Key Points
SRPRA and SRP19 are novel genes affected in congenital neutropenia and essential for granule protein processing.
Comparative proteomics in neutrophils of patients with defects in SRPRA, SRP19, SRP54, HAX1, and ELANE reveal genotype-specific alterations.
Abstract
The mechanisms of coordinated changes in proteome composition and their relevance for the differentiation of neutrophil granulocytes are not well studied. Here, we discover 2 novel human genetic defects in signal recognition particle receptor alpha (SRPRA) and SRP19, constituents of the mammalian cotranslational targeting machinery, and characterize their roles in neutrophil granulocyte differentiation. We systematically study the proteome of neutrophil granulocytes from patients with variants in the SRP genes, HAX1, and ELANE, and identify global as well as specific proteome aberrations. Using in vitro differentiation of human induced pluripotent stem cells and in vivo zebrafish models, we study the effects of SRP deficiency on neutrophil granulocyte development. In a heterologous cell–based inducible protein expression system, we validate the effects conferred by SRP dysfunction for selected proteins that we identified in our proteome screen. Thus, SRP-dependent protein processing, intracellular trafficking, and homeostasis are critically important for the differentiation of neutrophil granulocytes.
Introduction
Neutrophil granulocytes play sophisticated roles in the regulation of antimicrobial host defense, cancer, and chronic inflammation.1,2 They execute their main function by the use of proteins stored in a heterogeneous set of granules, including primary, secondary, ficolin-rich, and tertiary granules.3
Granule protein synthesis is tightly linked to the highly ordered differentiation of pluripotent hematopoietic stem cells into mature neutrophil granulocytes.3
Studying patients with rare genetic diseases has proven powerful in highlighting novel genes and pathways orchestrating the development and function of neutrophil granulocytes.4,5 Severe congenital neutropenia (SCN) comprises a clinically and genetically heterogeneous spectrum of rare inherited disorders characterized by impaired maturation of neutrophil granulocytes.4 Originally identified by Kostmann,6 patients with autosomal recessive congenital neutropenia have mutations in the mitochondrial protein HCLS1-associated protein X 1 (HAX1).7 The genes encoding the endoplasmic reticulum (ER)–resident proteins glucose-6-phosphate translocase (G6PC3) and Jagunal homolog 1 (JAGN1) are also involved in maintaining physiological neutrophil differentiation.8,9 Monoallelic mutations affecting ELANE10 or the signal recognition particle 54 (SRP54)11,12 are common causes of SCN and involve homeostasis of the ER, as evidenced by increased ER stress in mutated myeloid cells.13,14
Here, we discover 2 novel human monogenic defects in SRP receptor alpha (SRPRA) subunit and SRP19 affecting the differentiation of neutrophil granulocytes. SRPRA encodes the soluble subunit of the eukaryotic SRP receptor (SRPR) that recognizes the SRP, a universally conserved protein machinery. The SRP is composed of 7 subunits (consisting of the 6 polypeptides: SRP9, SRP14, SRP19, SRP54, SRP68, and SRP72 and a noncoding RNA) and couples the synthesis of nascent proteins, which emerge from the ribosome, to the ER.15 SRPR is a heterodimeric complex composed of a cytosolic SRPRA that interacts with the SRP and a transmembrane SRPR beta subunit that localizes SRPRA to the ER.16 The SRP subunit SRP54 (mutated in a Shwachman-Diamond–like syndrome [Carapito et al])11 and SRPRA coordinate their GTPase activity in concert to ensure the precise targeting of nascent polypeptides into the ER.16,17 Misfolded proteins that are unable to enter the ER are recognized and targeted for proteasomal degradation.18,19
We compare the protein contents of neutrophil granulocytes from patients with SCN with variants in SRPRA, SRP19, SRP54, ELANE, and HAX1, and identify genotype-specific differences as well as an essential and nonredundant role of the SRP complex and its receptor, SRPRA, in human granulopoiesis.
Methods
Human participants
We present clinical, genetic, and biological data from 6 pedigrees with 11 patients carrying variants in SRP complex subunits. A novel heterozygous de novo mutation in SRPRA was found in 1 pedigree with 1 patient (family A: II.3). A novel homozygous mutation in SRP19 was identified in 2 related pedigrees with 5 patients affected (family B: IV.2, IV.3, and IV.5 and V.1 and V.2). SRP54 variants were found in 3 pedigrees with 5 patients. A detailed description of gene variant identification is given in the supplemental Methods, available on the Blood website.
Maintenance and differentiation of human iPSCs
Induced pluripotent stem cells (iPSCs) were maintained on a tissue culture dish coated with growth factor–reduced Matrigel (Corning, catalog 356231) in mTeSR1 serum-free medium (Stemcel, catalog #5850). Differentiation toward neutrophil-like granulocytes was initiated according to our outlined protocol in supplemental Figure 4A.
Proteome analysis
Proteome analysis was conducted as described by Grabowski et al20 (supplemental Figure 6A). Primary neutrophil granulocytes were isolated from fresh venous blood by negative selection (Miltenyi Biotec, catalog #130-104-434) and erythrocyte depletion (Miltenyi Biotec, catalog #130-098-196), yielding a final purity of >96%. Pellets of 1 × 106 cells were frozen in 5 μL of 25× protease inhibitor (Roche, catalog #04693159001) and stored in liquid nitrogen. Peptides were extracted from the pellets using filter-aided sample preparation (according to 30259475) and trypsin digestion (Thermo Fisher Scientific, catalog #90057). Using 2 μg of peptides per sample, spectra were measured by a Q Exactive HF Mass Spectrometer (Thermo Fisher Scientific) employing a data-independent acquisition approach.21 The protein identification and quantification from raw data was based on Biognosys Spectronaut version 14, resulting in a total of 5494 proteins, 3624 of which were reliably quantified in each sample. A detailed description is outlined in the supplemental Methods.
Results
Discovery of human SRPRA and SRP19 deficiency
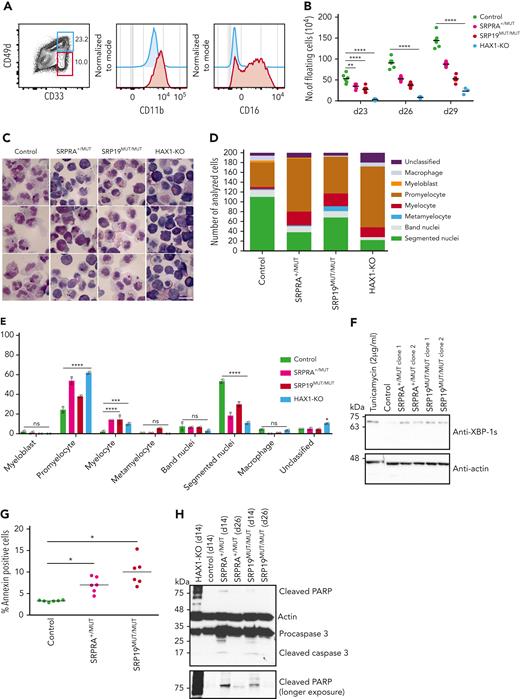
Our index patient (A.II-3), a 5-year-old Romanian girl, suffered from failure to thrive and recurrent pulmonary infections (see supplemental Table 2 for further clinical details). She presented with growth failure, bronchiectasis (Figure 1A), pancreatic insufficiency, and congenital neutropenia associated with myeloid maturation arrest in the bone marrow (supplemental Figure 1A). Electron microscopy studies of her neutrophil granulocytes showed a significant reduction of electron-dense granules (Figure 1B-C). In search of an underlying mutation, we performed whole-genome sequencing of the core family (Figure 1D). Approximately 6 million genetic variants were found in the whole-genome sequencing data of 7 family members. Variant filtering and prioritization revealed only 2 variants with combined annotation-dependent depletion scores) higher than 25 (supplemental Table 1). These de novo variants were identified in METTL26 [C16orf13, chr16: g.686265C>T, p. (Arg9Gln)] and in SRPRA [chr11:g.126134989G >C, p. (Gln464Glu)]. Because METTL26 is not expressed in neutrophil granulocytes,22 we focused on SRPRA, which is highly expressed in neutrophil granulocytes (see also Figure 3Ec). For validation, we performed Sanger sequencing for SRPRA in 6 family members (Figure 1D) and confirmed the mutation to be heterozygous and de novo in the patient. Of note, the G>C point mutation in SRPRA results in an amino acid exchange (glutamine to glutamic acid, position 464) in an alpha helix loop close to the GTPase active center known to mediate interaction with the cognate-binding partner, SRP54. Modeling the side chain substitution with PyMOL indicates that a hydrogen bond to guanosine triphosphate is lost, which might impair guanosine triphosphate hydrolysis and/or SRP complex function (Figure 1E-F). In search of additional patients with SCN with variants in SRP subunits (SRP9, SRP14, SRP19, SRP54, SRP68, and SRP72) and in the SRPRs (SRPRA and SRPRB), we screened our in-house database (including a total of 278 patients at the date of search) and identified 5 patients with rare heterozygous variants in SRP54 as well as 5 patients with a homozygous splice site variant in SRP19 (Figure 1G; supplemental Table 1). The Human Splicing Finder predicted that the variant causes an alteration of the splice donor site with a high likelihood of causing an effect on protein splicing.23
Identification of SRPRA and SRP19 novel gene variants. (A) Chest computed tomography scan of index patient II-3 (family A). (B) Transelectron microscopy sections of neutrophils from index patient A.II-3 compared with unaffected family members (brother A.II-2, father A.I-2) and healthy donors. Non-adherent Neutrophils have a size of ca. 9 μm. (C) Quantification of neutrophil granule content from healthy donor, A.I-2, A.II-2, and A.II-3. Group differences with P < .0001. Single-group differences via Student t test. ∗P < .01, ∗∗P < .001, and ∗∗∗∗P < .00001. (D) Pedigree of family A and Sanger sequencing chromatograms of wild-type (WT) and the SRPRA mutation site. Ribbon representation of the 3-dimensional structure of the human SRPRA WT (E) and mutated SRPRA (Q464E) (F). (G) Pedigree of family B and Sanger sequencing chromatograms of the WT and the SRP19 mutation site. (H) Reverse transcription polymerase chain reaction (RT-PCR) results documenting WT and mutated (mut) SRP19_2 to 4 minigene transcripts in HeLa cells. F1-R1 and F1-R2 are the primer pairs used to amplify the indicated SRP19 exons. The primer pair F1-R2 amplifies SRP19 exon 2 to 4 as indicated with a red arrow (281 base pair [bp]). The SRP19 variant results in a PCR product of only 209 base pair (blue arrow). The primer pair F1-R1 amplifies parts of the SRP19 exon 2 and is used as a transfection efficiency control. (I) Immunoblot analysis of EBV-LCL lysates from healthy donor, patient (Pat.) B.V-1, and a heterozygous family (fam.) member (B.IV-6). Experiment performed in triplicate.
Identification of SRPRA and SRP19 novel gene variants. (A) Chest computed tomography scan of index patient II-3 (family A). (B) Transelectron microscopy sections of neutrophils from index patient A.II-3 compared with unaffected family members (brother A.II-2, father A.I-2) and healthy donors. Non-adherent Neutrophils have a size of ca. 9 μm. (C) Quantification of neutrophil granule content from healthy donor, A.I-2, A.II-2, and A.II-3. Group differences with P < .0001. Single-group differences via Student t test. ∗P < .01, ∗∗P < .001, and ∗∗∗∗P < .00001. (D) Pedigree of family A and Sanger sequencing chromatograms of wild-type (WT) and the SRPRA mutation site. Ribbon representation of the 3-dimensional structure of the human SRPRA WT (E) and mutated SRPRA (Q464E) (F). (G) Pedigree of family B and Sanger sequencing chromatograms of the WT and the SRP19 mutation site. (H) Reverse transcription polymerase chain reaction (RT-PCR) results documenting WT and mutated (mut) SRP19_2 to 4 minigene transcripts in HeLa cells. F1-R1 and F1-R2 are the primer pairs used to amplify the indicated SRP19 exons. The primer pair F1-R2 amplifies SRP19 exon 2 to 4 as indicated with a red arrow (281 base pair [bp]). The SRP19 variant results in a PCR product of only 209 base pair (blue arrow). The primer pair F1-R1 amplifies parts of the SRP19 exon 2 and is used as a transfection efficiency control. (I) Immunoblot analysis of EBV-LCL lysates from healthy donor, patient (Pat.) B.V-1, and a heterozygous family (fam.) member (B.IV-6). Experiment performed in triplicate.
To provide functional proof of the significance of the SRP19 splice site variant, we generated an artificial minigene with exons 2, 3, and 4 (supplemental Figure 1C). Upon expression in HeLa or HEK293T cells, the vector with the SRP19 WT minigene gave rise to only 1 transcript, whereas the patient-specific SRP19 mutation resulted in partial skipping of exon 3 (Figure 1H; supplemental Figure 1D). To confirm aberrant splicing in patients’ cells, we next designed a set of primer pairs to amplify distinct SRP19 transcripts (supplemental Figure 2A) from complementary DNA. Whereas EBV-LCL cells from healthy individuals predominantly expressed SRP19 isoform 1, EBV-LCL cells from patients with homozygous splice site variants in SRP19 showed reduced expression of SRP19 isoform 1 and instead additionally expressed isoform 3 (supplemental Figure 2B). Similar skewing of SRP19 transcripts was observed in primary neutrophil granulocytes from patients with SRP19 mutations (supplemental Figure 2C). Anti-SRP19 immunoblot studies confirmed decreased SRP19 protein expression in EBV-LCL cells from patient B.V-1 (Figure 1I). To validate the effects of aberrant SRP19 splicing, we generated expression vectors of green fluorescent protein (GFP)–SRP19 fusion constructs and transfected HeLa cells. Cells were costained with eIF6 (to visualize nucleoli), the ER marker calnexin, and the DNA marker 4’,6-diamidino-2-phenylindole. Whereas GFP-SRP19-isoform 1 was detected in nucleoli>nuclei>cytoplasm, GFP-SRP19-isoform 3 was not found in the cytoplasm (supplemental Figure 3). These studies confirm aberrant expression of SRP19 isoforms resulting from the identified splice site variant.
Functional validation of genomic studies
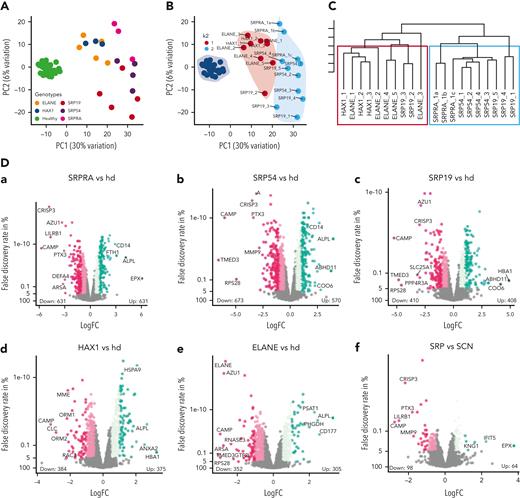
Clinical and molecular studies strongly suggest a pathogenic role for novel variants in SRPRA and SRP19. To provide functional evidence, we set up an in vitro differentiation system allowing us to model the genetic defects in human SRP complex subunits and its receptor SRPRA on granulopoiesis. We refined a previously published protocol,24 allowing us to differentiate neutrophil granulocytes in vitro from iPSCs. Hematopoietic cytokines (supplemental Figure 4A) induced differentiation into neutrophil-like neutrophil granulocytes, as indicated by fluorescence-activated cell sorter–based cell-surface marker analysis (Figure 2A) and microscopy studies of Giemsa-stained cells (Figure 2C left panel). In contrast to undifferentiated iPSCs, in vitro–differentiated neutrophil granulocytes expressed myeloid cell–specific genes such as ELANE, RUNX1, MPO, AZU1, and CSF3R (supplemental Figure 4B). They displayed reduced NADP oxidase activity (supplemental Figure 4C) and showed bactericidal activity similar to neutrophil granulocytes isolated from healthy volunteers (supplemental Figure 4D). They also revealed adhesion and migration characteristics indistinguishable from peripheral blood neutrophil granulocytes isolated from healthy volunteers (supplemental Figure 4E-H). Having established this modeling system, we introduced patient-specific variants (monoallelic SRPRA c.1390C>G and biallelic SRP19 c.189 +5G>A) into WT iPSCs (supplemental Figure 5B-C). As a positive control, we generated iPSCs deficient in HAX1 expression (HAX1-KO) (supplemental Figure 5D). WT and mutant iPSC clones that had similar expression of Oct4 and SSEA4 were chosen for in vitro differentiation (supplemental Figure 5E). SRPRA- and SRP19-mutated iPSCs were characterized by a maturation defect of developing neutrophil granulocytes (Figure 2B), evident by an increase in myeloid progenitor cells such as promyelocytes (Figure 2C-E). A series of colony-forming unit assays with iPSC-derived CD34+/CD45+ cells confirmed a decreased ability of SRP-mutant cells to differentiate into neutrophil granulocytes (supplemental Figure 4I). Next, we examined whether the decrease in neutrophil granulocyte formation was associated with unfolded protein response (UPR) activation and increased cell death. We analyzed the presence of spliced BiP and XBP1, key regulators in UPR signaling during ER stress. On day 14 of the neutrophil granulocyte differentiation protocol, SRP-mutant cells displayed activated UPR as shown by an increased BiP protein expression in SRP-mutated cells (supplemental Figure 4J) and a significant enrichment of the spliced form of XBP-1 (Figure 2F; supplemental Figure 5A). Activated UPR was concomitant with an increase in annexin V+ cells in SRP-mutated iPSCs (Figure 2G). Moreover, we observed an increase in cleaved PARP and cleaved caspase 3 in SRP-mutated progenitor cells (Figure 2H), suggesting that a defect in SRP19 and SRPRA leads to increased susceptibility to apoptosis.
Characterization of iPSC-derived neutrophil granulocytes. (A) Flow cytometric analysis of control iPSC-derived neutrophil granulocytes at day 29. Experiment performed in triplicate. (B) Quantification of live floating cells per 6 iPSC colonies (per well), determined at indicated time points during differentiation. For control, SRPRA+/MUT, and SRP19MUT/MUT, 2 biological replicates (2 different clones) are presented as mean of 3 independent experiments. For HAX1-knockout (KO) the mean of 1 clone of 3 independent experiments is presented. Statistical analysis using 2-way analysis of variance (ANOVA) followed by Dunnett multiple comparisons test. ∗∗P < .0032 and ∗∗∗∗P < .0001. (C) Light microscopy of control, SRPRA+/MUT, SRP19MUT/MUT, and HAX1-KO iPSC-derived myeloid cells stained with May-Grünwald Giemsa stain at day 29. (D) Quantification of the distribution of precursor populations in iPSC-derived myeloid cells for the indicated genotypes. Floating cells were harvested and stained with May-Grünwald Giemsa stain at day 29 and classified by light microscopy. The quantification was performed for 2 independent experiments; a total of 200 cells per genotype were classified. (E) Statistical analysis of the quantification shown in panel D using 2-way ANOVA followed by Dunnett multiple comparisons test. ∗P < .03, ∗∗∗P < .0009, and ∗∗∗∗P < .0001. Error bars represent mean with standard deviation. Quantification of 2 independent experiments are shown. (F) Immunoblot analysis of XBP-1s (spliced form of XBP-1) expression in iPSC-derived myeloid cells at day14. Experiment performed in triplicate. (G) Quantification of annexin V+ cells in sorted (CD33 high/CD49d high) immature iPSC-derived neutrophil granulocytes. Cells were analyzed on day 26 after differentiation. (n = 3 wells per each clone, multiple t test). For control, SRPRA+/MUT and SRP19MUT/MUT, 2 biological replicates (2 different clones) are presented as mean of 3 independent experiments. (H) Immunoblot analysis of apoptosis-specific markers (cleaved PARP and cleaved caspase 3) in iPSC-derived myeloid cells at day14. Experiment performed in triplicate. ns, not significant.
Characterization of iPSC-derived neutrophil granulocytes. (A) Flow cytometric analysis of control iPSC-derived neutrophil granulocytes at day 29. Experiment performed in triplicate. (B) Quantification of live floating cells per 6 iPSC colonies (per well), determined at indicated time points during differentiation. For control, SRPRA+/MUT, and SRP19MUT/MUT, 2 biological replicates (2 different clones) are presented as mean of 3 independent experiments. For HAX1-knockout (KO) the mean of 1 clone of 3 independent experiments is presented. Statistical analysis using 2-way analysis of variance (ANOVA) followed by Dunnett multiple comparisons test. ∗∗P < .0032 and ∗∗∗∗P < .0001. (C) Light microscopy of control, SRPRA+/MUT, SRP19MUT/MUT, and HAX1-KO iPSC-derived myeloid cells stained with May-Grünwald Giemsa stain at day 29. (D) Quantification of the distribution of precursor populations in iPSC-derived myeloid cells for the indicated genotypes. Floating cells were harvested and stained with May-Grünwald Giemsa stain at day 29 and classified by light microscopy. The quantification was performed for 2 independent experiments; a total of 200 cells per genotype were classified. (E) Statistical analysis of the quantification shown in panel D using 2-way ANOVA followed by Dunnett multiple comparisons test. ∗P < .03, ∗∗∗P < .0009, and ∗∗∗∗P < .0001. Error bars represent mean with standard deviation. Quantification of 2 independent experiments are shown. (F) Immunoblot analysis of XBP-1s (spliced form of XBP-1) expression in iPSC-derived myeloid cells at day14. Experiment performed in triplicate. (G) Quantification of annexin V+ cells in sorted (CD33 high/CD49d high) immature iPSC-derived neutrophil granulocytes. Cells were analyzed on day 26 after differentiation. (n = 3 wells per each clone, multiple t test). For control, SRPRA+/MUT and SRP19MUT/MUT, 2 biological replicates (2 different clones) are presented as mean of 3 independent experiments. (H) Immunoblot analysis of apoptosis-specific markers (cleaved PARP and cleaved caspase 3) in iPSC-derived myeloid cells at day14. Experiment performed in triplicate. ns, not significant.
Proteome signatures of primary neutrophil granulocytes from patients with variants in SCN genes
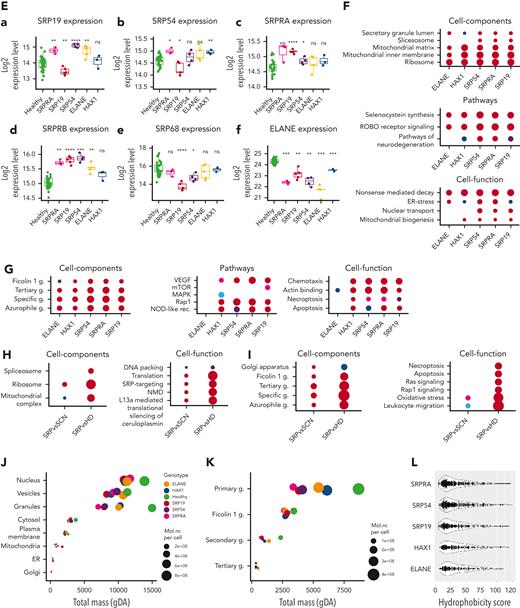
Adapting our previously described pipeline,20 we performed data-independent acquisition21 mass spectrometry–based deep proteome profiling of primary neutrophil granulocytes (supplemental Figure 6A). We analyzed primary neutrophil proteomes from patients with variants in SRPRA (n = 1 with 3 biological replicates [1a, 1b, 1c] sampled weeks apart), SRP19 (n = 5), and SRP54 (n = 4), as well as ELANE (n = 5) and HAX1 (n = 3) together with a healthy donor cohort of 48 individuals. Principal component (PC) analysis of PC1 and PC2 (Figure 3A) separates proteomes from patients with SCN and healthy individuals. We performed k-means cluster analysis to test grouping of patient samples (Figure 3B). Although k1 contains all proteomes from patients with mutations in ELANE and HAX1; k2 contains exclusively proteomes from patients with mutations in SRPRA, SRP54, and SRP19. For validation, we performed coefficient-controlled agglomerative hierarchical clustering (Figure 3C), resulting in a first cluster with all ELANE and HAX1 samples and a second cluster containing exclusively the SRP patient samples. We conclude that mutations in different subunits of the SRP result in largely overlapping proteome changes in human neutrophils that are clearly distinguishable from proteome aberrations caused by mutations in ELANE or HAX1.
Proteome analysis of patient-derived primary neutrophil granulocytes. (A) Principal component analysis showing PC1 and PC2 of neutrophil granulocyte proteomes. Each dot represents a sample and each color represents a genotype. (B) Principal component analysis showing PC1 and PC2 of neutrophil granulocyte proteomes with k-means cluster results. Each dot represents a sample, each color represents the cluster assignment to k1 or k2. Genotypes are shown as labels. (C) Hierarchical dendrogram showing ward-based agglomerative coefficient clustering of neutropenia samples with k = 2. Each tree leaf represents a single sample with the 2 clusters framed in red and green. (D) Volcano plots showing differentially expressed proteins of the comparisons SCN genotypes vs healthy donor (hd) (a-e) and SRP vs SCN (f) (ELANE/HAX1). Each dot represents a protein and is located in the plot according to its fold change (x-axis) and inverted P value (y-axis). Dot color intensity represents increasing fold change levels. The total amount of significantly overexpressed and underexpressed proteins (adjusted P < .05 and abs[fold change (FC)] > 0.5) is written in numbers above the axis. The most extreme proteins (maximum 12, dependent on maximum > FC rank) of each genotype are labeled. (E) Expression boxplots of significantly differentially expressed single SRP complex constituents (a-e) and ELANE (f) in SCN genotypes and healthy donor (x-axis). Differential expression was first evaluated using ANOVA (SRP19, P = 2.8e-9 [a]; SRP54, P = 4.9e-6 [b]; SRPRA, P = 6.3e-8 [c]; SRPRB, P = 2.2e-16 [d]; SRP68, P = 2.4e-7 [e]; and ELANE, P = 2.2e-16 [f]) and after t test using healthy donors as control (∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 or not significant). (F-I) Enrichment plots showing significant terms of each group subdivided into function groups. Overexpressed in genotype vs healthy (F), underexpressed in genotype vs healthy (G), overexpressed in SRP vs common SCN (ELANE, HAX1) compared with SRP vs healthy (H), and underexpressed in SRP vs common SCN (ELANE, HAX1) compared with SRP vs healthy (I). Significance threshold for protein inclusion is adjusted P < .05 and abs (FC) > 0.5, threshold of significant term enrichment is P < .05. (J-K) Bubble plot showing total mass (x-axis) and total molecular number (bubble size) of cellular compartments (y-axis) in healthy (green color) and diseased genotypes (other colors). Panel J shows main cellular compartments, whereas panel K shows the neutrophil granule subsets. (L) Hydrophobicity scores (y-axis) using amino acid hydrophobicity values of the last 9 N-terminal amino acids. Inclusion criteria per genotype (vs healthy donor, x-axis) was adjusted P < .05 and abs (FC) > 1. Each dot represents a protein, the violin plot shows the distribution of proteins with equal scores. NMD, nonsense-mediated decay. Abs, absolute.
Proteome analysis of patient-derived primary neutrophil granulocytes. (A) Principal component analysis showing PC1 and PC2 of neutrophil granulocyte proteomes. Each dot represents a sample and each color represents a genotype. (B) Principal component analysis showing PC1 and PC2 of neutrophil granulocyte proteomes with k-means cluster results. Each dot represents a sample, each color represents the cluster assignment to k1 or k2. Genotypes are shown as labels. (C) Hierarchical dendrogram showing ward-based agglomerative coefficient clustering of neutropenia samples with k = 2. Each tree leaf represents a single sample with the 2 clusters framed in red and green. (D) Volcano plots showing differentially expressed proteins of the comparisons SCN genotypes vs healthy donor (hd) (a-e) and SRP vs SCN (f) (ELANE/HAX1). Each dot represents a protein and is located in the plot according to its fold change (x-axis) and inverted P value (y-axis). Dot color intensity represents increasing fold change levels. The total amount of significantly overexpressed and underexpressed proteins (adjusted P < .05 and abs[fold change (FC)] > 0.5) is written in numbers above the axis. The most extreme proteins (maximum 12, dependent on maximum > FC rank) of each genotype are labeled. (E) Expression boxplots of significantly differentially expressed single SRP complex constituents (a-e) and ELANE (f) in SCN genotypes and healthy donor (x-axis). Differential expression was first evaluated using ANOVA (SRP19, P = 2.8e-9 [a]; SRP54, P = 4.9e-6 [b]; SRPRA, P = 6.3e-8 [c]; SRPRB, P = 2.2e-16 [d]; SRP68, P = 2.4e-7 [e]; and ELANE, P = 2.2e-16 [f]) and after t test using healthy donors as control (∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 or not significant). (F-I) Enrichment plots showing significant terms of each group subdivided into function groups. Overexpressed in genotype vs healthy (F), underexpressed in genotype vs healthy (G), overexpressed in SRP vs common SCN (ELANE, HAX1) compared with SRP vs healthy (H), and underexpressed in SRP vs common SCN (ELANE, HAX1) compared with SRP vs healthy (I). Significance threshold for protein inclusion is adjusted P < .05 and abs (FC) > 0.5, threshold of significant term enrichment is P < .05. (J-K) Bubble plot showing total mass (x-axis) and total molecular number (bubble size) of cellular compartments (y-axis) in healthy (green color) and diseased genotypes (other colors). Panel J shows main cellular compartments, whereas panel K shows the neutrophil granule subsets. (L) Hydrophobicity scores (y-axis) using amino acid hydrophobicity values of the last 9 N-terminal amino acids. Inclusion criteria per genotype (vs healthy donor, x-axis) was adjusted P < .05 and abs (FC) > 1. Each dot represents a protein, the violin plot shows the distribution of proteins with equal scores. NMD, nonsense-mediated decay. Abs, absolute.
To further quantify proteome aberrations, we performed differential expression analysis comparing patient-specific genotypes to healthy donors (Figure 3Da-e) as well as the ELANE/HAX1 (SCN) cluster and the SRP cluster (Figure 3Df). SRPRA and SRP54 showed the largest fraction of differentially expressed proteins. SRPRA-mutated neutrophil granulocytes had 631 proteins showing decreased abundance and 549 proteins showing increased abundance. Neutrophil granulocytes with mutations in SRP54 (down 673/up 570) and SRP19 (down 410/up 408) also revealed markedly disturbed proteome profiles. By contrast, the changes in protein expression in HAX1-deficient neutrophils (down 384/up 375) and ELANE-mutated neutrophils (down 352/up 305) appeared less prominent.
Proteins showing less abundant expression in SRP genotypes were CRISP3, PTX3, LILRB1, CAMP, and MMP9. Next, we analyzed the expression levels of the respective gene products in our proteome data set. Only SRP19 mutant neutrophil granulocytes showed a reduced protein abundance of SRP19, SRP54, and SRP68, whereas all other genotypes (SRPRA, SRP54, ELANE, and HAX1) showed an increased abundance of SRP proteins (Figure 3E). Neutrophil elastase protein levels were reduced in all genotypes (Figure 3Ef).
Gene ontology (GO) term enrichment analysis revealed that regardless of the specific phenotype, numerous subcellular compartments, pathways, and cellular functions are affected (Figure 3F-G). Whereas granule proteins showed reduced expression levels (Figure 3I), mitochondria, the translational apparatus, ROBO receptor signaling, ER stress, and NMD showed increased expression levels (Figure 3H). No genotype-specific GO term signature could be identified.
Next, we annotated every protein to its specific location by unifying information from uniprot, GO, and the human protein atlas in addition to previously published results on the proteome of specific neutrophil compartments,25,26 and made use of a proteome ruler approach27 to derive the total protein number and mass for each cellular compartment in patient and healthy donor neutrophils (Figure 3J-K).
This cellular compartment–specific view confirmed that protein expression level differences between neutrophils from patients are most striking in the granule compartment, whereas differences in other subcellular compartments were less pronounced (Figure 3J). Specifically, SRP genotypes were characterized by a marked reduction of primary granule proteins when compared with HAX1- and ELANE-mutated neutrophil granulocytes (Figure 3K).
Because SRP-deficient yeast cells are characterized by reduced expression of proteins with strongly hydrophobic N-terminals,28 we analyzed the N-terminal hydrophobicity scores of the underexpressed proteins in all genotypes (Figure 3L; supplemental Figure 6 B-C). In contrast to yeast, we could not document a specific underrepresentation of proteins with hydrophobic N-terminal domains in neutrophil proteomes from patients with SRP mutations.
Functional validation of proteomic studies
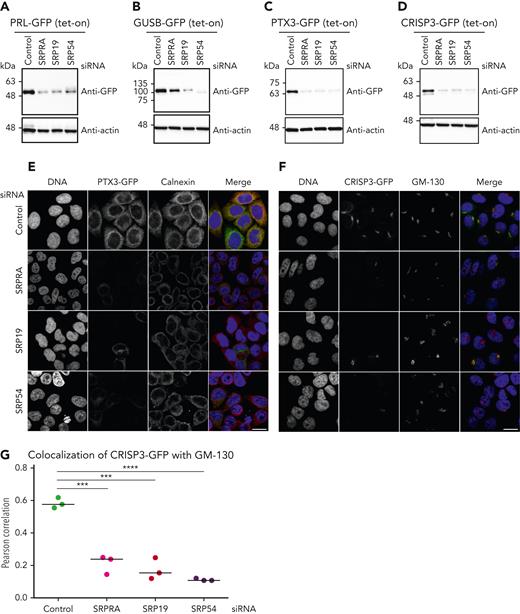
To provide functional proof that decreased expression of proteins in SRP/SRPRA-mutant neutrophil granulocytes is indeed a consequence of aberrant posttranscriptional protein dynamics, we generated a series of tet-responsive HeLa cell lines, allowing us to control the expression kinetics of newly synthesized GFP fusion proteins (HeLa-Flp-In/T-Rex)29 (supplemental Figure 7A). Upon tetracycline induction, reporters were expressed in their mature forms, as indicated by the glycosylated form of CRISP3 (supplemental Figure 7A, marked with an asterisk). We treated reporter cells with small interfering RNAs directed against SRPRA, SRP19 and SRP54 (supplemental Figure 7B-D) and subsequently induced expression of transgenic reporter constructs to monitor the effect of SRP depletion on the maturation of newly synthesized secretory proteins (Figure 4A-D). Preprolactin, a secretory protein containing a prototypical signal sequence, served as control. In accordance with previous studies,18,30 prolactin was expressed in its mature form in control cells (Figure 4A; supplemental Figure 7A), but was not appropriately processed in cells depleted for SRPRA, SRP19, and SRP54 (Figure 4A). Similarly, expression of newly synthesized GUSB-GFP and PTX3-GFP expression was markedly reduced upon knock down of SRP components (Figure 4B-C). We also studied CRISP3, a cysteine-rich secretory glycoprotein stored in secondary granules.31,32 As shown in Figure 4D, CRISP3 expressed with reduced abundance in primary neutrophil granulocytes from SRP-deficient patients (Figure 3B-C) was also decreased upon knock down of SRPRA, SRP19, or SRP54 in HeLa cells. The expression of the nuclear pore complex constituent GFP-Nup53 was not affected upon SRP depletion (supplemental Figure 7E). Finally, we studied the SRP dependency of intracellular trafficking of PTX3 and CRISP3 fusion proteins. The PTX3-GFP fusion protein was enriched throughout the entire ER with partial colocalization with calnexin in control cells (Figure 4E), whereas it was markedly reduced upon knock down of SRPRA, SRP19, or SRP54. In control cells, CRISP3-GFP accumulated in close proximity to the Golgi, as evidenced by colocalization with the cis-Golgi matrix protein GM130,33 however, siRNA-mediated depletion of SRPRA, SRP19, and SRP54 prevented proper trafficking (Figure 4F-G). Thus, these data indicate that the SRP is essential for targeting, processing, and distribution of the granule proteins PTX3 and CRISP3.
Maturation, processing, and targeting of newly identified SRP-dependent proteins (de novo expression). (A-D) Immunoblot analysis of HeLa cell lysates expressing tetracycline-induced PRL-GFP, GUSB-GFP, PTX-GFP, and CRISP-GFP. Cells were treated with control or SRP small interfering RNAs (siRNAs) and processed for immunoblotting with the indicated antibodies. Experiment performed in triplicate. (E) Confocal microscopy images of HeLa cells depleted of endogenous SRP proteins by siRNA and stably expressing PTX3-GFP. Cells were immunostained with an anticalnexin antibody, and DNA was visualized with 4′,6-diamidino-2-phenylindole (DAPI). Scale bar, 10 μm. Experiment performed in triplicate. (F) Confocal images of HeLa cells depleted of endogenous SRPs by siRNA and stably expressing CRISP3-GFP. Cells were immunostained with an anti-GM130 antibody and DNA was visualized with DAPI. Scale bar, 10 μm. Experiment performed in triplicate. (G) Quantification of the colocalization of CRISP3-GFP with GM130. Unpaired t test, 2-tailed (P < .0001 and P < .005). Quantitative analysis represents 3 independent experiments.
Maturation, processing, and targeting of newly identified SRP-dependent proteins (de novo expression). (A-D) Immunoblot analysis of HeLa cell lysates expressing tetracycline-induced PRL-GFP, GUSB-GFP, PTX-GFP, and CRISP-GFP. Cells were treated with control or SRP small interfering RNAs (siRNAs) and processed for immunoblotting with the indicated antibodies. Experiment performed in triplicate. (E) Confocal microscopy images of HeLa cells depleted of endogenous SRP proteins by siRNA and stably expressing PTX3-GFP. Cells were immunostained with an anticalnexin antibody, and DNA was visualized with 4′,6-diamidino-2-phenylindole (DAPI). Scale bar, 10 μm. Experiment performed in triplicate. (F) Confocal images of HeLa cells depleted of endogenous SRPs by siRNA and stably expressing CRISP3-GFP. Cells were immunostained with an anti-GM130 antibody and DNA was visualized with DAPI. Scale bar, 10 μm. Experiment performed in triplicate. (G) Quantification of the colocalization of CRISP3-GFP with GM130. Unpaired t test, 2-tailed (P < .0001 and P < .005). Quantitative analysis represents 3 independent experiments.
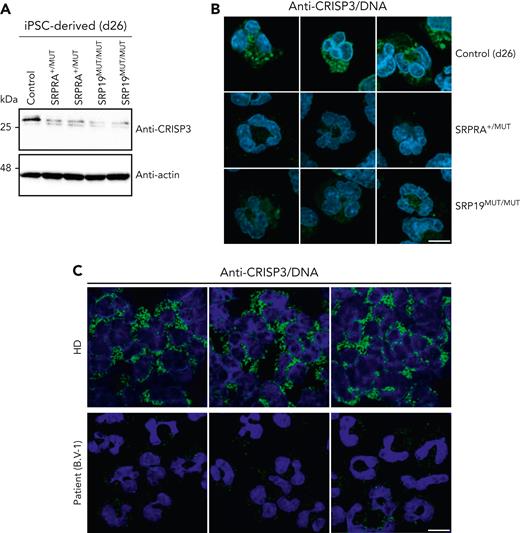
Next, we examined the expression and localization of CRISP3 in neutrophil granulocytes (day 26) derived from control, SRPRA+/MUT, and SRP19MUT/MUT iPSCs.
SRPRA+/MUT and SRP19MUT/MUT neutrophils cells showed a decrease of endogenous CRISP3 expression (Figure 5A-B). Of note, the mature, glycosylated form of CRISP3 was reduced in SRPRA+/MUT and SRP19MUT/MUT neutrophil-like granulocytes, whereas concurrently, the unglycosylated form of CRISP3 was more abundantly expressed (Figure 5A). Notably, a reduction of CRISP3 expression was also observed by immunofluorescence in primary neutrophil granulocytes from the SRP19 patient B.V-1 compared with a healthy donor (Figure 5C).
Expression and localization of CRISP3 in iPSC-derived neutrophil-like granulocytes and in primary cells. (A) Immunoblot analysis of lysates from iPSC-derived control and SRP-mutated iPSCs harvested at differentiation day 26. Cells were harvested at day 26, lysed, and processed for immunoblotting with the indicated antibodies. (B) Confocal images of iPSC-derived control and SRP-mutated cells stained for CRISP3. Cells were immunostained on differentiation day 26 with an anti-CRISP3 antibody and DNA was visualized with DAPI. Scale bar, 10 μm. (C) Confocal images of immunostaining of CRISP3 in a healthy donor and in a patient with the identified SRP19 gene variant. DNA was visualized with DAPI. Scale bar, 10 μm. The experiment in panels A and B was performed in triplicate, whereas the experiment in panel C twice.
Expression and localization of CRISP3 in iPSC-derived neutrophil-like granulocytes and in primary cells. (A) Immunoblot analysis of lysates from iPSC-derived control and SRP-mutated iPSCs harvested at differentiation day 26. Cells were harvested at day 26, lysed, and processed for immunoblotting with the indicated antibodies. (B) Confocal images of iPSC-derived control and SRP-mutated cells stained for CRISP3. Cells were immunostained on differentiation day 26 with an anti-CRISP3 antibody and DNA was visualized with DAPI. Scale bar, 10 μm. (C) Confocal images of immunostaining of CRISP3 in a healthy donor and in a patient with the identified SRP19 gene variant. DNA was visualized with DAPI. Scale bar, 10 μm. The experiment in panels A and B was performed in triplicate, whereas the experiment in panel C twice.
Zebrafish models of SRPRA and SRP19 deficiency
To confirm our observations in an in vivo model, we targeted the zebrafish SRPRA ortholog, srpra in zebrafish embryos by a CRISPR/Cas9 gene editing approach designed to disrupt srpra (supplemental Figure 8A).
F0 crispant embryos displayed severe developmental deformities and high embryonic lethality (Figure 6A-C), which precluded quantitative evaluation of neutrophil abundance. On-target srpra gene editing in these embryos was verified by Sanger sequencing, which demonstrated a complex pattern of assorted gene-edited alleles around the targeted PAM site, confirming that the gene editing approach was efficacious as designed (Figure 6D). To circumvent these general developmental effects of global srpra gene disruption in vivo, we selectively knocked down srpra in neutrophils using our neutrophil-specific gene editing line Tg(mpx:KalTA4) × (UAS:Cas9) × (UAS:NTR-mCherry), in which Cas9 expression is confined to mCherry-marked neutrophils by a myeloperoxidase promoter–driven compound transgenic system34 (Figure 6E). Srpra-gRNA delivery to these embryos significantly reduced neutrophil abundance at 3 dpf by 35% (Figure 6F-G). The mCherry+ neutrophils remaining in these embryos were purified by fluorescence-activated cell sorter for Sanger sequencing (Figure 6H; supplemental Figure 8B), which confirmed on-target gene editing in these residual neutrophils. Next-generation sequencing demonstrated the commonest variant to be an 8-nucleotide deletion (variant allele frequency = 13%), resulting in a frameshift mutation (supplemental Figure 8C). Although the gene editing strategy used provided an opportunity for generating a zebrafish equivalent of the candidate missense disease allele by homology-directed repair following CRISPR/Cas9 editing (supplemental Figure 8A), this did not occur at a detectable frequency in the F0 crispants.
Zebrafish models of SRP component loss-of-function effects on neutrophil abundance. (A) Schematic of global CRISPR/Cas9 srpra knockdown in zebrafish by microinjection of srpra gRNA and Cas9 into 1-cell embryos. (B) Impaired survival of global srpra crispant embryos at 3 days post fertilization (dpf) compared with control noninjected siblings (n = 3 independent experiments; total embryos in each experiment [n control/n crispant]: 179/127, 195/184, and 133/113). Unpaired 2-tailed t test. (C) Representative dysmorphic surviving srpra Tg(mpx:EGFP) crispant embryo at 5 dpf. Panels show bright field (i), rhodamine dextran (tracing reagent delivery) (ii), and enhanced GFP (EGFP) (neutrophil reporter gene) images (iii). (D) Sanger sequencing chromatogram confirming on-target srpa gene editing in global crispants. (i) WT sequence from noninjected control. (ii) Sequence from severely deformed dead embryo, showing multiple superimposed heterogeneous traces starting in the vicinity of the guide RNA (gRNA)–targeted PAM site (red line). (E) Schematic of neutrophil-specific CRISPR/Cas9 srpra knockdown in zebrafish by microinjection of synthetic srpra gRNA into 1-cell mpx-cas9 zebrafish embryos. (F) Depleted trunk neutrophil numbers in mpx-cas9 embryos with neutrophil-specific srpra gene knockdown (pooled embryos from n = 2 experiments, indicated by different color symbols). Unpaired 2-tailed t test. (G) Representative images corresponding to the 2 groups in panel F. White box shows area where trunk neutrophil numbers were enumerated. (H) Sanger sequencing chromatogram confirming on-target srpa gene editing in neutrophils of mpx-cas9 neutrophil-lineage crispants. (i) WT reference sequence from noninjected control. (ii) Sequence from DNA prepared from neutrophils of mpx-cas9 neutrophil-lineage crispants, showing sequence heterogeneity starting in the vicinity of the PAM site (red line). (I) Schematic of global srpr19 knockdown in zebrafish by microinjection of srpra splice-blocking morpholino into 1-cell Tg(mpx:EGFP) zebrafish embryos. (J) Trunk neutrophil numbers in srp19-MO–injected morphants scored at 2 dpf (n = 24-53 embryos per group; experiment 1). One-way ANOVA with Dunnett post hoc test. Corresponding embryo survival rates are shown in supplemental Figure 9E. (K) Trunk neutrophil numbers in srp19-MO–injected morphants scored at 3 dpf (n = 24 embryos per group; experiment 2). One-way ANOVA with Dunnett post hoc test. Corresponding embryo survival rates are shown in supplemental Figure 9F. Ctrl, control; FACS, fluorescence-activated cell sorter; MO, morpholino.
Zebrafish models of SRP component loss-of-function effects on neutrophil abundance. (A) Schematic of global CRISPR/Cas9 srpra knockdown in zebrafish by microinjection of srpra gRNA and Cas9 into 1-cell embryos. (B) Impaired survival of global srpra crispant embryos at 3 days post fertilization (dpf) compared with control noninjected siblings (n = 3 independent experiments; total embryos in each experiment [n control/n crispant]: 179/127, 195/184, and 133/113). Unpaired 2-tailed t test. (C) Representative dysmorphic surviving srpra Tg(mpx:EGFP) crispant embryo at 5 dpf. Panels show bright field (i), rhodamine dextran (tracing reagent delivery) (ii), and enhanced GFP (EGFP) (neutrophil reporter gene) images (iii). (D) Sanger sequencing chromatogram confirming on-target srpa gene editing in global crispants. (i) WT sequence from noninjected control. (ii) Sequence from severely deformed dead embryo, showing multiple superimposed heterogeneous traces starting in the vicinity of the guide RNA (gRNA)–targeted PAM site (red line). (E) Schematic of neutrophil-specific CRISPR/Cas9 srpra knockdown in zebrafish by microinjection of synthetic srpra gRNA into 1-cell mpx-cas9 zebrafish embryos. (F) Depleted trunk neutrophil numbers in mpx-cas9 embryos with neutrophil-specific srpra gene knockdown (pooled embryos from n = 2 experiments, indicated by different color symbols). Unpaired 2-tailed t test. (G) Representative images corresponding to the 2 groups in panel F. White box shows area where trunk neutrophil numbers were enumerated. (H) Sanger sequencing chromatogram confirming on-target srpa gene editing in neutrophils of mpx-cas9 neutrophil-lineage crispants. (i) WT reference sequence from noninjected control. (ii) Sequence from DNA prepared from neutrophils of mpx-cas9 neutrophil-lineage crispants, showing sequence heterogeneity starting in the vicinity of the PAM site (red line). (I) Schematic of global srpr19 knockdown in zebrafish by microinjection of srpra splice-blocking morpholino into 1-cell Tg(mpx:EGFP) zebrafish embryos. (J) Trunk neutrophil numbers in srp19-MO–injected morphants scored at 2 dpf (n = 24-53 embryos per group; experiment 1). One-way ANOVA with Dunnett post hoc test. Corresponding embryo survival rates are shown in supplemental Figure 9E. (K) Trunk neutrophil numbers in srp19-MO–injected morphants scored at 3 dpf (n = 24 embryos per group; experiment 2). One-way ANOVA with Dunnett post hoc test. Corresponding embryo survival rates are shown in supplemental Figure 9F. Ctrl, control; FACS, fluorescence-activated cell sorter; MO, morpholino.
To mimic the biallelic exon 3 splice site mutation of SRP19 found in patients, the exon 3 splice donor site of zebrafish srp19 was targeted by a splice-blocking MO for global disruption of srp19 splicing (Figure 6I; supplemental Figure 9A). On-target srp19-MO action was confirmed by RT-PCR, which demonstrated reduced levels of the PCR product corresponding to the normal transcript at all MO doses (supplemental Figure 9B-C), and also a MO dose–dependent increase in an aberrant PCR product corresponding to intron 3 retention, which was confirmed by Sanger sequencing (supplemental Figure 9D). At the highest MO dose injected, srp19-MO–injected embryos demonstrated mild developmental defects not seen with control MO (absent swim bladder, curved tails, and reduced 2 dpf survival; supplemental Figure 9E-F), but were scorable for neutrophil numbers at all MO doses. srp19-MO–injected embryos had significantly reduced neutrophil numbers in a MO dose–dependent fashion, both at 2 dpf for injectate concentrations of 500 μM (where development and survival were normal) and at 2 and 3 dpf for 1000 uM (where development was perturbed and survival was reduced) (Figure 6J-K).
Collectively, these 2 animal models of transient loss of function support the hypothesis that normal levels of srpra and srp19 function are required for normal granulopoiesis in vivo and add to the genetic evidence that the SRPA and SRP19 mutations in these patients are responsible for their neutropenia.
Discussion
Here, we describe 2 novel human genetic defects in SRP19 and SRPRA, components of the cotranslational targeting machinery. Clinically, both SRP19 and SRPRA deficiencies are characterized by SCN associated with a myeloid maturation arrest resembling the neutrophil phenotype of SRP54 deficiency.11,12
During the initial steps of SRP biogenesis, SRP19 interacts with specific sites on the SRP RNA in the nucleolus and promotes the association of other SRP components. The assembly of this nuclear export competent, pre-SRP is a prerequisite for the binding of SRP54 to helix 8 of the SRP RNA in the cytoplasm (reviewed in Luirink and Sinning35). Consistent with a previous study,36 we observed GFP-SRP19 not only in the cytoplasm where the mature SRP resides, but also in the nucleoplasm and the nucleolus, whereas the identified splice site variant in SRP19 abolished the cytoplasmic localization of the expressed isoform. Moreover, the expression of SRP19 was reduced in patients’ cells.
It could be hypothesized that the identified splice site variant causes a reduction of SRP19 and hence affects the stable formation of the pre-SRP complex or its export into the cytoplasm, as also seen in studies of yeast SRP biogenesis that critically depend on adequate endogenous levels of Sec65p (SRP19 homolog of yeast). Levels of endogenous SRP19 might also be critical for robust core SRP assembly by guiding the efficient binding of SRP54 and its stable association with the SRP.
To study the effects of the SRPRA and SRP19 variants on granulopoiesis, we engineered iPSCs using CRISPR/Cas9 gene editing and thus modeled the monoallelic SRPRA variant (Q464E) and the biallelic splice site variants in SRP19.
iPSCs with variants in either SRPRA and SRP19 or deficient in HAX1 expression had a significantly reduced capacity to differentiate into neutrophil granulocytes compared to WT iPSCs, providing evidence that both SRPRA and SRP19 are required for granulopoiesis. Recently, a comprehensive gene editing strategy in human hematopoietic stem and progenitor cells associated ELANE mutations with their efficiency to restore neutrophil maturation and hematopoietic stem and progenitor cell function.37
The neutrophil differentiation process is tightly linked to the regulation of granule protein synthesis.3 Mass spectrometry–based deep proteome analysis allows the characterization of cellular protein composition in high detail, including neutrophil granulocytes.26,38 Rieckmann et al22 have recently published an ultradeep analysis of peripheral blood neutrophil granulocytes, among other leucocytes. They quantified 6007 proteins in 3 samples, whereas our study quantified 3624 proteins with at least 2 peptides in each of our 68 samples. The overlapping data show a Pearson correlation coefficient of 0.7, indicating a strong correlation between their data and ours (data not shown). We have previously shown that proteome profiles of neutrophil granulocytes promise to help unravel the effects and specificities of monogenic defects. Here, we provide the first comprehensive proteomic analysis of neutrophil granulocytes from patients with variants in SRP/SRPRA, ELANE, and HAX1.
Independent of the genetic variant, all neutrophil granulocytes were characterized by strong imbalances in their proteome. Whereas proteins of the translational apparatus, mitochondrial proteins, and stress response proteins (ER stress and NMD) were more abundantly expressed in patient-derived neutrophils, proteins from all granule subsets were less abundantly expressed. These global changes may reflect incomplete terminal differentiation and/or cellular adaption to the consequences of the underlying mutation (eg, unfolded protein response to variants in neutrophil elastase39). Even though no pathognomonic proteome aberrations could be established, our proteome analysis revealed a pronounced loss of proteins of neutrophil granules, including CRISP3 and PTX3. Proper SRP function might emerge as a rate-limiting step to secure timely and efficient production of defined granule proteins, a prerequisite for physiological neutrophil maturation.
A requirement for intact SRP function to sustain normal granulopoiesis in larval zebrafish in vivo has been established in studies examining the global genetic requirement of srp54 in zebrafish development11,14 and modeling specific SRP54 disease alleles.14 We, therefore, used zebrafish granulopoiesis as a bioassay to evaluate the requirement for srpra and srp19 in neutrophil production. Crispants of the srpra locus (with assorted disruptive gene edits) phenocopied the embryonic lethality of the zebrafish srp54 knockout.14 To circumvent this, we gene-edited srpra in a neutrophil-lineage restricted manner using our system that limits Cas9 expression to maturing neutrophils using the myeloperoxidase promoter.34 The neutrophil deficiency of these lineage-specific crispants demonstrated a requirement for normal srpra function in granulopoiesis in vivo. Similarly, srp19 morphants with confirmed intron 3 retention (predicted to result in a nonsense transcript) confirmed a requirement for normal srp19 splicing in sustaining normal granulopoiesis in vivo. Although these strategies did not replicate the exact disease allele variants, these data represent a formal reverse genetic test of gene requirement in vivo and functionally support the candidacy of SRPRA and SRP19 as SCN disease genes. Collectively, these observations are consistent with mutation of SRP54 and several other SRP components being causative of an SCN disease phenotype.
Acknowledgments
The authors thank all the patients and their families for allowing us to study their cells and all clinical staff for taking excellent care. The authors are particularly indebted to Sorin Iurian (Romania) for referring family A. The authors acknowledge Sebastian Hollizeck and Isabelle Plonner for outstanding expertise in the C4R sequencing and computational facility. The authors are very grateful to Thomas U. Mayer for providing the HeLa-Flp-In/T-Rex cell line. The authors thank Tanja Vlaovic for isolating neutrophils from healthy volunteers and Diana Laverde for her guidance in performing the phagocytosis assay. The authors acknowledge Manfred Rohde (Department of Medical Microbiology, Helmholtz Centre for Infection Research, Braunschweig, Germany) for the acquisition and processing of electron microscopy studies. The authors are very grateful to the Pediatric Radiology team at Ludwig-Maximilians-Universität for acquisition of the computed tomography scan. The authors thank Lucy Fox and Ella Thompson for the help with the zebrafish next-generation sequencing analysis.
This work was supported by grants from the DFG, Gottfried Wilhelm Leibniz Program CRC914 (project A08 and A02); the TRR332 (project B1); the BMBF (PIDNET); and the Care-for-Rare Foundation. The Peter MacCallum Cancer Centre Molecular Hematology Laboratory is supported by funding from Wilson Centre for Lymphoma Genomics through the Snowdome Foundation. The Australian Regenerative Medicine Institute is supported by grants from the State Government of Victoria and the Australian Government. This work was supported by grants from the National Health and Medical Research Council (1044754, 1086020, and 1159278) (G.J.L.) and Maddie Riewoldt’s Vision (ARMI-MRV-2018G) (G.J.L., C.K., and P.B.). Members of the Lieschke Laboratory are supported by grants of the Monash University (Graduate Scholarship and International Tuition Scholarship). Whole-genome sequencing on family A was supported by the Junior Researcher Fund of Ludwig-Maximilians-Universität Excellence Initiative (N.Z.).
Authorship
Contribution: C.K. conceived the original idea; C.K., M.I.L., Y.M., S.H., and R.Z. conceptualized and developed the methodology; C.K., S.H., Z.A., M.K., N.R., E.U., and F.H. performed clinical and patient-related investigations and performed the experiments; S.H., G.C., K.A., T.J., M.H., and R.Z. performed computational analysis; M.S., V.P., A.I.I., B.B.M., P.B. and G.J.L. conceptualized and performed the zebrafish experiments; M.D., B.W., J.R., R.Z., C.K.; P.B., and G.J.L. provided resources; C.G., M.R., R.Z., and M.S. curated data; C.K., M.I.L., S.H., Y.M., and G.J.L. wrote the original draft; C.K., B.W., V.P., R.Z., J.R. and G.J.L. provided supervision; and all authors wrote, reviewed and edited the draft and read and approved the manuscript before submission.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.L. is the Department of Pediatrics and Adolescent Medicine, University Medical Center Ulm, Ulm, Baden-Württemberg, Germany.
The current affiliation for N.Z. is Cancer Immunology and Immune Modulation, Boehringer Ingelheim Pharma GmbH & Co KG, Biberach an der Riss, Baden-Württemberg, Germany.
Correspondence: Christoph Klein, Department of Pediatrics, Dr. von Hauner Children’s Hospital, Ludwig-Maximilians-Universität München, Lindwurmstraße 4, 80337 Munich, Germany; e-mail: christoph.klein@med.uni-muenchen.de.
References
Author notes
Data are available on request from the corresponding author, Christoph Klein (christoph.klein@med.uni-muenchen.de).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
M.I.L., Y.M., and S.H. contributed equally to this study.

![Identification of SRPRA and SRP19 novel gene variants. (A) Chest computed tomography scan of index patient II-3 (family A). (B) Transelectron microscopy sections of neutrophils from index patient A.II-3 compared with unaffected family members (brother A.II-2, father A.I-2) and healthy donors. Non-adherent Neutrophils have a size of ca. 9 μm. (C) Quantification of neutrophil granule content from healthy donor, A.I-2, A.II-2, and A.II-3. Group differences with P < .0001. Single-group differences via Student t test. ∗P < .01, ∗∗P < .001, and ∗∗∗∗P < .00001. (D) Pedigree of family A and Sanger sequencing chromatograms of wild-type (WT) and the SRPRA mutation site. Ribbon representation of the 3-dimensional structure of the human SRPRA WT (E) and mutated SRPRA (Q464E) (F). (G) Pedigree of family B and Sanger sequencing chromatograms of the WT and the SRP19 mutation site. (H) Reverse transcription polymerase chain reaction (RT-PCR) results documenting WT and mutated (mut) SRP19_2 to 4 minigene transcripts in HeLa cells. F1-R1 and F1-R2 are the primer pairs used to amplify the indicated SRP19 exons. The primer pair F1-R2 amplifies SRP19 exon 2 to 4 as indicated with a red arrow (281 base pair [bp]). The SRP19 variant results in a PCR product of only 209 base pair (blue arrow). The primer pair F1-R1 amplifies parts of the SRP19 exon 2 and is used as a transfection efficiency control. (I) Immunoblot analysis of EBV-LCL lysates from healthy donor, patient (Pat.) B.V-1, and a heterozygous family (fam.) member (B.IV-6). Experiment performed in triplicate.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/6/10.1182_blood.2022016783/6/m_blood_bld-2022-016783-gr1.jpeg?Expires=1770990329&Signature=pJQ8Fb6RyA0hGq9OVkre7KAPfvP5lpXzNytj4Hbc-MrdmeM5gIgFL0HmM78hcbdW3hpnG7k9bh8NHKHcEVkqqT26H3ZqIFbC8P2d3pBZkea6MWSgVxK3nzx8re-~sjSYq7nstbF-JP~~NkFTi~vovucUpuByeAuJpf46s94mNP4DCF3NKt9Ur5s2Lg1bNxhykVpw9~Njm35iCKAHM3gpT1bE~nb2J6poBa57HTg4suO2k-VodNr6I~YVvga3veioQ5Epu5Ii4utFpd6zbzKQkw9Ag7sfhvxyu2yIT~4QkohiShmXK35yH7R6b7rxU6D3qdhziJAgd5vxAthIS5rpBQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Zebrafish models of SRP component loss-of-function effects on neutrophil abundance. (A) Schematic of global CRISPR/Cas9 srpra knockdown in zebrafish by microinjection of srpra gRNA and Cas9 into 1-cell embryos. (B) Impaired survival of global srpra crispant embryos at 3 days post fertilization (dpf) compared with control noninjected siblings (n = 3 independent experiments; total embryos in each experiment [n control/n crispant]: 179/127, 195/184, and 133/113). Unpaired 2-tailed t test. (C) Representative dysmorphic surviving srpra Tg(mpx:EGFP) crispant embryo at 5 dpf. Panels show bright field (i), rhodamine dextran (tracing reagent delivery) (ii), and enhanced GFP (EGFP) (neutrophil reporter gene) images (iii). (D) Sanger sequencing chromatogram confirming on-target srpa gene editing in global crispants. (i) WT sequence from noninjected control. (ii) Sequence from severely deformed dead embryo, showing multiple superimposed heterogeneous traces starting in the vicinity of the guide RNA (gRNA)–targeted PAM site (red line). (E) Schematic of neutrophil-specific CRISPR/Cas9 srpra knockdown in zebrafish by microinjection of synthetic srpra gRNA into 1-cell mpx-cas9 zebrafish embryos. (F) Depleted trunk neutrophil numbers in mpx-cas9 embryos with neutrophil-specific srpra gene knockdown (pooled embryos from n = 2 experiments, indicated by different color symbols). Unpaired 2-tailed t test. (G) Representative images corresponding to the 2 groups in panel F. White box shows area where trunk neutrophil numbers were enumerated. (H) Sanger sequencing chromatogram confirming on-target srpa gene editing in neutrophils of mpx-cas9 neutrophil-lineage crispants. (i) WT reference sequence from noninjected control. (ii) Sequence from DNA prepared from neutrophils of mpx-cas9 neutrophil-lineage crispants, showing sequence heterogeneity starting in the vicinity of the PAM site (red line). (I) Schematic of global srpr19 knockdown in zebrafish by microinjection of srpra splice-blocking morpholino into 1-cell Tg(mpx:EGFP) zebrafish embryos. (J) Trunk neutrophil numbers in srp19-MO–injected morphants scored at 2 dpf (n = 24-53 embryos per group; experiment 1). One-way ANOVA with Dunnett post hoc test. Corresponding embryo survival rates are shown in supplemental Figure 9E. (K) Trunk neutrophil numbers in srp19-MO–injected morphants scored at 3 dpf (n = 24 embryos per group; experiment 2). One-way ANOVA with Dunnett post hoc test. Corresponding embryo survival rates are shown in supplemental Figure 9F. Ctrl, control; FACS, fluorescence-activated cell sorter; MO, morpholino.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/6/10.1182_blood.2022016783/6/m_blood_bld-2022-016783-gr6.jpeg?Expires=1770990329&Signature=a8FHnukVwlNTn2n-sBY-eMbmGdKaYxZ7j3oHzPwQGpyED1XlUIvPfLAoMRfW80Nw5jqQqwMhSTMGGvC5vkD7qxtY0LAFQpWmTnnfobGupVzVKayTQ7Matx5rpCLeOOI3Ff7iWOAQ-i-BrsPTN-tSWGklNiGQqi9s4bYnpG0uk2eO8LXjpFTa1YN4pwYvEf3OV~vSDTpwwhOCYwV8srClS7KhV~UPGff-R7g465OkYGHfcZdF2Cbd9wLIZM9Lm9ADcrhV-e4zdcplxSkgx1kLMAJozCth6FJ71SMA7aneNrRBRynwfg7qpXY7F9LelW1ixLuTaF7dkRjsufylQel7qA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal