symptomatic disease in terms of neutropenia (54.2% vs 38.8%, P = .0161), anemia (49.6% vs 11.6%, P < .0001), severe anemia (21% vs 8.5%, P = .0065), thrombocytopenia (15.1% vs 5.4%, P = .0187), and concurrent autoimmune/inflammatory diseases (41.5% vs 21.7%, P = .0009) (Table 4).
The markedly different observation times of Tγδ-LGLL and Tαβ-LGLL cases prevented use of a Fisher exact test for the comparison of time-dependent factors since this could lead to major bias due to lack of consideration of the time variable. Consequently, for SPMs and need for treatment, the data and the related P value were not available.
Survival analysis
All demographic, clinical, and biological features were evaluated for association with OS in Tγδ LGLL cases. After a median follow-up of 48 months, the median OS of our cohort was not reached. Splenomegaly was the only variable significantly associated with a shortened OS (log-rank test P = .0012), with an HR = 0.18 (95% CI: 0.06-0.59) (Figure 2A), and other clinical and biological features of the disease had no significant impact on patient OS, including those previously found to be relevant for Tαβ LGLL patients7 (ie, STAT3 and STAT5b mutation status or the presence of severe neutropenia or anemia) (supplemental Figure 2).
OS analysis of Tγδ LGLL patients. (A) OS analysis of the Tγδ LGLL cohort with respect to presence/absence of splenomegaly. Survival curves were estimated using the Kaplan-Meier method and compared by log-rank test. (B) OS comparison between Tαβ and Tγδ cohorts. With a median follow-up of 108 months (Tαβ) and of 48 months (Tγδ), median OS was not reached in both the cohorts. Survival curves were estimated using the Kaplan-Meier method and compared by log-rank test.
OS analysis of Tγδ LGLL patients. (A) OS analysis of the Tγδ LGLL cohort with respect to presence/absence of splenomegaly. Survival curves were estimated using the Kaplan-Meier method and compared by log-rank test. (B) OS comparison between Tαβ and Tγδ cohorts. With a median follow-up of 108 months (Tαβ) and of 48 months (Tγδ), median OS was not reached in both the cohorts. Survival curves were estimated using the Kaplan-Meier method and compared by log-rank test.
Direct comparison of patients’ OS between Tγδ LGLL and the more common Tαβ LGLL is likely to prove a poorer overall outcome for Tγδ LGLL cases vs Tαβ LGLL cases (log-rank test P = .017) (Figure 2B). This result must be interpreted with caution, since the 2 cohorts have different median follow-up times (Tγδ LGLL, 4 years, vs Tαβ LGLL, 9 years), and the proportional hazards assumption seems not to be fully satisfied due to the lack of events in the Tγδ cohort from 143 months onward. Given the rarity of Tγδ LGLL, it was not possible to increase the cohort size; consequently, we provided a supplementary analysis using a different measure of the effect that does not require the proportional hazards assumption (ie, the RMST). This analysis confirms a significant disadvantage in terms of survival of Tγδ LGLL patients with respect to Tαβ LGLL (supplemental Table 2).
Discussion
Here we report on the largest cohort of Tγδ LGL leukemia patients described so far in the literature with data collected between 1997 and 2020, as the result of a collaborative study involving 8 LGLL referral centers across the world. For the first time, we evaluated the clinical and biological features of this rare subset of T-LGLL on a large number of patients, screened for STAT3 and STAT5b mutations. Overall, our results showed that Tγδ LGLL represents a variant with higher frequency of symptomatic disease and reduced survival compared with the most common Tαβ LGLL subtype, despite a similar frequency of STAT3 and to a less extent of STAT5b mutations.
In the past, LGLL was considered a unique chronic and indolent disease, except for a few patients presenting with very aggressive disease.23 In recent years, however, a better understanding of this disorder has been achieved, pointing out the need for therapy in a significant fraction of LGLL patients.6,7,24 Data provided in this study further encourage distinguishing Tγδ LGLL from Tαβ LGLL, since Tγδ LGLL patients showed unique clinical and biological features. Even though characterized by lower LGL counts, Tγδ LGLs more frequently express the CD16 and CD94 receptors, and the CD56 adhesion molecule and the CD57 immunosenescence-associated protein are less commonly expressed. Most important, Tγδ LGLL patients more frequently displayed symptomatic disease due to anemia (often transfusion dependent), potentially partially explained by an increased frequency of autoimmune hemolytic anemia and PRCA,25 and concomitant autoimmune diseases. Altogether, this translates into a poorer outcome as compared with that from the more common Tαβ subtype of LGLL.
Overall these results are not consistent with previously reported data that did not show clear clinical differences between Tγδ LGLL and Tαβ LGLL26; however, the T-LGLL cohort reported by Bourgault-Rouxel et al included only a small number of Tγδ patients (20 cases) compared with the almost 200 Tαβ reported cases, which limits the robustness of the conclusions raised.26 A possible limitation to be considered in the explanation of the worst outcome in Tγδ LGLL could be related to a high frequency of late-stage diseases due to the challenging diagnosis. As a matter of fact, in our series Tγδ patients showed lower LGL counts and CD57 expression as compared with the those in the more common Tαβ patients. The high frequency of symptomatic patients herein reported within the Tγδ LGLL cohort may account for the reduced OS in this LGLL subtype. Aside from this potential bias in survival analysis, our data point to the recommendation to include the Tγδ immunophenotype in the diagnostic workup of unexplained cytopenia.
Despite the comparable size, the Tγδ and Tαβ LGLL cohorts we studied are characterized by different median follow-up (48 vs 108 months, respectively); moreover, the Tγδ LGLL cohort, due to its retrospective nature, suffers for the presence of several censored data. These findings led to certain limitations in the interpretation of results. For this reason, an additional RMST analysis has been provided to mitigate these limitations, confirming a significant survival disadvantage for Tγδ LGLL patients with respect to Tαβ LGLL. In future perspective studies aimed at comparing the 2 cohorts, it could be interesting to carefully plan the data collection to analyze variables that may depend on observation time (eg, SPM or need for treatment) with a more appropriate time-to-event approach, thus minimizing any bias due to different follow-up lengths.
According to retrospective studies including few and heterogenous series of patients,27-29 treatment of LGLL still relies on immunosuppressive therapy, where MTX and CTX are used upfront, and CyA is generally reserved for relapsed or refractory patients.1,2,21 To date, only 1 published prospective trial evaluating the efficacy of immunosuppressive therapy in LGLL is available,30 and 1 prospective and randomized trial comparing MTX and CTX as first-line therapy in LGLL is currently ongoing (NCT01976182). However, all these studies do not report on the frequency of Tγδ LGLL analyzed and their specific response to therapy. Unexpectedly, MTX treatment led to unsatisfactory response rates in our series of Tγδ LGLL patients, with ORR being observed in less than a third of patients, including CR in a very limited number of cases (7.7%). In contrast, first-line therapy with CyA turned out to provide higher efficacy, with almost half the patients responding, of whom 23.1% reached CR.
An association between Tγδ LGLL and PRCA has been widely described, and it is also known that PRCA patients benefit from CyA treatment. In our cohort, treatment indication for the CyA cohort was available for 14 patients, and 12 patients started therapy due to anemia, in 8 cases transfusion dependent; the remaining 2 patients had a concomitant diagnosis of PRCA. It can be argued that PRCA has been underestimated in Tγδ LGLL with anemia or severe anemia, thus explaining the high overall and CR rates obtained with CyA in this subgroup of patients. Of notice, the choice of the appropriate therapy is of utmost clinical relevance since we demonstrated here that responding patients were also characterized by a prolonged PFS and an improved OS. These data could offer a rationale for investigating CyA in the first-line treatment of Tγδ LGLL (eg, in new prospective trials).
Altogether, the results indicate that, besides the distinction between T-LGLL and natural killer–LGLL, further dissection of T-LGLL into the Tαβ and Tγδ LGLL disease variants is of clinical relevance due to the poorer outcome and distinct treatment response profile of the latter patients. It is also worth noting that Tγδ LGLL cases did not appear as a homogeneous disease entity. Vδ2 positivity was associated with an immunophenotype characterized by Vγ9, CD56, KIR, and CD94/NKG2A expression and, on clinical grounds, by lower frequency of symptomatic disease in terms of neutropenia, anemia, splenomegaly concomitant autoimmune/inflammatory disease, and need of treatment compared with that of Vδ2− patients. Furthermore, the Vδ2 expression profile also correlated with the STAT mutational status since all STAT3-mutated cases were Vδ2−, and the 3 patients with STAT5b mutations were Vδ2+. Interestingly, the 2 subsets of Tγδ LGLL defined by the Vδ2 expression profile are likely to identify distinct cells of origin of Tγδ LGLL.22 In line with this hypothesis, Vδ2+ Tγδ LGLL might represent the neoplastic counterpart of blood circulating Tγδ cells, and Vδ2− Tγδ LGLL might mostly originate from tissue-derived Tγδ cells, with potential pathogenic implications.
Accumulating evidence indicates that the association between STAT3 mutation and symptomatic disease is already recognized in Tαβ LGLL.6,7,31 Recent data also support a reduced survival for STAT3-mutated vs STAT3 wild-type cases.7 In contrast, the clinical impact of STAT5b mutations is still matter of debate; this mutation is present in the rare aggressive variants of LGLL8 as well as in indolent CD4+ T-LGLL.7,32 In the Tγδ LGLL setting, the real incidence of STATs gene mutations is still unknown, being studied up to now only in small cohorts of patients.7,16,33 In our study, mutations in STAT3 and STAT5b were screened in nearly 100 Tγδ LGLL cases, and a frequency of STAT3 mutations was found to be comparable with previously reported data in LGLL.4-6 Moreover, we also detected 3 Tγδ LGLL cases harboring STAT5b mutations who displayed an indolent disease as observed in CD4+ Tαβ LGLL. In our cohort, we confirm the association between STAT3 mutation and symptomatic disease, particularly with neutropenia, and increased need for therapy, although we did not observe a reduced OS for STAT3-mutated cases. These results support a more aggressive disease behavior of Tγδ LGLL, particularly for cases who do not show Vδ2 expression, independently from the STAT3 mutational status.
In conclusion, data from this large multicentric cohort of Tγδ LGLL highlight the unique biological and clinical hallmarks of this rare variant of T-LGLL, likely associated with a discrete treatment response profile. Despite the similar frequency of STAT3 and STAT5b, Tγδ LGLL cases in general, and Vδ2− Tγδ LGLL in particular, showed more symptomatic disease and a poorer outcome compared with those with Tαβ LGLL. At the same time, Tγδ LGLL patients appear to mostly benefit from CyA as first-line therapy. Altogether, these results underly the relevance of a precise characterization and subclassification of LGLL.
Acknowledgment
The authors thank the Associazione Italiana per la Ricerca sul Cancro (grant IG 2017-20216).
Authorship
Contribution: G.B. designed the research, analyzed data, and wrote the manuscript; A.G. analyzed data, performed statistical analysis, and wrote the manuscript; H.J.C., A.T., G.C., J.C., C.V., B.C.S., V.R.G., N.M.-G., H.N., and C.P. provided patient samples and patient data; J.A., M.S., K.O., L.S., F.I., T.P.L., A.O., W.G.M., and T.L. participated in the analysis of data and critically reviewed and edited the manuscript; G.S. provided funding, participated in the analysis of data, and critically reviewed and edited the manuscript; R.Z. designed the study, analyzed data, wrote the manuscript, and supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Renato Zambello, Padua University School of Medicine, Department of Medicine, Via Giustiniani 2, 35128 Padua, Italy; e-mail: r.zambello@unipd.it; and Gianpietro Semenzato, University of Padua, Veneto Institute of Molecular Medicine, Via Orus 2, 35129 Padua, Italy; e-mail: g.semenzato@unipd.it.
References
Author notes
For original data, please contact r.zambello@unipd.it or g.semenzato@unipd.it. In this research article, we compared the international Tγδ LGLL cohort with a recently published Tαβ LGLL cohort of comparable size (Barilà et al7).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal