In this issue of Blood, Hengeveld et al describe an openly available, highly sensitive next-generation sequencing approach developed to track minimal residual disease (MRD) in chronic lymphocytic leukemia (CLL).1 This tool improved patients’ outcome prediction in comparison with the traditionally available MRD tools.
Allele-specific oligonucleotide quantitative polymerase chain reaction (ASO-qPCR) targeting immunoglobulin (IG) gene rearrangements has been for years the criterion standard for monitoring minimal residual disease (MRD) in several lymphoproliferative diseases. Its wide use relied on strict standardization among centralized laboratories by defining common strategies for patient-specific assay designs and strict guidelines for data analysis.2 This standardization process was undertaken by the international EuroMRD group, which since 2001 has been coordinating quality control rounds twice a year among the 69 MRD-PCR laboratories spread across 26 countries in Europe, Asia, Australia, and North and South America (www.euromrd.org).
However, while ASO-qPCR has considerable sensitivity (up to 10−5, which means detection of 1 clonal cell out of 100 000 analyzed cells) it also has several shortcomings, specifically, technical complexity, labor-intensiveness, and the need for patient-tailored primer design. Next-generation sequencing (NGS) technology is currently used to overcome the limitations of MRD ASO-qPCR by using high-throughput consensus primers and deeper analysis, able to achieve superimposable sensitivity levels as the gold standard method.3 Recently, a company-dependent IG-based NGS platform (ClonoSEQ) was developed for MRD monitoring and risk stratification in lymphoproliferative disorders, which claimed a sensitivity up to 10−6, a claim that has been questioned. Several papers have extolled the potential advantage of the approach but ignored the large DNA input required to achieve such a cutoff.4-6 The ClonoSEQ platform has recently received regulatory approval for the detection of MRD in acute lymphoblastic leukemia, multiple myeloma, and CLL. However, this approach is available only commercially, so the protocols and primer sequences are not at the disposal of the scientific community.7 Alternatively, many research groups have implemented different NGS-based approaches for MRD: for example, international development and standardization efforts in this field are ongoing within the EuroClonality-NGS working group.8
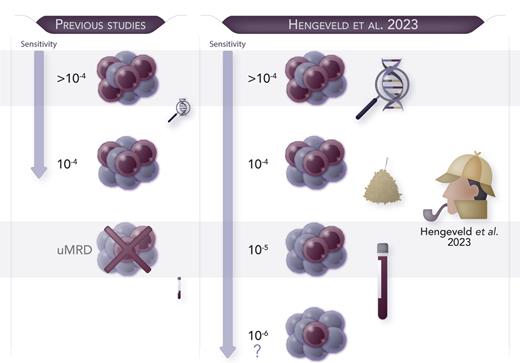
In this issue of Blood, Hengeveld et al address several unresolved issues in this field. Starting from the EuroClonality-NGS experience, they showed the feasibility of an academically developed, IG-based NGS assay for the quantification of MRD in CLL. They demonstrated that a 10−5 MRD cutoff was easily achievable using a considerable DNA amount (8 μg).1 Notably, a linear range of MRD monitoring beyond this threshold was associated with the most favorable progression-free survival (PFS) and time to next treatment in the context of the CLL11 trial (clinicaltrials.gov, number NTC01010061). A new patient group in this trial was identified, characterized by intermediate MRD levels (between 10−5 and 10−4) and intermediate PFS risk, thus supporting the clinical utility of this high-throughput assay for refining patients’ prognostic stratification. Of note, the authors also demonstrated concordance with standardized ASO-qPCR and droplet digital PCR results, an alternative technique largely used for MRD in lymphoproliferative diseases.9 Interestingly, the authors demonstrated that further MRD monitoring down to 10−6 is theoretically possible only by increasing the input amount of DNA up to 27 μg, a quantity rarely achievable in clinical practice (see figure). Thus, inevitably, the feasibility of very deep MRD quantification is conditional on the availability of a large amount of input DNA and carries significant financial costs.
Hengeveld et al developed an academic NGS-based MRD approach “to find a needle in the haystack of CLL.” This highly sensitive tool is able to identify 1 tumoral cell among 100 000 healthy cells and may reach even deeper sensitivity if adequate amount of DNA is provided. uMRD, undetectable MRD. Professional illustration by Somersault18:24.
Hengeveld et al developed an academic NGS-based MRD approach “to find a needle in the haystack of CLL.” This highly sensitive tool is able to identify 1 tumoral cell among 100 000 healthy cells and may reach even deeper sensitivity if adequate amount of DNA is provided. uMRD, undetectable MRD. Professional illustration by Somersault18:24.
Overall, the performance of Hengeveld et al’s described tool is similar to that of the ClonoSEQ platform, but the authors committed to open publication of their protocols and primer sequences. This will allow for implementation at multiple centers and includes the ability to have interlaboratory proficiency testing to ensure stringent quality control. Moreover, the employment of Leader IG primers, allowing sequencing of the entire variable region, increases the feasibility of this NGS assay, for both IG heavy-chain variable region mutational status determination and MRD monitoring, further optimizing a single general workflow for CLL patients.
This paper highlights the importance of supporting the development of collaborative academic translational research within large prospective clinical trials. It supports the free and open availability of protocols and open-source bioinformatics tools to improve NGS implementation at multiple centers. International collaborations, as represented by the EuroClonality-NGS working group, are essential to building interlaboratory standardization networks, and to integrating both wet lab and bioinformatics procedures to promote feasible, robust, and clinically meaningful MRD monitoring tools.10 In our opinion, such efforts are the optimal way to promote MRD tracking with the use of NGS as a reliable primary objective for use in large prospective clinical trials in CLL and other hematologic diseases.
Conflict-of-interest disclosure: S.F. reports consultancy and advisory board participation for and speaker honoraria and research funding from Janssen; consultancy and advisory board participation for and speaker honoraria from EUSA Pharma; research funding from Gilead and Morphosys; advisory board participation for Incyte and Clinigen; and speaker honoraria from Servier and Gentili. E.G. reports speaker honoraria from Werfen.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal