Key Points
Natural variation in proteins controlling blood stem cell genes leads to imbalances that accumulate with age until malignant disease arises.
Abstract
MYB plays a key role in gene regulation throughout the hematopoietic hierarchy and is critical for the maintenance of normal hematopoietic stem cells (HSC). Acquired genetic dysregulation of MYB is involved in the etiology of a number of leukemias, although inherited noncoding variants of the MYB gene are a susceptibility factor for many hematological conditions, including myeloproliferative neoplasms (MPN). The mechanisms that connect variations in MYB levels to disease predisposition, especially concerning age dependency in disease initiation, are completely unknown. Here, we describe a model of Myb insufficiency in mice that leads to MPN, myelodysplasia, and leukemia in later life, mirroring the age profile of equivalent human diseases. We show that this age dependency is intrinsic to HSC, involving a combination of an initial defective cellular state resulting from small effects on the expression of multiple genes and a progressive accumulation of further subtle changes. Similar to previous studies showing the importance of proteostasis in HSC maintenance, we observed altered proteasomal activity and elevated proliferation indicators, followed by elevated ribosome activity in young Myb-insufficient mice. We propose that these alterations combine to cause an imbalance in proteostasis, potentially creating a cellular milieu favoring disease initiation.
Introduction
The relationship between aging and stem cells has considerable implications, not least for chronic disease occurrence. Both intrinsic and extrinsic stem cell regulation are susceptible to the accumulation of changes with age, including acquired DNA mutations or protein aggregates in the cell itself or an altered environment, such as compositional variations in the extracellular matrix (ECM) or the profile of inflammatory cells and mediators.1,2
Myeloproliferative neoplasm (MPN) or myelodysplastic syndrome (MDS) arises within hematopoietic stem cells (HSC)3,4 and occurs predominantly in old age. Aging of HSCs is characterized by multiple changes including a more proliferative state and skewing of lineage commitment,5-7 paralleled by changes in RNA expression for genes involved in inflammatory responses, and altered proteostasis and metabolism.6,8 These changes are probably compounded by variations in the gene regulatory network, which together could predispose to disease. Indeed, genome-wide association studies (GWAS) show that the age-dependent risk of MPN is linked with genes encoding epigenetic or transcriptional regulators. MYB is one such protein with the risk of MPN being associated with single nucletide polymorphisms (SNP) upstream of the promoter that results in lower expression.9
Although lower Myb activity in model systems has negative impacts on HSC function,10,11 we observed that it can also confer stem cell-like properties on committed myeloid progenitors, producing an MPN-like condition.12,13 In this study, we have used a model with a less pronounced reduction in Myb expression, thereby better reflecting human-inherited variation, to explore how such changes can influence myeloid disease throughout the lifespan. Our findings show that constitutive Myb haploinsufficiency at high frequency leads to age-related–MPN/MDS, with features analogous to human disease. Using a conditional haploinsufficiency approach, we demonstrate that Myb levels determine a time-dependent intracellular environment for proteostasis, thereby explaining the age-dependency of disease in Myb-haploinsufficient animals. We propose that this drives malignant transformation or provides conditions favoring clonal expansion after driver gene mutation. We show that disturbances in proteostasis can be the result of life-long effects on the proteasome, which are because of the action of Myb on specific target genes, combined with secondary effects, particularly around ribosome activity, which accumulate over time because of the primary effects.
Materials and methods
Mouse models
Animal experiments were performed in accordance with UK legislation. Mice were controlled using wild-type (WT) littermates and maintained on a C57BL/6J background. Transplantation hosts were male B6.SJL-Ptprca/BoyJ. A cohort of male Myb+/F:CreERT2:mTmG and Myb+/+:CreERT2:mTmG mice were randomly assigned for conditional deletion using 75mg/kg Tamoxifen at either 2 or 12 months of age. Primer sequences for genotyping are listed in supplemental Table 1, available on the Blood website.
Engraftment potential of HSC
Transplantation of HSCs from WT and Myb+/− mice was performed as previously described.13 Test donor cells carried the vWf-Tomato transgene for the assessment of donor platelets.
RNA-seq
Refer to the supplemental Information.
Flow cytometric functional assays and intracellular Myb protein measurements
The mixed population of HSC and progenitors were defined using prestaining for Kit+Sca+Lin− (KSL) markers (refer to the supplemental Information) before staining for intracellular markers after fixation in 1.6% paraformaldehyde and permeabilization in ice-cold acetone. For proteasome activity assay, cells were treated with 5μM Me4bodipyFL-Ahx3Leu3VS probe (R&D Systems, Minneapolis, MN) for 1 hour at 37°C with or without prior incubation with 10 μM of proteasome inhibitor MG-132 (Millipore Sigma, St Louis, MO). Protein synthesis was assayed using O-propargyl-puromycin followed by Click-iT technology (Invitrogen, Waltham, MA) and assayed by flow cytometry. Proliferation and autophagy were assessed by intracellular staining with anti-Ki67 antibody (clone SolA15; ThermoFisher, Waltham, MA) or anti-LC3B (clone 1251A). Intracellular Myb protein expression was determined using anti-c-Myb FITC (clone D-7; Santa Cruz Biotechnology, Dallas, TX).
Human SNP analysis (see supplemental Methods)
Human bone marrow was obtained with informed consent through the Human Biomaterials Resource Centre, Birmingham. Genomic DNA from bone marrow hematopoietic progenitors was isolated and Taqman SNP genotyping for SNP rs9376092 (Applied Biosystems, Waltham, MA) was performed.
Gene expression analysis
RNA was extracted using TRIzol and converted to complementary DNA using Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI). Quantitative polymerase chain reaction (PCR) was performed using Taqman (Applied Biosystems) for mouse B2m (Mm00437762), Psmb3 (Mm04210356), Rps8 (Mm01187191), human GAPDH (Hs02758991), and MYB (Hs00920556).
Quantification and statistical analysis
Significant changes were calculated using a 2-tailed t test except for the Kaplan-Meier survival analysis, which was calculated using the log rank test. The significance threshold was <.05.
Results
Myb haploinsufficiency represents a model for transcription factor-dependent–age-related development of myeloid disease
Myb haploinsufficiency has minor effects on the hematological status of young mice,14 although long-term consequences are unknown. To model human MYB insufficiency, cohorts of WT and Myb+/− mice were aged for up to 22 months. Myb haploinsufficiency was confirmed in KSL cells from 2-month-old mice using intracellular staining with an antibody against Myb (supplemental Figure 1A). To confirm that the Myb haploinsufficiency model was representative of human MYB insufficiency, bone marrow Lin−CD38−CD45RA−CD34+CD49f+ HSC were assayed for their SNP rs9376092 status. MYB expression was assayed in samples designated as either CC (normal) or AC (heterozygous for variant A allele with lower expression), confirming that it was reduced to a similar level as seen in Myb+/− mice (supplemental Figure 1B). This is the first time, to our knowledge, that the effect of this SNP on MYB expression has been determined in HSC rather than the mature progenitors analyzed previously in the study by Tapper et al,9 which presumably means that the effect of the SNP variant on MYB expression is more profound in progenitors.
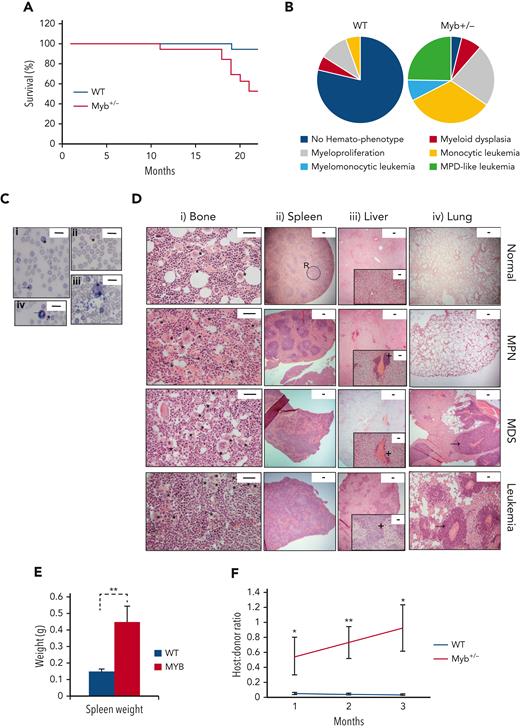
The hematological disease features in the WT and Myb+/− cohorts were monitored by the monthly measurement of blood values (supplemental Figure 2). The disease was classified using the Bethesda criteria for nonlymphoid hematopoietic neoplasms.15 Survival plots are shown in Figure 1A. All except 1 WT animal survived for 22 months, with signs of hematological disease in 4 animals (1 MDS, 2 MPN, and 1 myeloid leukemia). In contrast, all except 1 Myb+/− animal developed the myeloid disease with 52% surviving till the end of the experiment (P = .0035). Eight Myb+/− mice were killed because of myeloid leukemia (50% monocytic, 38% MPN-like, and 13% myelomonocytic) and 1 because of myeloproliferation. The profile of nonfatal myeloid disease among the Myb+/− cohort included 2 animals with MDS and 5 animals with MPN (Figure 1B).
Myb deficiency leads to age-related hematological disease. (A) Kaplan-Meier curve showing the percentage survival of a cohort of aging WT and Myb+/− mice (n = 19 and 17, respectively) over 22 months. Significance was calculated using the log rank test with P = .0035. (B) Pie charts depicting the types of myeloid disease arising with age. (C) Blood smears from diseased mice showing examples of abnormal peripheral blood cells. (i) Target cell erythrocyte (codocyte) (∗). Polychromatic erythrocyte (→). (ii) Teardrop erythrocyte (dacrocyte) (∗). (iii) Micromegakaryocte (∗). (iv) Howell-Jolly body in erythrocytes (∗). Ring-neutrophil (→). Scale bar = 20 μm. (D) Representative sections from tibial bone, spleen, liver, and lung from mice characterized as WT, MPN, MDS, or myeloid leukemia. Scale bars are indicated. (i) Bone section with megakaryocytes (∗) showing an increase in diseased mice. Scale bar = 50 μm. (ii) Spleen sections showing disrupted red and white pulp structure in diseased mice. An example area of white (circle) and red (R) pulp is indicated. Scale bar = 100 μm. (iii) Liver sections showing infiltration of myeloid cells (+). Scale bar = 100 μm (inset: scale bar = 50 μm). (iv) Lung sections showing infiltration of myeloid cells (→). Scale bar = 100 μm. (E) Spleen weight (g) of aging WT and Myb+/− mice at the point of culling (n = 19 and 17, respectively). Significance was calculated using t test P = .006. Data are represented as the mean and SEM. (F) Ratio of host:donor bone marrow cells from WT (n = 4) and mice with myeloid leukemia (n = 6). Whole bone marrow was transplanted into sublethally irradiated (450 Gy) B6.SJL-Ptprca/BoyJ mice. Ratios are calculated by antibody staining of peripheral blood against CD45.1 and CD45.2 antigens at 1, 2, and 3 months after transplant (P = .043, .007, and 0.011, respectively). Data points represent the mean ratios with SEM. SEM, standard error of the mean.
Myb deficiency leads to age-related hematological disease. (A) Kaplan-Meier curve showing the percentage survival of a cohort of aging WT and Myb+/− mice (n = 19 and 17, respectively) over 22 months. Significance was calculated using the log rank test with P = .0035. (B) Pie charts depicting the types of myeloid disease arising with age. (C) Blood smears from diseased mice showing examples of abnormal peripheral blood cells. (i) Target cell erythrocyte (codocyte) (∗). Polychromatic erythrocyte (→). (ii) Teardrop erythrocyte (dacrocyte) (∗). (iii) Micromegakaryocte (∗). (iv) Howell-Jolly body in erythrocytes (∗). Ring-neutrophil (→). Scale bar = 20 μm. (D) Representative sections from tibial bone, spleen, liver, and lung from mice characterized as WT, MPN, MDS, or myeloid leukemia. Scale bars are indicated. (i) Bone section with megakaryocytes (∗) showing an increase in diseased mice. Scale bar = 50 μm. (ii) Spleen sections showing disrupted red and white pulp structure in diseased mice. An example area of white (circle) and red (R) pulp is indicated. Scale bar = 100 μm. (iii) Liver sections showing infiltration of myeloid cells (+). Scale bar = 100 μm (inset: scale bar = 50 μm). (iv) Lung sections showing infiltration of myeloid cells (→). Scale bar = 100 μm. (E) Spleen weight (g) of aging WT and Myb+/− mice at the point of culling (n = 19 and 17, respectively). Significance was calculated using t test P = .006. Data are represented as the mean and SEM. (F) Ratio of host:donor bone marrow cells from WT (n = 4) and mice with myeloid leukemia (n = 6). Whole bone marrow was transplanted into sublethally irradiated (450 Gy) B6.SJL-Ptprca/BoyJ mice. Ratios are calculated by antibody staining of peripheral blood against CD45.1 and CD45.2 antigens at 1, 2, and 3 months after transplant (P = .043, .007, and 0.011, respectively). Data points represent the mean ratios with SEM. SEM, standard error of the mean.
The Myb+/− MPN phenotypes included thrombocytosis and leukocytosis; whereas mice with MDS exhibited combinations of thrombocytopenia, neutropenia, lymphopenia, and anemia (supplemental Figure 3A), with significant dyserythropoiesis, which include polychromasia, macrocytes, target cells, Howell-Jolly bodies, dacrocytes, and helmet cells, together with the presence of micromegakaryocytes (Figure 1C). Neutrophil dysplasia involved an excess of immature (band) neutrophils and hypersegmented granulocytes (Figure 1C). Bone sections from Myb+/− MPN mice revealed hypercellularity with increased megakaryocytes, typically occurring in clusters and possessing characteristic stag-horn nuclei (Figure 1D). MDS was distinguishable by increased cellularity and dysplastic megakaryocytes, including micromegakaryocytes and hypolobulated cells.
Most Myb+/− diseased mice had splenomegaly (Figure 1E); those with MPN exhibited enlarged red pulp indicative of extramedullary hematopoiesis. Minor hepatic infiltration was also observed, but no infiltration in lungs (Figure 1D). Analysis of sections from Myb+/− MDS mice revealed disrupted splenic architecture with extramedullary megakaryocytes, as well as hepatic and lung infiltration (Figure 1D).
Myb+/− mice with myeloid leukemia possessed features of MPN or MDS suggesting disease progression. Bone marrow and spleen sections showed marked increases in megakaryocytes with highly disrupted architecture. The liver and lungs exhibited extensive hematopoietic infiltration (Figure 1D). Leukemia progression was confirmed by transplantation of bone marrow into sublethally irradiated recipients. The leukemic Myb+/− bone marrow engrafted effectively and transferred the phenotype to the recipients (Figure 1F).
KSL cells in Myb+/− that developed MPN were confirmed to be haploinsufficient for Myb protein to a similar degree to that observed when comparing young WT with Myb+/− cells (supplemental Figure 3B). HSCs from Myb+/− mice that developed either MDS or leukemia had increased Myb as expected for aberrant blast proliferation or in proliferative-transformed leukemic cells.
The age dependency of Myb insufficiency–associated disease is largely intrinsic to HSCs
Most animals in the Myb+/− cohort exhibited a hematological phenotype after 1 year of age (average, 13.8 months), which might be an effect intrinsic to HSC or could be a consequence of the interaction between lower levels of Myb and age-related changes in the environment. To discriminate between these possibilities, we combined a floxed Myb allele16 with Tamoxifen-inducible Cre recombinase (CreERT2)17 and a Cre activity reporter (mTmG18). One Myb allele was deleted at either 2 or 12 months of age followed by tracking of the incidence of myeloid disease (Figure 2A). Cohort 1 consisted of 10 Myb+/+:CreERT2:mTmG and 11 Myb+/F:CreERT2:mTmG mice treated with Tamoxifen at 2 months of age, whereas cohort 2 consisted of 10 Myb+/+:CreERT2:mTmG and 10 Myb+/F:CreERT2:mTmG mice in which Myb deletion was induced at 12 months. The efficiency of deletion (∼50%) was confirmed by quantitative PCR of the relevant genomic interval using DNA prepared from blood cells (Figure 2B). A small cohort of Myb+/− mice was injected with Tamoxifen to demonstrate that the agonist does not accelerate the hematopoietic phenotype previously observed with 80% of the Myb+/− mice surviving up to 18 months (the average time of onset of disease in aged Myb+/− mice) (supplemental Figure 4A).
Conditional deletion of Myb at 2 or 12 months determines that the disease is HSC intrinsic. (A) Schematic depicting the Tamoxifen treatment of Myb+/+:CreERT2:mTmG (Myb+/+) and Myb+/F:CreERT2:mTmG (Myb+/F) mice at either at 2 or 12 months of age. (B) Quantitative PCR on peripheral blood genomic DNA, detecting the relative levels of Mybex5 product relative to an internal control GpIIb calculated using the ΔCT method. Values represent the mean expression plotted as 2−ΔCT with SEM (n = 10 P = .020). (C) Kaplan-Meier curve of the survival over 22 months of Myb+/+ and Myb+/F mice after Myb deletion at 2 or 12 months of age. The arrows signify the average time from Tamoxifen deletion until disease appearance. Significance was calculated using the log rank test with P = .026 and .006 for the 2– and 12–month deleted Myb, respectively. n = 10 for Myb+/+ 2- and 12-months, n = 11 for Myb+/F 2-months, n = 10 for Myb+/F 12-months. (D) Classification of myeloid disease arising in the aging cohorts of Myb-deleted mice. (E) Absolute bone marrow cell numbers based on antibody staining and total bone marrow count. n = 10 for Myb+/+ 2- and 12-months, n = 11 for Myb+/F 2-months, n = 10 for Myb+/F 2- months. Significance was calculated by t test. (i) KSL12mP = .032. (ii) MEP2mP = .052, CMP2mP =.023, CMP12mP = .014, GMP12mP = .031. (iii) Prog2mP = .052, Prog12mP = .047. (iv) Monocytes12mP = .028, (v) Granulocytes2mP = .032. CMP, common myeloid progenitor; GMP, granulocyte-monocyte progenitor; granulocytes, CD11b+Gr1+; MEP, megakaryocyte-erythroid progenitor; myeloid progenitors, Kit+Sca−Lin−; monocytes, CD11b+Gr1−; SEM, standard error of the mean.
Conditional deletion of Myb at 2 or 12 months determines that the disease is HSC intrinsic. (A) Schematic depicting the Tamoxifen treatment of Myb+/+:CreERT2:mTmG (Myb+/+) and Myb+/F:CreERT2:mTmG (Myb+/F) mice at either at 2 or 12 months of age. (B) Quantitative PCR on peripheral blood genomic DNA, detecting the relative levels of Mybex5 product relative to an internal control GpIIb calculated using the ΔCT method. Values represent the mean expression plotted as 2−ΔCT with SEM (n = 10 P = .020). (C) Kaplan-Meier curve of the survival over 22 months of Myb+/+ and Myb+/F mice after Myb deletion at 2 or 12 months of age. The arrows signify the average time from Tamoxifen deletion until disease appearance. Significance was calculated using the log rank test with P = .026 and .006 for the 2– and 12–month deleted Myb, respectively. n = 10 for Myb+/+ 2- and 12-months, n = 11 for Myb+/F 2-months, n = 10 for Myb+/F 12-months. (D) Classification of myeloid disease arising in the aging cohorts of Myb-deleted mice. (E) Absolute bone marrow cell numbers based on antibody staining and total bone marrow count. n = 10 for Myb+/+ 2- and 12-months, n = 11 for Myb+/F 2-months, n = 10 for Myb+/F 2- months. Significance was calculated by t test. (i) KSL12mP = .032. (ii) MEP2mP = .052, CMP2mP =.023, CMP12mP = .014, GMP12mP = .031. (iii) Prog2mP = .052, Prog12mP = .047. (iv) Monocytes12mP = .028, (v) Granulocytes2mP = .032. CMP, common myeloid progenitor; GMP, granulocyte-monocyte progenitor; granulocytes, CD11b+Gr1+; MEP, megakaryocyte-erythroid progenitor; myeloid progenitors, Kit+Sca−Lin−; monocytes, CD11b+Gr1−; SEM, standard error of the mean.
The deletion of 1 Myb allele at 2 or 12 months caused a significant reduction in the survival of Myb+/F:CreERT2:mTmG mice compared with the controls (P = .026 and P = .0062, respectively) (Figure 2C). As for the aged Myb+/− animals, not all mice subjected to conditional deletion of Myb succumbed to the disease; however, by 21 months, a significant proportion had developed the myeloid disease. In cohort 1, 10 Myb+/F:CreERT2:mTmG mice exhibited disease (1 MDS, 8 MPN, 1 myeloid leukemia) compared with just 2 Myb+/+:CreERT2:mTmG animals (1 MDS, 1 MPN). In cohort 2, 8 Myb+/F:CreERT2:mTmG mice developed the myeloid disease, with a slightly higher incidence of myeloid leukemia compared with the 1 Myb allele-deleted animals at 2 months of age (20% and 9%, respectively), along with 1 developing MDS and 5 MPN (Figure 2D). Tissue sections from the conditionally haploinsufficient aged mice had features like those described for the aged Myb+/− mice, enabling the determination of disease phenotype (supplemental Figure 4B). Deletion of Myb at both 2 and 12 months led to an increase in the absolute number of HSC and Kit+ myeloid progenitors (Figure 2E), the increase in granulocyte-monocyte progenitor being somewhat greater when this occurred at 12 months, paralleled by an increase in monocytes. These results indicate that the age-related disease onset in Myb+/− animals requires a time-dependent accumulation of events downstream of Myb, and that any aged environment effects are of less significance.
Myb-insufficient HSCs are functionally compromised
Because both MPN and MDS are thought to arise through acquired mutations in HSC, we compared the profile and function of stem cells from young and aged WT and Myb+/− mice. Two-month-old Myb+/− mice exhibited no distinct hematological pathology (supplemental Figure 5), however increased numbers of KSL HSC were observed (P = .0009), including long-term HSC (KSL CD48−CD150+) (P = .013) (Figure 3A). By 12 months of age, the relative number of long-term HSC was reduced (P = .005).
Myb-deficient HSCs are compromised in their function. (A) Bone marrow absolute counts of KSL and LT-HSC (KSLCD48-CD150+) from 2- (n = 8) and 12-month-old (n = 8) WT and Myb+/− mice. KSL2mP = .0009, KSL12mP = .053, LT-HSC2mP = .013, LT-HSC12mP = .005. (B and D) Sorted KSL cells from 2- (n = 9) and 12-month-old (n = 9) WT or Myb+/− donors (carrying vWF-tdTomato transgene) plus reference bone marrow were transplanted into lethally irradiated recipients (900 Gy). Reference:donor cell ratios were calculated using flow cytometry. P2m = 0.040, 0.038 and P12m = 0.030, 0.006, 0.055. (C) Analysis of donor cell percentage in the periphery of recipient mice (n = 3) 4 months after transplant receiving 2-month-old KSL. Antibody staining gated on CD45.2 donor cells: myeloid (CD11b+), T-lymphoid (CD4+CD8a+), B-lymphoid (B220+), and platelet (vWF+). Myeloid P = .001, B-lymphoid P = .011.
Myb-deficient HSCs are compromised in their function. (A) Bone marrow absolute counts of KSL and LT-HSC (KSLCD48-CD150+) from 2- (n = 8) and 12-month-old (n = 8) WT and Myb+/− mice. KSL2mP = .0009, KSL12mP = .053, LT-HSC2mP = .013, LT-HSC12mP = .005. (B and D) Sorted KSL cells from 2- (n = 9) and 12-month-old (n = 9) WT or Myb+/− donors (carrying vWF-tdTomato transgene) plus reference bone marrow were transplanted into lethally irradiated recipients (900 Gy). Reference:donor cell ratios were calculated using flow cytometry. P2m = 0.040, 0.038 and P12m = 0.030, 0.006, 0.055. (C) Analysis of donor cell percentage in the periphery of recipient mice (n = 3) 4 months after transplant receiving 2-month-old KSL. Antibody staining gated on CD45.2 donor cells: myeloid (CD11b+), T-lymphoid (CD4+CD8a+), B-lymphoid (B220+), and platelet (vWF+). Myeloid P = .001, B-lymphoid P = .011.
To assess stem cell potential, transplantation assays were performed on KSL HSC from 2-month-old WT and Myb+/− mice, incorporating the vWF-tdTomato transgene19 to facilitate the tracking of platelet differentiation. Engraftment rates of more than 5 months revealed that Myb+/− HSC are significantly impaired in reconstitution capacity (P < .05) (Figure 3B). Analysis of the donor-derived cells in blood showed that Myb+/− HSCs give rise to proportionately more myeloid cells (CD11b+) (P = .001) and that reduced engraftment is not an artifact of increased platelet numbers, because the CD45− vWF+ fraction is not different for WT and Myb+/− donor cells (Figure 3C). A similar difference was observed using HSC from 12-month-old WT mice although the reduction in engraftment was evident much earlier compared with the 2-month-old donor cells (2- vs 4-months, respectively) (Figure 3D). As expected, the 12-month-old WT KSL cells engrafted at a lower efficiency compared with the younger WT cells.
Myb insufficiency causes distinct gene expression differences in young vs old HSCs
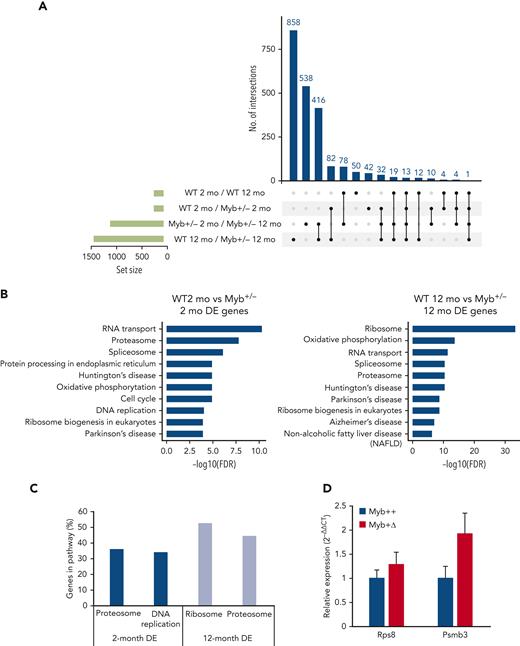
RNA-Seq analysis was performed on KSL HSCs derived from 2- and 12-month-old WT and Myb+/− mice. Considering expression differences with a standard false discovery rate cutoff of <0.1, 247, the following 1108, 188, and 1433 genes were identified as differentially expressed: comparing WT 2- vs 12-month old, Myb+/− 2- vs 12-month-old, 2-month -old WT vs Myb+/−, and 12-month-old WT vs Myb+/−, respectively. The overlap of genes uniquely shared as differences in 2-month-old Myb+/− and aged WT HSC was just 4 (Figure 4A), implying that young Myb+/− HSC are not prone to premature aging.
RNA-seq analysis determines key pathways that are altered by low levels of Myb. (A) UpSet plot showing the significantly (adjusted P < .10) DE genes from RNA-seq comparisons of WT and Myb+/− KSL from young (2-month) vs older (12-month) animals. Each row represents the different conditions where points to the right of the sets provide information about the intersecting sets of the different conditions. The vertical bars indicate the number of intersecting DE genes in each combination of conditions. The set size below the horizontal bars represents the number of DE genes within each set (condition). The UpSet plot was generated using Intervene (version 0.6.4). (B) KEGG pathway analysis from the comparison of 2-month-WT vs Myb+/− and 12-month-old WT and Myb+/− KSL cells. The top 10 most significant pathways are depicted based on their FDR. (C) The percentage of genes in each depicted pathway that are significantly DE in 2- and 12-month-old KSL. (D) Quantitative PCR of RNA expression in aged Myb+/+:CreERT2:mTmG and Myb+/Δ:CreERT2:mTmG sorted GFP+ KSL cells. Values represent the mean expression plotted as 2−ΔΔCT with SEM n = 3. DE, differentially expressed; FDR, false discovery rate; SEM, standard error of the mean.
RNA-seq analysis determines key pathways that are altered by low levels of Myb. (A) UpSet plot showing the significantly (adjusted P < .10) DE genes from RNA-seq comparisons of WT and Myb+/− KSL from young (2-month) vs older (12-month) animals. Each row represents the different conditions where points to the right of the sets provide information about the intersecting sets of the different conditions. The vertical bars indicate the number of intersecting DE genes in each combination of conditions. The set size below the horizontal bars represents the number of DE genes within each set (condition). The UpSet plot was generated using Intervene (version 0.6.4). (B) KEGG pathway analysis from the comparison of 2-month-WT vs Myb+/− and 12-month-old WT and Myb+/− KSL cells. The top 10 most significant pathways are depicted based on their FDR. (C) The percentage of genes in each depicted pathway that are significantly DE in 2- and 12-month-old KSL. (D) Quantitative PCR of RNA expression in aged Myb+/+:CreERT2:mTmG and Myb+/Δ:CreERT2:mTmG sorted GFP+ KSL cells. Values represent the mean expression plotted as 2−ΔΔCT with SEM n = 3. DE, differentially expressed; FDR, false discovery rate; SEM, standard error of the mean.
The analysis of Kyoto encyclopedia of genes and genomes (KEGG) pathways by functional class scoring20 of the comparison between 2-month-old WT vs Myb+/− revealed enrichment in genes encoding proteasome-associated proteins, which were similarly predominant in the differences distinguishing 12-month-old WT from Myb+/− HSC (Figure 4B-C; supplemental Figure 6). Myb+/− HSC at 2 and 12 months showed a higher expression of 17 and 33 proteosome-associated genes (using a P value of .05), respectively, encoding 19S and 20S proteasome components (Figure 4B), crucial for the normal functioning of HSC.21 In addition, the 12-month-old Myb+/− HSC showed highly significant enrichment of genes encoding proteins linked with rRNA processing and ribosome biogenesis (Figure 4B-C). A total of 93 ribosome component genes and 34 ribosomal biogenesis genes are increased in the Myb+/− HSC at 12 months of age (supplemental Figure 7). Another interesting pathway enrichment characterizing 2-month-old Myb+/− HSC involves genes connected to DNA replication and cell cycle, including components of the DNA polymerases and minichromosome maintenance (MCM) complexes (Figure 4B-C; supplemental Figure 8). The same DNA replication-associated genes are also enriched in 12-month-old Myb+/− HSCs compared with their WT equivalents.
Selected gene expression differences were also compared between aged WT and Myb+/F mice rendered haploinsufficient by Cre-mediated deletion at 2 months (Myb+/Δ). HSCs in which deletion had occurred (GFP+) were sorted and quantitative reverse transcription-PCR was performed on representative genes highlighted in the comparison between WT and constitutively Myb+/− mice. HSCs were collected at 14 months of age, that is, at the average point of disease incidence after Myb allele deletion. Myb+/Δ HSC showed an increased expression of genes associated with the ribosome (Rps8) and the proteasome (Psmb3) at levels consistent with the RNA-seq data derived from the aged cohort of Myb+/− mice (Figure 4D).
Some proteostasis-associated protein genes are directly regulated by Myb
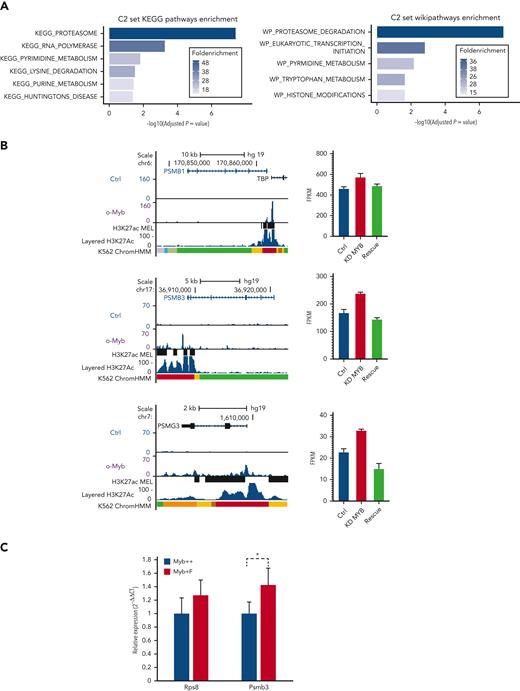
To investigate if the Myb-dependent gene expression changes are conserved and whetherthey involve direct target genes, we analyzed transcriptome and chromatin immunoprecipitation (ChIP)-seq data for MYB from human K562 cells.22,23 In total, 525 genes were defined as direct targets of MYB based on MYB knockdown and rescue and their association with 1 or several MYB ChIP-peaks using the STITCHIT algorithm.24 We investigated the overlap of this gene set with the differentially expressed mouse genes but with a stringent filter (P < .05, Log2FC ≥ 0.5, n = 662). This resulted in n = 32 genes, which were then analyzed using the clusterProfiler R package (version 3.12.0).25 The C2 curated list of genes from MSigDB was used to investigate relevant pathways and functional terms (Figure 5A). Among the 32 genes, 8 were proteasome component-encoding (PSM) genes. To further support the functional link between the top-ranked proteasome-associated genes and MYB, we visualized the MYB ChIP-seq peak occupancy at the loci of selected PSM genes (Figure 5B left, and data not shown). We also integrated these gene sets with data on genes affected by MYB knockdown and rescue in K562,22 revealing that the selected proteasome-associated genes showed a significant increase in expression when MYB was knocked down (Figure 5B right, and data not shown).
A subset of genes encoding proteostasis-associated proteins are directly regulated by Myb in both mouse HSC and a human hematopoietic progenitor line. (A) Functional enrichment of the subset of mouse Myb insufficiency–responsive genes where their human homologs are direct target genes of MYB. Results from Wikipathways (upper panel) and KEGG pathway (lower panel) are shown. (B) A subset of Myb insufficiency–responsive genes are direct target genes of MYB in K562 cells. Left panels: UCSC tracks showing MYB occupancy at the PSMB1, PSMB3, and PSMG3. In addition to the K562 ChIP-seq tracks, we incorporated H3K27Ac tracks (accession ID: ENCFF117MIF) from the MEL mouse cell line, which is described as analogous to K562 cells in the ENCODE database. To make the MEL cell line–derived H3K27Ac ChIP-seq peaks compatible with human hg19 coordinates, we used the UCSC liftOver tool26 before uploading it to the MYB UCSC session and visualizing the tracks using the UCSC genome browser.27 Right panels: expression profiles of PSMB1, PSMB3, and PSMG3, respectively, in K562 cells after small interfering RNA knockdown of MYB and rescue by the transient expression of 3✕TY1-MYB. (C) Quantitative PCR of RNA expression in Myb+/+:CreERT2:mTmG and Myb+/F:CreERT2:mTmG sorted GFP+ KSL cells 24 hours after deletion. Values represent the mean expression plotted as 2−ΔCT with SEM. n = 4 P = .047. SEM, standard error of the mean.
A subset of genes encoding proteostasis-associated proteins are directly regulated by Myb in both mouse HSC and a human hematopoietic progenitor line. (A) Functional enrichment of the subset of mouse Myb insufficiency–responsive genes where their human homologs are direct target genes of MYB. Results from Wikipathways (upper panel) and KEGG pathway (lower panel) are shown. (B) A subset of Myb insufficiency–responsive genes are direct target genes of MYB in K562 cells. Left panels: UCSC tracks showing MYB occupancy at the PSMB1, PSMB3, and PSMG3. In addition to the K562 ChIP-seq tracks, we incorporated H3K27Ac tracks (accession ID: ENCFF117MIF) from the MEL mouse cell line, which is described as analogous to K562 cells in the ENCODE database. To make the MEL cell line–derived H3K27Ac ChIP-seq peaks compatible with human hg19 coordinates, we used the UCSC liftOver tool26 before uploading it to the MYB UCSC session and visualizing the tracks using the UCSC genome browser.27 Right panels: expression profiles of PSMB1, PSMB3, and PSMG3, respectively, in K562 cells after small interfering RNA knockdown of MYB and rescue by the transient expression of 3✕TY1-MYB. (C) Quantitative PCR of RNA expression in Myb+/+:CreERT2:mTmG and Myb+/F:CreERT2:mTmG sorted GFP+ KSL cells 24 hours after deletion. Values represent the mean expression plotted as 2−ΔCT with SEM. n = 4 P = .047. SEM, standard error of the mean.
To confirm that mouse Myb similarly regulates proteasomal genes, we performed conditional deletion in Myb+/F:CreERT2:mTmG mice, using a Myb+/+:CreERT2:mTmG control. HSCs that transitioned from tomato-FP to GFP were sorted and quantitative reverse transcription-PCR was performed to measure the expression of Psmb3 RNA. Expression of Rps8 RNA was used as a control to determine if there was any change in ribosomal genes. This revealed that loss of 1 Myb allele resulted in significantly higher expression of Psmb3 (P = .047), with no change in the expression of the ribosomal gene, Rps8 (Figure 5C).
Myb-dependent differences in gene expression are reflected in HSC functional processes
To confirm that observed changes in gene expression are reflected in alterations in the corresponding cellular processes, assays were performed relating to the collective activity of the relevant encoded proteins concerning proteostasis and DNA replication.
Proteasome activity was assessed in 2-month-old HSCs using staining with a Me4bodipyFL-Ahx3Leu3VS probe, with or without pretreatment with the proteasome inhibitor MG132, followed by flow cytometric analysis (Figure 6A). The median fluorescent intensity (MFI) analysis demonstrated a significant 15% higher proteasome activity (P = .007) in the Myb+/− HSC. We also wanted to assess whether the Myb+/− cells exhibited increased autophagy. Using intracellular antibody staining of the autophagosome marker LC3B, we identified a significant (P = .025) increase in MFI in HSCs from Myb+/− compared with WT bone marrow (supplemental Figure 9).
Myb deficiency affects protein production and degradation. (A) KSL cells from 2-month-old WT and Myb+/− mice were stained with Me4BodipyFL-Ahx3Leu3VS proteasome activity probe either with or without prior incubation with MG-132. MFI was calculated. n = 3, P = .007. (B) Protein synthesis was assessed in 12-month-old KSL cells from WT and Myb+/− mice, either with or without cycloheximide inhibition, using the OPP assay. MFI was calculated. n = 3, P = .016. (C) Proliferation rate of WT vs Myb+/− HSC from 2-month-old mice. Bone marrow from WT and Myb+/− mice (n = 5) were fixed and permeabilized for intracellular anti-Ki67 staining. KSL HSC and LT-HSC (KSL48-150+) cells were gated for the analysis of Ki67. Data represents the average percentage of Ki67+ cells with SEM. For KSL HSC P = .024. OPP, O-propargyl-puromycin; SEM, standard error of the mean.
Myb deficiency affects protein production and degradation. (A) KSL cells from 2-month-old WT and Myb+/− mice were stained with Me4BodipyFL-Ahx3Leu3VS proteasome activity probe either with or without prior incubation with MG-132. MFI was calculated. n = 3, P = .007. (B) Protein synthesis was assessed in 12-month-old KSL cells from WT and Myb+/− mice, either with or without cycloheximide inhibition, using the OPP assay. MFI was calculated. n = 3, P = .016. (C) Proliferation rate of WT vs Myb+/− HSC from 2-month-old mice. Bone marrow from WT and Myb+/− mice (n = 5) were fixed and permeabilized for intracellular anti-Ki67 staining. KSL HSC and LT-HSC (KSL48-150+) cells were gated for the analysis of Ki67. Data represents the average percentage of Ki67+ cells with SEM. For KSL HSC P = .024. OPP, O-propargyl-puromycin; SEM, standard error of the mean.
Similarly, ribosome activity was measured in 12-month-old HSC by labeling using O-propargyl-puromycin together with Click-iT technology and flow cytometric analysis (Figure 6B). Twelve-month-old Myb+/− HSC showed a significantly two-fold higher MFI (P = .016) compared with equivalent WT cells, indicating an increase in protein synthesis in the aged Myb-insufficient cells above the low levels observed (and expected) in normal HSC.
As an indicator of proliferation rate, we stained cells with an antibody specific to Ki67 and analyzed using flow cytometry. This analysis revealed a higher rate of proliferation in Myb-insufficient HSC, which was significant in the broad KSL population but less pronounced in the LT-HSC (Figure 6C).
Overall, the functional assays show that the alterations in proteostasis- and proliferation-associated gene expression as a consequence of Myb insufficiency have a measurable impact on the corresponding biological processes.
Discussion
Transcription factor networks enable HSCs to respond to external signals reflecting changes in the requirements for differentiated cells.28 Such networks encompass multiple crossregulatory interactions. Any single acquired or inherited alteration in an individual transcription factor might be expected to disturb the network. Although such a disturbance may be subtle, the consequence could be a significant change in cell function, resulting in disease initiation or facilitation.29
Myb plays a critical role in hematopoiesis, with perturbations of its activity leading to cell deficits, cellular dysfunction, and malignancy.30,31 Our studies, using models of Myb deficiency, have illustrated that the requirement of Myb has both loss- and gain-of-function consequences when the normal level is disturbed.12,13 A natural inherited variation in the regulation of MYB gene expression is associated with a range of hematological conditions, including MPN.9,32 The MPN risk–associated variants cause lower expression of MYB, but it is unclear how this leads to disease in older age.
Myb-haploinsufficient mice are in most respects hematologically indistinguishable from their WT counterparts, at least up to 1 year of age, although more profound deficiency results in significant perturbations with features of MPN from an early age.10,13 This study shows that the aging of Myb-haploinsufficient animals leads to myelomonocytic neoplastic phenotypes, MPN/MDS predominating with a significant proportion of leukemia, some exhibiting extramedullary invasion.
The fact that constitutive Myb insufficiency only reveals as a hematological disease in later life raises the question of the reason behind the delay and whether this is a consequence of cell intrinsic or extrinsic aging effects. Studies on HSCs have shown that age-related decline in stem cell capability and consequent risk of hematological disease, results from a combination of intrinsic and extrinsic changes.6 The intrinsic changes involve DNA damage, cell cycle regulation, epigenetic and transcriptional regulation of gene expression, metabolism, proteostasis, and autophagy, many of which are interrelated. Age-dependent extrinsic influences include the composition and architecture of the niche and both local and systemic changes in cytokines and chemokines. Overall, these changes lead to increased proliferation, diminished self-renewal potential, skewed commitment bias toward myeloid lineages, and limited clonal diversity.5,33-35
By inducing loss of 1 Myb allele in young or older animals, we have shown that signs of MDS/MPN/leukemia arise after a considerable time lag, irrespective of the age at the time of deletion, implying that the “clock” relating to the effects of Myb insufficiency has a major cell intrinsic component. Given the time-dependent intrinsic effect of Myb deficiency, we looked for cell function differences in Myb+/− HSC at 2 and 12 months of age before any signs of hematological changes. HSC numbers seemed affected by both age and Myb insufficiency, the increase in WT stem cells being in line with previous studies,36 whereas in young Myb+/−, the absolute number of HSC was higher than in WT mice. Assaying HSC function revealed that both young and old Myb+/− HSC are compromised, exhibiting profoundly decreased self-renewal potential and skewing toward myeloid commitment. Because Myb operates through most stages of hematopoiesis, it might be expected that Myb insufficiency would have effects in different lineages. However, the lineage skewing we observed appears to be a consequence of applying stress to the HSCs (ie, by transplantation) because the steady-state blood values for WT and Myb+/− mice are comparable, at least before disease phenotypes arise (supplemental Figure 2).
KEGG pathway analysis suggests that in young adults, Myb insufficiency has an impact on DNA replication, metabolism, and especially the proteasome, changes in each of which have been linked with aging.6 Crucially, the effect on the genes associated with these processes is relatively small, being of the order of a 10% to 20% change in expression. Although similar differences in proteasome-associated genes persist in older animals, Myb+/− HSC additionally exhibits a striking increase in the expression of ribosome biogenesis –associated genes, which is reflected by an increase in protein synthesis. Although our findings suggest that life-long imbalance in proteasomal activity because of Myb insufficiency is a major driver for perturbed proteostasis, it is also quite possible that other aspects of cellular function, like proliferation and metabolism, could also have a contribution.
Proteostasis is key in stem cell aging,37 the quiescent state in HSC in normal circumstances being characterized by low levels of protein synthesis that serve to reduce the accumulation of misfolded proteins, thereby minimizing the need for protein clearance.38 HSC exhibits lower proteasome activity compared with the downstream progenitors,21 applying a tight control of protein synthesis, at least in part through targets of mTOR.39 The Myb-dependent increase in proteasome activity in HSC could cause an imbalance in the proteome, which in turn stimulates ribosome biogenesis. The consequent upturn in protein synthesis, which accelerates with age, could result in damaged protein accumulation, already known to be a feature of leukemogenesis originating from HSC, thereby creating an intracellular environment that compromises HSC function and possibly facilitating the selection of disease driver mutations.
The elevated expression of proteasome-associated genes in young Myb+/− mice suggests that this is a direct consequence of Myb insufficiency. This is supported by the observation that in human K562 cells, some proteasome-associated (PSM) genes are bound by MYB and respond to MYB knockdown23 and furthermore are rapidly affected by conditional Myb deletion in mouse HSCs. Most Psm genes seem to be influenced by Myb haploinsufficiency, including those encoding the α and β components of the 20S subunit and many of the 19S cap elements. Proteasome-associated genes seem to be coordinately regulated,40 although the mechanisms involved are unclear, and Myb could be a common PSM-gene regulatory component. Analysis of the 8 human PSM-gene homologs of the Myb insufficiency–responsive mouse genes showed that all are negatively regulated by MYB and are bound by MYB in their promoter region.
The link between Myb, proteasome-associated gene regulation, and myeloid disease propensity is further supported by GWAS traits for the key highlighted PSM genes . Hence, Psm genes elevated in Myb+/− HSC and in K562 cells are bound by MYB and respond to MYB knockdown and have been linked to variations in red cell numbers (PSMA3, PSMA7, PSMB1, and PSMG3), platelet numbers (PSMA7, PSMB1, PSMD13, and PSMG1), or the occurrence of acute myeloid leukemia (PSMB3).
It is clear that susceptibility to myeloid neoplastic disease can have an inherited component, as is apparent from the occurrence of disease in close relatives of individuals who have already died,41,42 and that variations in a number of genes can be identified as risk factors.43 Aside from SNPs upstream of the MYB gene,9 other proteins that influence MYB expression, such as GATA2 and KMT2a, are also associated with myeloid disease susceptibility.44-46 Whatever the inherited variation affecting MYB levels, what we have shown is that the impact on HSC can be a broad range of small molecular changes, particularly affecting proteostasis, which collectively and progressively shifts the cellular milieu, potentially providing the conditions for clonal expansion, especially in the circumstance when an acquired driver mutation arises.
Acknowledgments
The authors thank Paloma García for help in classifying the hematological disease phenotypes and to past members of the Frampton and García groups for their assistance and discussions. The authors are also grateful to the staff in Genomics Birmingham for next-generation sequencing support and the Biomedical Services Unit for animal care. Computational analysis of the RNA-seq data was performed on the CaStLeS infrastructure at the University of Birmingham. Part of the bioinformatic analysis was performed on the Saga supercomputing resources (Project nn9374k) provided by UNINETT Sigma2, the National Infrastructure for High Performance Computing and Data Storage in Norway.
This work was supported by Blood Cancer UK through a Specialist Programme Grant (ref 12030) and a Continuity Grant (ref 17008), and relied upon a MoFlo XDP cell sorter funded through a Wellcome Trust Equipment Grant (084330/Z/07/Z). B.N. was funded through the Cancer Research UK Birmingham Centre award C17422/A25154.
Authorship
Contribution: M.L.C. designed and conducted experiments, analyzed data, and wrote the manuscript; R.B.L. and O.S.G. provided RNA-seq and ChIP-seq data sets and their analysis and edited the manuscript; B.N. performed bioinformatic analysis; G.V. collected and processed samples; D.S.W. provided technical support; and J.F. designed the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: M. L. Clarke, Institute of Cancer & Genomic Sciences, College of Medical & Dental Sciences, University of Birmingham, Birmingham B15 2TT, United Kingdom; e-mail: m.l.clarke@bham.ac.uk.
References
Author notes
All RNA-seq data have been deposited in the European Nucleotide Archive (ENA) (# PRJEB47199) and are publicly available as of the date of publication.
Data are available on request from the corresponding author, M.L. Clarke (m.l.clarke@bham.ac.uk).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal