Abstract
BCR::ABL1-negative myeloproliferative neoplasms (MPNs) are clonal diseases originating from a single hematopoietic stem cell that cause excessive production of mature blood cells. The 3 subtypes, that is, polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF), are diagnosed according to the World Health Organization (WHO) and international consensus classification (ICC) criteria. Acquired gain-of-function mutations in 1 of 3 disease driver genes (JAK2, CALR, and MPL) are the causative events that can alone initiate and promote MPN disease without requiring additional cooperating mutations. JAK2-p.V617F is present in >95% of PV patients, and also in about half of the patients with ET or PMF. ET and PMF are also caused by mutations in CALR or MPL. In ∼10% of MPN patients, those referred to as being “triple negative,” none of the known driver gene mutations can be detected. The common theme between the 3 driver gene mutations and triple-negative MPN is that the Janus kinase–signal transducer and activator of transcription (JAK/STAT) signaling pathway is constitutively activated. We review the recent advances in our understanding of the early events after the acquisition of a driver gene mutation. The limiting factor that determines the frequency at which MPN disease develops with a long latency is not the acquisition of driver gene mutations, but rather the expansion of the clone. Factors that control the conversion from clonal hematopoiesis to MPN disease include inherited predisposition, presence of additional mutations, and inflammation. The full extent of knowledge of the mutational landscape in individual MPN patients is now increasingly being used to predict outcome and chose the optimal therapy.
Introduction
The clonal origin of myeloproliferative neoplasms (MPNs) had already been recognized in 1976 using X-chromosome inactivation patterns in the peripheral blood of female MPN patients,1 and it was later firmly established using somatic gene mutations as the clonal markers.2JAK2-p.V617F is detectable in purified human hematopoietic stem cells (HSCs),3 and the stem cell origin of MPNs was demonstrated experimentally in mouse models of MPN disease. by transplanting a single HSC that expresses JAK2-p.V617F into recipient mice, which then developed MPN disease.4 Recent studies using whole-genome sequencing (WGS) of DNA from single hematopoietic colonies confirmed the single-HSC origin in human MPN disease and established a timeline for the preclinical initiation and expansion of the JAK2-mutant clone (Figure 1).5,6 With the use of next-generation sequencing (NGS), our knowledge of the mutational landscape in MPN disease has greatly improved, and we now have an advanced understanding of the genetic basis of MPNs. Until recently, the risk factors used to predict prognosis in MPN disease were mostly based on clinical parameters, but information on the mutational landscape of MPNs is now increasingly being added to improve and individualize the prognostic scores.
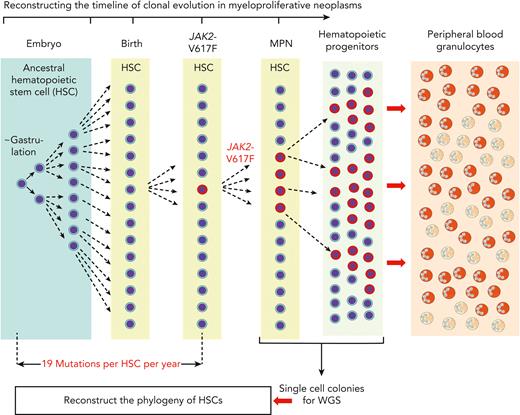
Reconstructing the timeline of clonal evolution in myeloproliferative neoplasms. Schematic drawing of the timeline (not to scale) and the different hematopoietic compartments during the clonal evolution of MPNs caused by JAK2-p.V617F. Starting in embryogenesis with the first division of an ancestral HSC, the daughter cells acquire a number of mutations in their genomes that can be used as markers to distinguish them from all other cells that underwent cell division. New mutations are added during each of the next cell divisions, and by comparing the sequence similarities and differences of individual HSCs, a phylogenetic tree can be reconstructed. This analysis relies on taking bone marrow cells after MPN has been diagnosed and depositing single HSCs or progenitor cells into wells where they can be expanded in liquid culture to obtain single cell–derived colonies. DNA from each of these colonies is then analyzed by WGS. By comparing the sequence similarities and differences of individual HSCs, a phylogenetic tree can be reconstructed that originates in a common ancestor HSC that first divided at the time of gastrulation. The estimate of the time when JAK2-p.V617F mutation was acquired is calculated by assuming a constant mutational rate of 19 mutations per HSC per year. This estimate is not confounded by the increased cell division rate of JAK2-p.V617F mutant HSCs, because only sequence alterations that occurred before JAK2-p.V617F was acquired were used for deriving the estimate.
Reconstructing the timeline of clonal evolution in myeloproliferative neoplasms. Schematic drawing of the timeline (not to scale) and the different hematopoietic compartments during the clonal evolution of MPNs caused by JAK2-p.V617F. Starting in embryogenesis with the first division of an ancestral HSC, the daughter cells acquire a number of mutations in their genomes that can be used as markers to distinguish them from all other cells that underwent cell division. New mutations are added during each of the next cell divisions, and by comparing the sequence similarities and differences of individual HSCs, a phylogenetic tree can be reconstructed. This analysis relies on taking bone marrow cells after MPN has been diagnosed and depositing single HSCs or progenitor cells into wells where they can be expanded in liquid culture to obtain single cell–derived colonies. DNA from each of these colonies is then analyzed by WGS. By comparing the sequence similarities and differences of individual HSCs, a phylogenetic tree can be reconstructed that originates in a common ancestor HSC that first divided at the time of gastrulation. The estimate of the time when JAK2-p.V617F mutation was acquired is calculated by assuming a constant mutational rate of 19 mutations per HSC per year. This estimate is not confounded by the increased cell division rate of JAK2-p.V617F mutant HSCs, because only sequence alterations that occurred before JAK2-p.V617F was acquired were used for deriving the estimate.
Genetic basis and clonal evolution of MPNs
A large number of genes carrying mutations in hematopoietic cells from MPN patients have been identified by targeted NGS and whole exome sequencing (WES; Table 1). No general consensus has been reached regarding the nomenclature that should be used to classify these mutations, although the distinction between “driver” vs “passenger” mutations is widely used. In the context of MPNs, we propose to further distinguish between “disease driver” mutations and “clonal driver” mutations.
Recurrent somatic gene mutations in myeloproliferative neoplasms (MPNs) and secondary acute myeloid leukemia (AML)
| Gene . | Function . | Location and type of mutation . | Frequency, % . | Clinical impact . | |||
|---|---|---|---|---|---|---|---|
| PV . | ET . | PMF . | Post-MPN AML . | ||||
| Disease driver mutations | |||||||
| JAK2 | JAK-STAT signaling | V617F or exon 12 | 98% | 55% | 60% | — | WHO/ICC criterion for MPN diagnosis |
| MPL | TPO receptor JAK-STAT signaling | Exon 10 | 0% | 5%-7% | 7%-10% | — | WHO/ICC criterion for MPN diagnosis |
| CALR | Chaperone protein CALR-mut binds and activates MPL | Frameshift in exon 9 | 0% | 25%-30% | 20%-30% | — | WHO/ICC criterion for MPN diagnosis |
| Clonal driver mutations | |||||||
| TET2 | Epigenetic regulation | All exons | 10%-20% | 3%-10% | 10%-20% | 19%-25% | No prognostic impact reported |
| DNMT3A | Epigenetic regulation | R882 and all exons | 5%-10% | 1%-5% | 8%-12% | 3%-17% | No prognostic impact reported |
| IDH1 | Epigenetic regulation | R132 | 1%-2% | 1%-2% | 5%-6% | 13% | HMR in PMF and adverse prognostic impact in all MPN subtypes |
| IDH2 | Epigenetic regulation | R140 or R172 | 1%-2% | 1%-2% | 5%-6% | 7%-15% | HMR in PMF and adverse prognostic impact in all MPN subtypes |
| ASXL1 | Epigenetic regulation | Mostly nonsense / frameshift in the last exon | 2%-7% | 5%-10% | 15%-35% | 17%-47% | HMR in PMF and adverse prognostic impact in all MPN subtypes |
| EZH2 | Epigenetic regulation | All exons | 1%-2% | 1%-2% | 7%-10% | 7%-13% | HMR in PMF and adverse prognostic impact in all MPN subtypes |
| NRAS | ERK/MAPK signaling | G12, G13, or Q61 | <2% | <2% | 2%-4% | 11% | Adverse prognostic impact in all MPN subtypes |
| KRAS | ERK/MAPK signaling | G12, G13, or Q61 | <2% | <2% | 2% | 4%-7% | Adverse prognostic impact in all MPN subtypes |
| SH2B3 | JAK signaling regulation | Exon 2 | 2%-9% | 1%-3% | 2%-4% | 6%-11% | Rare mutations in JAK2-negative MPNs |
| CBL | JAK signaling regulation | Exons 8 and 9 | <2% | <2% | 4% | 4% | Adverse prognostic impact in all MPN subtypes |
| SRSF2 | mRNA splicing | P95 | <2% | <2% | 6%-14% | 7%-22% | HMR in PMF and adverse prognostic impact in all MPN subtypes |
| SF3B1 | mRNA splicing | Exon 14-16 | 2%-3% | 2%-5% | 5%-7% | 7%-11% | Adverse prognostic impact in ET |
| U2AF1 | mRNA splicing | S34 or Q157 | <2% | <2% | 7%-10% | 5%-12% | Adverse prognostic impact in all MPN subtypes |
| NFE2 | Transcriptional factor | All exons | 3%,6% | 1%-7% | 3%-5% | ? | Increased risk of leukemic transformation |
| RUNX1 | Transcriptional factor | All exons | <2% | <2% | 2%-3% | 20% | Adverse prognostic impact in all MPN subtypes |
| TP53 | Transcriptional factor | All exons | <2% | 4%-5% | 16%-50% | Adverse prognostic impact in all MPN subtypes | |
| Gene . | Function . | Location and type of mutation . | Frequency, % . | Clinical impact . | |||
|---|---|---|---|---|---|---|---|
| PV . | ET . | PMF . | Post-MPN AML . | ||||
| Disease driver mutations | |||||||
| JAK2 | JAK-STAT signaling | V617F or exon 12 | 98% | 55% | 60% | — | WHO/ICC criterion for MPN diagnosis |
| MPL | TPO receptor JAK-STAT signaling | Exon 10 | 0% | 5%-7% | 7%-10% | — | WHO/ICC criterion for MPN diagnosis |
| CALR | Chaperone protein CALR-mut binds and activates MPL | Frameshift in exon 9 | 0% | 25%-30% | 20%-30% | — | WHO/ICC criterion for MPN diagnosis |
| Clonal driver mutations | |||||||
| TET2 | Epigenetic regulation | All exons | 10%-20% | 3%-10% | 10%-20% | 19%-25% | No prognostic impact reported |
| DNMT3A | Epigenetic regulation | R882 and all exons | 5%-10% | 1%-5% | 8%-12% | 3%-17% | No prognostic impact reported |
| IDH1 | Epigenetic regulation | R132 | 1%-2% | 1%-2% | 5%-6% | 13% | HMR in PMF and adverse prognostic impact in all MPN subtypes |
| IDH2 | Epigenetic regulation | R140 or R172 | 1%-2% | 1%-2% | 5%-6% | 7%-15% | HMR in PMF and adverse prognostic impact in all MPN subtypes |
| ASXL1 | Epigenetic regulation | Mostly nonsense / frameshift in the last exon | 2%-7% | 5%-10% | 15%-35% | 17%-47% | HMR in PMF and adverse prognostic impact in all MPN subtypes |
| EZH2 | Epigenetic regulation | All exons | 1%-2% | 1%-2% | 7%-10% | 7%-13% | HMR in PMF and adverse prognostic impact in all MPN subtypes |
| NRAS | ERK/MAPK signaling | G12, G13, or Q61 | <2% | <2% | 2%-4% | 11% | Adverse prognostic impact in all MPN subtypes |
| KRAS | ERK/MAPK signaling | G12, G13, or Q61 | <2% | <2% | 2% | 4%-7% | Adverse prognostic impact in all MPN subtypes |
| SH2B3 | JAK signaling regulation | Exon 2 | 2%-9% | 1%-3% | 2%-4% | 6%-11% | Rare mutations in JAK2-negative MPNs |
| CBL | JAK signaling regulation | Exons 8 and 9 | <2% | <2% | 4% | 4% | Adverse prognostic impact in all MPN subtypes |
| SRSF2 | mRNA splicing | P95 | <2% | <2% | 6%-14% | 7%-22% | HMR in PMF and adverse prognostic impact in all MPN subtypes |
| SF3B1 | mRNA splicing | Exon 14-16 | 2%-3% | 2%-5% | 5%-7% | 7%-11% | Adverse prognostic impact in ET |
| U2AF1 | mRNA splicing | S34 or Q157 | <2% | <2% | 7%-10% | 5%-12% | Adverse prognostic impact in all MPN subtypes |
| NFE2 | Transcriptional factor | All exons | 3%,6% | 1%-7% | 3%-5% | ? | Increased risk of leukemic transformation |
| RUNX1 | Transcriptional factor | All exons | <2% | <2% | 2%-3% | 20% | Adverse prognostic impact in all MPN subtypes |
| TP53 | Transcriptional factor | All exons | <2% | 4%-5% | 16%-50% | Adverse prognostic impact in all MPN subtypes | |
Only gene mutations occurring with a frequency of >2% are listed.
CALR-mut, calreticulin mutant; ERK, extracellular signal-regulated kinase; ET, essential thrombocythemia; HMR, high molecular risk category for PMF; ICC, international consensus classification; JAK, Janus kinase; MAPK, mitogen-activated protein kinase; MPL, thrombopoeitin receptor; PMF, primary myelofibrosis; PV, polycythemia vera; TPO, thrombopoietin; WHO, World Health Organization.
“Disease driver” mutations recapitulate the human MPN phenotype when introduced into model organisms such as mice. A large proportion of MPN patients carry mutations solely in 1 of the 3 disease driver genes JAK2, CALR, or MPL,7-15 without additional clonal driver mutations detectable by targeted NGS.16,17 This finding was also confirmed by WES and WGS, which found that some sequence alterations can be detected, but these were not in any of the genes known to have a functional impact on hematopoiesis.14,15
“Clonal driver” mutations do not cause the MPN phenotype when expressed in mouse models, but they modify the phenotype when they are combined with one of the “disease driver” mutations. Some of these “clonal driver” mutations alter blood counts, increase HSC fitness or proliferation rate, and induce pre-malignant changes in hematopoiesis not directly related to MPN. The “clonal driver” genes that are most frequently mutated in MPNs include TET2, DNMT3A, ASXL1, and EZH2. These gene mutations are not MPN-specific and can be found across all myeloid malignancies. The MPN “disease driver” mutations, in particular JAK2-p.V617F, can also be “clonal drivers” in the context of clonal hematopoiesis, without resulting in MPNs.
Finally, “passenger” mutations are sequence alterations without a functional consequence. They can be useful as markers to determine and follow the subclonal structure in MPN. In many cases, whether a gene mutation alters function is difficult to predict, because many mutations have not yet been analyzed functionally. Such mutations are considered “variants of unknown significance”. Recurrence of such mutations can be used as a criterion in favor of a clonal advantage, but functional assays, such as those in cell lines, will be required in order to assign variants of unknown significance to either the “passenger” or “clonal driver” category.
Given that MPNs originate from a single HSC, early events after the acquisition of a driver gene mutation that lead to the expansion of the clone have become the focus of intense research. This focus is particularly relevant for JAK2-p.V617F, because JAK2 is also one of the genes most frequently mutated in healthy individuals with clonal hematopoiesis. Clonal hematopoiesis in the context of MPNs can be defined as the overproportional presence of blood cells that originate from a single HSC. Detecting clonality relies on finding markers that allow these clonal cells to be distinguished from the progeny of other HSCs.
In a seminal study, DNA from single hematopoietic colonies derived from bone marrow of a healthy donor was analyzed by WGS to reconstruct the phylogeny of HSCs.18 The method relies on the fact that during each cell division, the 2 daughter cells acquire a number of mutations in their genomes, owing to the imperfect fidelity of DNA replication and DNA repair. The vast majority of these sequence alterations are functionally silent, but they can be used as markers that distinguish the daughter cells from all other cells that underwent cell division. With a sufficient number of sequence alterations, markers can be found that distinguish very early HSC ancestors, from which a large proportion of adult HSCs are derived. By WGS of 140 single cell–derived colonies, all blood cells were mapped to originate from a common ancestor at the time of gastrulation (Figure 1).18 Markers that identify very early cell divisions, when applied to adult hematopoiesis, would define large “clones” in normal hematopoiesis, and would falsely suggest clonality, because the markers are shared by a large number of adult HSCs. The most useful markers of clonality in hematopoiesis are gene alterations that distinguish between HSCs at the time when the clonal driver gene mutation was acquired—that is, the best clonality marker is the actual “clonal driver” mutation itself. A second aspect to consider when assessing clonality is the threshold percentage of contribution of a single HSC to peripheral blood that is sufficient to be considered “overproportional.” Unfortunately, the number of different HSCs that at any given time are contributing to peripheral blood cells under healthy steady state conditions is not known. Age-dependent constriction of the HSC pool was recently described, and this could decrease the threshold for “clonality.”19
Clonal hematopoiesis of indeterminate potential and the evolution of MPNs
The term “clonal hematopoiesis of indeterminate potential” (CHIP) was created to denote a potentially “pre-malignant” state in individuals without a hematologic malignancy, and the most frequently found markers in CHIP, not surprisingly, also represent mutations in known cancer-associated genes, in particular the epigenetic regulators DNMT3A, TET2, and ASXL1, and at lower frequencies also JAK2. Recurrent somatic mutations in TET2 were the first to be described in elderly individuals with clonal hematopoies.20 The threshold for CHIP in most studies was set at variant allele fraction (VAF) >2% (ie, >4% of peripheral blood cells carry this heterozygous mutation). The measured %VAF is also influenced by the source of cells used for the analysis. DNA from unfractionated total white blood cells, which represent a mixture of B and T-cells with a long half-life, will decrease the VAF of most CHIP-markers, compared with granulocytes that have a short half-life and high turnover.
In the context of JAK2-p.V617F, several studies used highly sensitive polymerase chain reaction (PCR) methods and expanded the definition of CHIP to include individuals with JAK2-p.V617F VAF as low as VAF 0.01%,21 which means that 1 in 5000 cells was positive for the JAK2-p.V617F. With this expanded definition of CHIP, the authors characterized a cohort of 19 958 probands and found 613 that carried JAK2-p.V617F (3.1%) and 32 that carried CALR mutations (0.2%). With a threshold of 1% VAF, the prevalence of JAK2-p.V617F was still 0.46%.21 Within these 613 patients, 16 previously unrecognized cases of MPN were found, but the vast majority of individuals with JAK2-p.V617F CHIP did not fulfill the diagnostic criteria for MPN. The cumulative incidence of evolution from JAK2-p.V617F CHIP to MPN remains to be determined, but VAF JAK2-p.V617F >2%, or an increase in VAF during follow-up, was associated with a high rate of conversion from CHIP to MPN.22,23 Some patients with very low VAF in purified granulocytes can still display MPN phenotypes, due to selective expansion of the JAK2-mutant clone in late stages of erythropoiesis and thrombopoiesis.24
The time required for the conversion from CHIP to MPN was estimated, in a study that analyzed blood donors who progressed to MPN, and from whom earlier blood samples were available for analysis, to be 5 to 15 years.25 WGS of single cell colonies was also applied to reconstruct the phylogeny and timing of acquisition of the JAK2-p.V617F mutation in MPNs.5,6 Both studies concluded that the JAK2-p.V617F mutation occurred several decades before the diagnosis of MPN. Thus, the conversion from acquisition of JAK2-p.V617F to manifestation of MPN appears to take several decades, and from CHIP to MPN takes 5 to 15 years. Therefore, the time from acquisition of JAK2-p.V617F until it is detectable as CHIP is also likely to be in the range of decades (Figure 1).26
In the context of MPN, a noteworthy point is that the prevalence of JAK2-p.V617F CHIP is 15-fold higher than that of mutated CALR. This difference could be due to more frequent acquisition of JAK2-p.V617F. Alternatively, CALR mutations, by conferring a higher rate of clonal expansion compared to JAK2-p.V617F, may shorten the transition from acquisition of the mutation through the CHIP phase to MPN, resulting in a lower prevalence of CHIP.27 Reconstructing the phylogeny of CALR mutations by WGS will likely clarify these issues. The likelihood that an HSC carrying a driver gene mutation will escape immune surveillance, expand, and produce peripheral blood cells detectable as CHIP could also contribute to the observed differences. As many healthy individuals have been found to carry memory T-cells that recognize the mutated CALR-neoantigen without any evidence for CALR-mutated CHIP,28 the lower prevalence of CALR-mutated CHIP could be due to a more effective elimination of CALR-mutated cells at the pre-CHIP phase by the immune system in an HLA-restricted manner. In contrast, mutant CALR protein was also shown to decrease major histocompatibility complex-I expression and cause defects in peptide loading,29,30 providing a possible explanation for why immune checkpoint blockade and mutant CALR-derived peptide vaccination failed to generate clinical responses in MPN patients.31,32 Peptides containing the JAK2-p.V617F mutation also are unlikely to form high-affinity interactions with any HLA allotype,33 and activation of JAK/STAT signaling by JAK2-p.V617F increases programmed death ligand 1 (PD-L1) expression.34,35 Therefore, cells expressing JAK2-p.V617F are expected to evade immune surveillance, which could explain why JAK2-p.V617F CHIP is so frequent. Conversely, JAK2-p.V617F CHIP only rarely progresses to MPN, suggesting that some mechanisms are active mainly at this later conversion stage (Figure 2).
Evolution of MPNs originating from a single HSC that acquired a disease driver mutation. Model summarizing the events from the acquisition of JAK2-p.V617F until the development of MPN disease. The early events occur inside the bone marrow. A single HSC with JAK2-p.V617F can divide to yield 2 HSC daughter cells that carry the mutation, which leads to persistence, and later limited expansion of the mutated HSCs. Alternatively, the mutated HSC can differentiate into committed progenitors that produce a wave of mutant hematopoietic cells, but eventually are exhausted, due to loss of stemness. The mutant HSCs can also be eliminated at this early stage by cells of the immune system. During this phase, cells carrying JAK2-p.V617F are not yet detectable in peripheral blood (“pre-CHIP phase”). After expansion of the mutated HSCs and with a latency of years or decades, mature hematopoietic cells carrying JAK2-p.V617F are produced that become detectable in peripheral blood as “clonal hematopoiesis of undetermined potential (CHIP).” In only a minority of cases, the JAK2-p.V617F mutant HSC clone expands and produces committed progenitors that become dominant in bone marrow and can be diagnosed as MPN with elevated blood counts in peripheral blood. The factors favoring this conversion from CHIP to MPN are listed under the red arrow. The size of the JAK2-p.V617F mutant HSC clone can be reduced by interferon-α (IFNα), which acts by pushing the mutant HSCs into the cell cycle and thereby exhausting them,
Evolution of MPNs originating from a single HSC that acquired a disease driver mutation. Model summarizing the events from the acquisition of JAK2-p.V617F until the development of MPN disease. The early events occur inside the bone marrow. A single HSC with JAK2-p.V617F can divide to yield 2 HSC daughter cells that carry the mutation, which leads to persistence, and later limited expansion of the mutated HSCs. Alternatively, the mutated HSC can differentiate into committed progenitors that produce a wave of mutant hematopoietic cells, but eventually are exhausted, due to loss of stemness. The mutant HSCs can also be eliminated at this early stage by cells of the immune system. During this phase, cells carrying JAK2-p.V617F are not yet detectable in peripheral blood (“pre-CHIP phase”). After expansion of the mutated HSCs and with a latency of years or decades, mature hematopoietic cells carrying JAK2-p.V617F are produced that become detectable in peripheral blood as “clonal hematopoiesis of undetermined potential (CHIP).” In only a minority of cases, the JAK2-p.V617F mutant HSC clone expands and produces committed progenitors that become dominant in bone marrow and can be diagnosed as MPN with elevated blood counts in peripheral blood. The factors favoring this conversion from CHIP to MPN are listed under the red arrow. The size of the JAK2-p.V617F mutant HSC clone can be reduced by interferon-α (IFNα), which acts by pushing the mutant HSCs into the cell cycle and thereby exhausting them,
Factors influencing the appearance of CHIP and conversion to MPNs
Genetic predisposition to MPNs has been documented by many studies and could play an important role in increasing the likelihood of conversion from CHIP to MPNs. On a population scale, relatives of MPN patients have an approximately 6- to 8-fold higher risk of developing MPNs.36 The "strength" (penetrance) with which germline genetic factors exert their effect on developing MPNs inversely correlates with their prevalence in the general population (Figure 3). Rare families with autosomal dominant inheritance and a massively increased odds ratio (∼500×) for acquiring MPN disease have been described,37,38 but the mutations responsible for the predisposition have been identified in only very few cases. The best-studied cases are the duplication on chromosome 14q32,39 and mutations in RBBP6.40 The identification of mutations in other families with MPNs has been hampered by the low penetrance and also by the genetic heterogeneity among the different pedigrees. A mutation in EPOR (EPOR-p.P488S) that increases the risk of acquiring MPNs in one family was recently described.41
Frequencies of germline variants and the associated likelihood for developing a phenotype. Graph depicting the frequencies of germline gene mutations in the general population (x-axis) and the likelihood that they will promote the manifestation of MPN or MPN-like disease (y-axis). The highest penetrance (close to 100%) is observed in rare families with erythrocytosis or thrombocytosis caused by mutations in a single gene that are most frequently inherited as a Mendelian trait with autosomal dominant transmission (EPOR, EPO, THPO, and MPL, etc), or autosomal recessive transmission (Chuvash polycythemia due to mutations in VHL). Rare familial cases with inherited predisposition to MPN require the acquisition of somatic disease driver mutations for disease manifestation, most frequently JAK2-p.V617F, or mutations in CALR or MPL. The predisposing germline mutations are inherited as autosomal dominant traits with reduced penetrance, in most cases ∼20% to 40%. Inherited predisposition to MPN due duplication of chr.14q32 is an exception, reaching higher penetrance around 80%. Finally, common polymorphisms found at high frequencies in the general populations located in, for example, the JAK2, MECOM, and TERT genes, are associated with a much weaker predisposing effect and also require the acquisition of somatic disease driver mutations for MPN.
Frequencies of germline variants and the associated likelihood for developing a phenotype. Graph depicting the frequencies of germline gene mutations in the general population (x-axis) and the likelihood that they will promote the manifestation of MPN or MPN-like disease (y-axis). The highest penetrance (close to 100%) is observed in rare families with erythrocytosis or thrombocytosis caused by mutations in a single gene that are most frequently inherited as a Mendelian trait with autosomal dominant transmission (EPOR, EPO, THPO, and MPL, etc), or autosomal recessive transmission (Chuvash polycythemia due to mutations in VHL). Rare familial cases with inherited predisposition to MPN require the acquisition of somatic disease driver mutations for disease manifestation, most frequently JAK2-p.V617F, or mutations in CALR or MPL. The predisposing germline mutations are inherited as autosomal dominant traits with reduced penetrance, in most cases ∼20% to 40%. Inherited predisposition to MPN due duplication of chr.14q32 is an exception, reaching higher penetrance around 80%. Finally, common polymorphisms found at high frequencies in the general populations located in, for example, the JAK2, MECOM, and TERT genes, are associated with a much weaker predisposing effect and also require the acquisition of somatic disease driver mutations for MPN.
A growing number of single-nucleotide polymorphisms (SNPs) or haplotypes (genetically linked sets of SNPs) have been associated with an increased likelihood of acquiring MPNs. A haplotype located in the JAK2 gene (46/1 or GGCC) contributes to a 2 to 6 times increased risk of developing MPNs.42-44 Large genome-wide studies also identified SNPs in TERT, MECOM, CHEK2, and several additional loci that weakly increase the risk of acquiring MPNs on the population scale.45-48 The mechanisms of how these SNPs influence the likelihood of developing MPNs is unknown, but we suspect that they increase the conversion rate by synergizing with the driver gene mutations and facilitating the expansion of the mutant HSC clone. In most familial and population-based cases with germline predisposition to MPNs, the predominant acquired disease driver mutation is JAK2-p.V617F, and less frequently CALR or MPL.49 Interestingly, some patients acquired the JAK2-p.V617F mutation twice, located on the 2 homologous chromosomes.42
The fitness of JAK2-p.V617F CHIP clones showed interindividual variability that did not correlate with the presence or absence of known predisposition polymorphisms.19,26 The number of additional somatic mutations was among nonhereditary factors that increased the fitness of JAK2-p.V617F clones in human MPNs (Figure 2).6 Increased efficiency of MPN disease initiation of HSCs carrying JAK2-p.V617F and Ezh2 loss-of-function mutations was also shown in limiting dilution transplantations in mouse models of MPNs.50 Furthermore, the promoting effects of inflammation on the early steps of CHIP and MPN evolution are increasingly recognized. Loss of interleukin-1β (IL-1β) decreased the frequency of MPN disease initiation in limiting dilution transplantations in mouse models of MPN.51 The presence of CHIP was found to be associated with an increased risk for hematologic malignancies and unexpectedly also for cardiovascular disease,52 resulting in reduced survival.53-55 The cardiovascular risk was particularly high for JAK2-p.V617F CHIP (hazard ratio ∼12), and it was lower for other frequent CHIP mutations (∼2).52 Whether CHIP is a cause of the increased cardiovascular risk or just an associated symptom remains unclear. Inflammation could promote atherosclerosis and at the same time favor the expansion of mutated HSCs resulting in clonal hematopoiesis.56 In some patients with polycythemia vera (PV), complete molecular remission has been achieved by treatment with pegylated interferon-α (pegIFNα). A recent meta-analysis found a lower rate of thromboembolic events in patients treated by pegIFNα.57 Type I IFN was found to inhibit IL-1β production and inflammasome activation,58 providing a possible link between the 2 observations. When pegIFNα was discontinued in patients with complete molecular response, blood counts remained in the normal range, but JAK2-p.V617F remained detectable at low VAFs (<1%-5%), reminiscent of CHIP. The thrombotic risk in these patients is currently unknown, but an elevated basal level of inflammation could continue to increase the risk for cardiovascular disease, despite very low JAK2-p.V617F VAFs.
Determining the mutational landscape of MPN at diagnosis and during follow-up
Detecting disease driver gene mutations is a mandatory step in the diagnostic workup according to the World Health Organization and International Consensus Classification guidelines.59,60 NGS techniques allow detection of a large number of additional gene mutations in MPNs that occur in lower frequencies than they do in other hematologic malignancies.15,61,62 The median number of somatic mutations detected per patient by WES was 6.5 for essential thrombocythemia (ET) and PV, and 13 for primary myelofibrosis (PMF),15 but most of these were passenger mutations.17 Sequential analysis revealed that most mutations were already present at MPN diagnosis, and very few were acquired during follow-up.16,63 This finding is unexpected, given that JAK2-p.V617F has been reported to induce genetic instability.64,65 Therefore, a useful approach is to perform in-depth mutational profiling by NGS at diagnosis. Targeted NGS panels of 30 to 50 genes are now commonly used because they provide high coverage at low cost with rapid turnaround time.66 However, WES and WGS are becoming more widely available with decreasing costs, higher throughput, and improved sensitivities due to proofreading. These genome-wide techniques will likely replace targeted NGS panels in the future.
Using and interpreting NGS data presents 2 major challenges—clarifying the somatic vs germline origin of the mutations and defining whether the mutations have functional consequences or represent functionally silent variants. A reliable distinction between somatic and germ-line mutations would be an important first step in filtering NGS results. Mutations with low VAF are likely to be somatic, but mutations with VAF closer to 50% require sequencing of DNA from non-hematopoietic tissues, such as hair roots, nails, or fibroblasts, to exclude germline origin. DNA from buccal swabs is frequently contaminated by leukocytes and is therefore less reliable as a non-hematopoietic control,66 unless strict optimization procedures are applied.67 Functional assays testing the consequences of gene mutations are ultimately required, but these constitute a bottleneck, as they are difficult to perform with high throughput. Eliminating nonfunctional variants and distinguishing between somatic and germline mutations would improve the prognostic value of mutational profiles in MPNs.
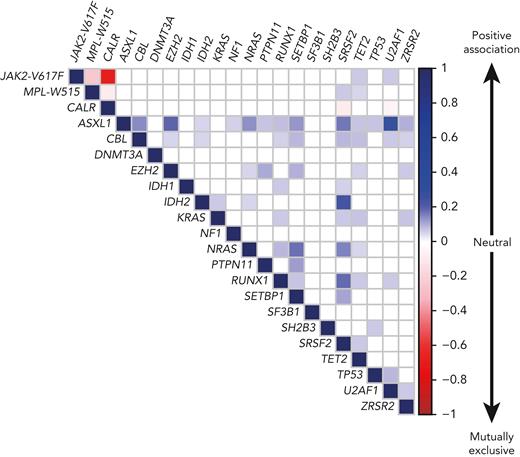
PMF is characterized by a complex molecular landscape, with enrichment of mutations in chromatin regulator and spliceosome genes.16,17,68,69 However, mutations in spliceosome genes were rarely found together with CALR mutations (Figure 4),15,16,70 due to reduced HSC fitness when co-expressed.71 Although JAK2-p.V617F and CALR mutations rarely occur in the same patients and usually represent separate clones, no negative impact on HSC function was observed when JAK2-p.V617F and CALR-del52 were co-expressed in the same cells.72,73 Post-PV and post-ET myelofibrosis had molecular profiles similar to those of PMF, but mutations in ASLX1 and SRSF2 were less frequent.74,75 Similar to gene mutations, chromosomal aberrations are more frequently found in PMF than in PV and ET.76,77 The most frequent karyotypic abnormalities involve chromosomes 9, 8, 20, 14, and 13. NGS technologies allow detection of large chromosomal abnormalities by monitoring of changes in the coverage of the affected regions. Similarly, uniparental disomy on chromosome 9p (9pUPD), which renders cells homozygous for JAK2-p.V617F,8,78 can be detected, whereas NGS currently cannot detect balanced translocations or minor subclonal chromosomal aberrations on DNA, but transcriptome sequencing can be used instead.33
Correlation plot showing the likelihood of co-occurrence of gene mutations in the same patient. The association between gene mutations is based on the compilation of data from 5 published cohorts comprising a total of 3002 MPN patients, including 1497 ET, 535 PV, and 970 PMF or secondary MF patients.16,17,79,80 The color code and numbers on the y-axis represent the Pearson R-coefficient, which indicates the strength of positive or negative associations between 2 gene mutations. Only the significant associations are depicted (P-value < .01).
Correlation plot showing the likelihood of co-occurrence of gene mutations in the same patient. The association between gene mutations is based on the compilation of data from 5 published cohorts comprising a total of 3002 MPN patients, including 1497 ET, 535 PV, and 970 PMF or secondary MF patients.16,17,79,80 The color code and numbers on the y-axis represent the Pearson R-coefficient, which indicates the strength of positive or negative associations between 2 gene mutations. Only the significant associations are depicted (P-value < .01).
In triple-negative ET patients, NGS covering the entire coding regions of JAK2 or MPL genes detected mutations located outside of mutational hot-spots in 20% to 30% of cases.17,81-84 These variants can be germline or acquired.85-88 Only a few of them have been tested functionally (Table 2), often showing either no or only weak oncogenic potential, indicating that other mutations must drive the phenotype.81,82 Sensitive functional tests for weak gain-of-function MPL mutations in cell lines are needed.81,82,89 Additional genes should be sequenced in suspected cases of hereditary erythrocytosis and thrombocytosis.90
Mutations in JAK2 and MPL genes described in myeloproliferative neoplasm (MPN) and MPN-like diseases
| Mutation . | Protein domain . | Somatic or germline . | Disease phenotype . | Found alone or together with another mutation . | Mean allele frequency . | Functional effect (gain-of-function) . |
|---|---|---|---|---|---|---|
| JAK2 mutations (ENST00000381652 transcript) | ||||||
| p.T108A | FERM | germ | PV | JAK2 p.V617F | 0.0007% | Weak |
| p.Y317H | FERM | NT | PMF | CALR | 0.002% | Weak |
| p.H345L | FERM | som | PMF | CALR | 0% | Weak |
| Exon 12 mutations between M535 to F547 | JH2 | som | PV | Alone | 0% | Strong driver gene mutations |
| p.F556V | JH2 | germ | ET | Alone | 0.001% | Weak |
| p.R564Q | JH2 | germ | HT | Alone | 0.003% | Intermediate |
| p.L583_A586delins | JH2 | som | PV | Alone | 0% | Intermediate |
| p.H608N | JH2 | germ | HT | Alone | 0% | Weak |
| p.L611S | JH2 | germ or NT | HT or PV | Alone or with JAK2 p.V617F | 0% | Weak |
| p.V617F | JH2 | som | PV, ET or PMF | Alone | 0.034% | Strong driver gene mutation |
| p.V617I | JH2 | germ or som | HT or ET | Alone | 0% | Weak |
| p.V625F | JH2 | germ | ET, PMF | Alone | 0% | Weak |
| p.E846D | JH1 | germ | HE | Alone | 0.045% | Need cooperating mutation |
| p.R867Q | JH1 | germ | HT | Alone | 0.0005% | Intermediate |
| p.T875N | JH1 | germ or som | HT or PMF | Alone | 0% | Strong |
| p.R755S/R938Q | JH2/1 | germ | HT | Alone | 0.003% | Intermediate |
| p.R1063H | JH1 | germ | HT | Alone | 0.47% | Need cooperating mutation |
| MPL mutations (ENST00000372470 transcript) | ||||||
| p.K39N | EC | germ | HT | Alone or with other MPL mutations | 0.46% and 4.6% in Africans | Reduced cell surface MPL |
| p.P106L | EC | germ | HT for hom | Alone | 0.038% | Reduced cell surface MPL |
| p.T119I | EC | som | ET or PMF | Alone | 0% | Minimal |
| p.S204P/F | EC | som | ET or PMF | Alone | 0% | Weak |
| p.E230G | EC | som | ET or PMF | Alone | 0% | Minimal |
| p.V285E | EC | germ | HT | Alone | 0% | Minimal |
| p.R321W | EC | germ | HT | Alone | 0% | Minimal |
| p.L498W | TM | som | ET | MPL p.S505N | 0% | Intermediate |
| p.L498_H499insVIAL | TM | som | ET | Alone | 0% | Intermediate |
| p.V501A | TM | som or NT | ET or PMF | MPL p.W515L/K or p.S505N | 0% | Intermediate |
| p.S505N | TM | som or germ | HT and ET, PMF | Alone | 0% | Strong driver gene mutation |
| p.W515K/L/A/R | TM | som | ET or PMF | Alone | 0% | Strong driver gene mutation |
| p.Y591D/N | IC | som | ET or PMF | Alone | 0% | Weak |
| Mutation . | Protein domain . | Somatic or germline . | Disease phenotype . | Found alone or together with another mutation . | Mean allele frequency . | Functional effect (gain-of-function) . |
|---|---|---|---|---|---|---|
| JAK2 mutations (ENST00000381652 transcript) | ||||||
| p.T108A | FERM | germ | PV | JAK2 p.V617F | 0.0007% | Weak |
| p.Y317H | FERM | NT | PMF | CALR | 0.002% | Weak |
| p.H345L | FERM | som | PMF | CALR | 0% | Weak |
| Exon 12 mutations between M535 to F547 | JH2 | som | PV | Alone | 0% | Strong driver gene mutations |
| p.F556V | JH2 | germ | ET | Alone | 0.001% | Weak |
| p.R564Q | JH2 | germ | HT | Alone | 0.003% | Intermediate |
| p.L583_A586delins | JH2 | som | PV | Alone | 0% | Intermediate |
| p.H608N | JH2 | germ | HT | Alone | 0% | Weak |
| p.L611S | JH2 | germ or NT | HT or PV | Alone or with JAK2 p.V617F | 0% | Weak |
| p.V617F | JH2 | som | PV, ET or PMF | Alone | 0.034% | Strong driver gene mutation |
| p.V617I | JH2 | germ or som | HT or ET | Alone | 0% | Weak |
| p.V625F | JH2 | germ | ET, PMF | Alone | 0% | Weak |
| p.E846D | JH1 | germ | HE | Alone | 0.045% | Need cooperating mutation |
| p.R867Q | JH1 | germ | HT | Alone | 0.0005% | Intermediate |
| p.T875N | JH1 | germ or som | HT or PMF | Alone | 0% | Strong |
| p.R755S/R938Q | JH2/1 | germ | HT | Alone | 0.003% | Intermediate |
| p.R1063H | JH1 | germ | HT | Alone | 0.47% | Need cooperating mutation |
| MPL mutations (ENST00000372470 transcript) | ||||||
| p.K39N | EC | germ | HT | Alone or with other MPL mutations | 0.46% and 4.6% in Africans | Reduced cell surface MPL |
| p.P106L | EC | germ | HT for hom | Alone | 0.038% | Reduced cell surface MPL |
| p.T119I | EC | som | ET or PMF | Alone | 0% | Minimal |
| p.S204P/F | EC | som | ET or PMF | Alone | 0% | Weak |
| p.E230G | EC | som | ET or PMF | Alone | 0% | Minimal |
| p.V285E | EC | germ | HT | Alone | 0% | Minimal |
| p.R321W | EC | germ | HT | Alone | 0% | Minimal |
| p.L498W | TM | som | ET | MPL p.S505N | 0% | Intermediate |
| p.L498_H499insVIAL | TM | som | ET | Alone | 0% | Intermediate |
| p.V501A | TM | som or NT | ET or PMF | MPL p.W515L/K or p.S505N | 0% | Intermediate |
| p.S505N | TM | som or germ | HT and ET, PMF | Alone | 0% | Strong driver gene mutation |
| p.W515K/L/A/R | TM | som | ET or PMF | Alone | 0% | Strong driver gene mutation |
| p.Y591D/N | IC | som | ET or PMF | Alone | 0% | Weak |
Mutations of JAK2 and MPL genes in MPN and MPN-like diseases were screened in PubMed publications and the Catalogue Of Somatic Mutations In Cancer (COSMIC) database. Only mutations with a functional impact on Janus kinase–signal transducer and activator of transcription (JAK-STAT) signaling and/or cell proliferation were retained. Functional effect was classified as being minimal to strong, as follows: minimal if only a constitutive STAT signaling was found; weak if a hypersensitivity to cytokine was found in transfected cell lines; intermediate if the mutation induced cytokine-independent growth; and strong if a mouse model had an MPN-like phenotype. MPL mutations inducing a reduction of the cell surface expression of thrombopoeitin receptor MPL lead to an increased level of plasmatic thrombopoietin (TPO) that stimulates megakaryopoiesis. The allele frequencies in the general population were extracted from the gnomAD v2.1 database. EC, extracellular; ET, essential thrombocythemia; FERM, protein 4.1R/ezrin/radixin/moesin; germ, germline; HE, hereditary erythropoiesis; hom, homozygous; HT, hereditary thrombocytosis; IC, intracellular; NT, not tested; PMF, primary myelofibrosis; PV, polycythemia vera; som, somatic; TM, transmembrane.
During follow-up of MPN patients, monitoring of VAF of mutations detected at diagnosis can be useful, but it is currently applied in only observational studies. An increase in VAF for JAK2-p.V617F was associated with higher risk of progression to myelofibrosis in ET and PV patients.91 Determining the VAF for JAK2-p.V617F and CALR mutations can be applied to monitor molecular response to treatment. Currently, only pegIFNα induces complete molecular responses in some patients.92-94 IFN is thought to act by exhausting HSCs that carry disease driver mutations,95-97 but not all patients respond similarly. Polymorphisms in the IFNL4 gene are correlated with the molecular response to pegIFNα.98 Overall, a higher rate of molecular responses to pegIFNα is observed in patients with JAK2-p.V617F than in patients with CALR mutations.99,100 Also, clones harboring additional mutations in TET2 or DNMT3A are less responsive to pegIFNα,101,102 and pegIFNα even promoted the expansion of subclones carrying both JAK2-p.V617F and DNMT3A mutations.103,104
Relevance of gene mutations for prognostic stratification
The type of disease driver mutations has been shown to influence prognosis in respect to thrombo-hemorrhagic complications and overall survival, with CALR mutations having a better prognosis than mutations in JAK2 or MPL, and triple-negative PMF having the least favorable outcome.105 The presence of additional mutations has not been reported to be associated with thrombosis. In contrast, the number of additional mutations per patient represents a risk factor for leukemic transformation and is inversely correlated with overall survival in MPN.16 This correlation was confirmed in PMF.106 Using a global pan-MPN approach based on clinical and molecular data in a cohort of 2035 patients, a personalized prognostic algorithm was developed that is accessible online.17 The presence of a mutation in one of 18 genes involved in chromatin or splicing regulation was associated in ET and PV with reduced overall survival and higher risk of progression to myelofibrosis or acute myeloid leukemia (AML). Some gene mutations often occur simultaneously in the same patient, whereas other combinations are mutually exclusive (Figure 4).
The impact of additional somatic mutations has been studied in great detail in patients with PMF or secondary myelofibrosis who were considered as candidates for allogeneic stem cell transplantation. Predicting prognosis in these patients primarily depends on estimating the risk of leukemic transformation and bone marrow failure with the associated complications due to cytopenia. Presence of mutations in ASXL1, EZH2, IDH1, IDH2, and SRSF2 has a stronger negative effect on prognosis, and these mutations were therefore designated to confer “high molecular risk” (HMR).107 HMR mutations have been incorporated into the International Prognostic Scoring System (IPSS)/Dynamic IPSS prognostic scores, resulting in MIPSS70.108 Recently, the U2AF1 gene has been added to the list of HMR genes (MIPSS70v2, GIPSS).109,110 Chromosomal aberrations are also associated with adverse prognosis and have been integrated into prognostic scores for PMF (MIPSS70+).108 Mutations in NRAS, KRAS, CBL, and TP53 are also associated with poor prognosis and were proposed to be added to the HMR list.17,75,111 The adverse impact of ASXL1 mutations alone was suggested to be weak, and the HMR effect of ASXL1 was shown to be dependent on the presence of other mutations.75,112
The order of acquisition of mutations may matter, and JAK2-p.V617F can be acquired as the first event, or after the acquisition of other mutations such TET2, ASXL1, and DNMT3A.16,113,114 The order of acquisition between JAK2-p.V617F and TET2 mutations was reported to influence the disease phenotype and the thrombotic risk.115 Clonal architecture can be determined by genotyping hematopoietic colonies derived from a single HSC or progenitor, but recently genotyping of individual single cells (single-cell DNA sequencing [scDNA-seq]) was described as an alternative method suited for high-throughput applications.116-118 A recent study defined the clonal architecture of 22 MPN patients that carried JAK2-p.V617F and at least one additional somatic mutation and found good agreement between the 2 methods that were used to analyze the same patient samples side-by-side.119 Unsupervised classification allowed defining clusters based only on clonal architecture data. One of the 4 clusters represented a risk factor for decreased survival independent of the MPN subtype or the age at diagnosis.119 These results suggest that deciphering the clonal architecture in patients with MPN can further improve and refine the molecular prognostic stratification.
Leukemic transformation
Transformation to secondary AML is a rare but devastating complication associated with a median survival of less than 6 months.120 The cumulative incidence of leukemic transformation at 10 years of follow-up was around 20% for PMF and 2.5% for both ET and PV.121-123 The mutational profile of post-MPN AML differs from de novo AML by the frequent occurrence of mutations in the ASXL1, SRSF2, IDH1/2, SH2B3, NRAS, RUNX1, and TP53 genes,124-132 In some patients, JAK2-p.V617F was present at MPN diagnosis, but absent in the leukemic blasts,133 indicating that AML either arose from a common mutated HSC, representing 2 branches of the same ancestral clone, or alternatively, represented a true bi-clonal disease originating from 2 different HSCs.134 Epigenetic dysregulation of gene expression also favors leukemic transformation and provides potential therapeutic targets.135,136
TP53 is the most frequently mutated gene in post-MPN AML.16,127 The frequency is particularly high (∼50%) when AML develops from PV or ET, typically following a long latency of >8 years after MPN diagnosis.137 In contrast, TP53 mutations are less frequent in post-PMF AML.130 Leukemic transformation with a short latency after MPN diagnosis often involves other oncogenic mechanisms characterized by a more complex molecular landscape and mutations in IDH1/2, EZH2, or DNMT3A genes, or acquisition of de novo mutations in RUNX1 that were not detectable during the chronic phase.137
Subclones carrying TP53 mutations are often detectable with low VAFs at MPN diagnosis. Using ultra-deep NGS, TP53 mutations were detected in 16% of MPNs, but the vast majority of these patients remained stable during follow-up.138 Leukemic transformation is frequently associated with an increase in TP53 VAF >50%.16,129,137,139 These results suggest that transformation of MPNs with TP53 mutations is a slow process requiring loss of the second TP53 allele or other oncogenic events. Indeed, in mouse models of Jak2-p.V617F, homozygous inactivation of Tp53 was required for leukemic transformation.129,136 Despite the known dominant negative and/or gain-of-function effect of some Tp53 missense mutations,140 no difference in phenotype was found for the heterozygous Tp53-p.R172H mutation, compared to that for the heterozygous Tp53+/−.136 These conclusions are supported by a recent study using single-cell genomics in post-MPN AML patients, which found that AML clones often displayed loss or another mutation in the second TP53 allele.141,142
Outlook and conclusions
MPN is a genetically heterogeneous disease in which germline and somatic mutations contribute to disease initiation and the course of disease. NGS-based techniques capture this heterogeneity, and we are at the beginning of understanding how to utilize this information for improving clinical management. To reach this goal, large international registries of MPN patients fully characterized by NGS for somatic and germline mutations will be needed to allow correlation of the disease course, complications, and response to therapies with patients' mutational profiles.
Acknowledgments
This work was supported by grants from the Swiss National Science Foundation (31003A_166613 and 310030_185297; R.C.S.), Swiss Cancer Research Foundation (KFS-3655-02-2015 and KFS-4462-02-2018; R.C.S.), and the Austrian Science Fund (FWF P34451-B; R.K.).
Authorship
Contribution: D.L.P., R.C.S., and R.K. analyzed data and wrote the manuscript.
Conflict of interest disclosure: R.C.S. is a scientific advisory board member for and has equity in Ajax Therapeutics; and consulted for and received honoraria from Novartis and BMS/Celgene. R.K. is a scientific advisory board member for AOP Orphan Pharmaceuticals; and is an equity holder in/scientific advisor for MyeloPro D&R GmbH. D.L.P. declares no competing financial interests.
Correspondence: Radek C. Skoda, Department of Biomedicine, Experimental Hematology, University Hospital Basel and University of Basel, Hebelstrasse 2, 4054 Basel, Switzerland; e-mail: radek.skoda@unibas.ch; Robert Kralovics, Department of Laboratory Medicine, Medical University of Vienna, Währinger Gürtel 18-20, 1090 Vienna, Austria; e-mail: robert.kralovics@meduniwien.ac.at; and Damien Luque Paz, Laboratory of Hematology, Angers University Hospital, 4 Rue Larrey, 49100 Angers, France; e-mail: damien.luquepaz@chu-angers.fr.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal