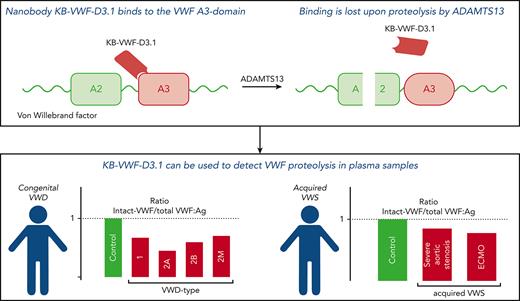
Key Points
Nanobody KB-VWF-D3.1 binds to the collagen-binding site in the VWF A3 domain, and it loses its binding upon proteolysis of VWF by ADAMTS13.
KB-VWF-D3.1 identified VWF degradation in patients with VWD, which correlated with a loss of larger VWF multimers.
Abstract
von Willebrand factor (VWF) is a multimeric protein, the size of which is regulated via ADAMTS13-mediated proteolysis within the A2 domain. We aimed to isolate nanobodies distinguishing between proteolyzed and non-proteolyzed VWF, leading to the identification of a nanobody (designated KB-VWF-D3.1) targeting the A3 domain, the epitope of which overlaps the collagen-binding site. Although KB-VWF-D3.1 binds with similar efficiency to dimeric and multimeric derivatives of VWF, binding to VWF was lost upon proteolysis by ADAMTS13, suggesting that proteolysis in the A2 domain modulates exposure of its epitope in the A3 domain. We therefore used KB-VWF-D3.1 to monitor VWF degradation in plasma samples. Spiking experiments showed that a loss of 10% intact VWF could be detected using this nanobody. By comparing plasma from volunteers to that from congenital von Willebrand disease (VWD) patients, intact-VWF levels were significantly reduced for all VWD types, and most severely in VWD type 2A–group 2, in which mutations promote ADAMTS13-mediated proteolysis. Unexpectedly, we also observed increased proteolysis in some patients with VWD type 1 and VWD type 2M. A significant correlation (r = 0.51, P < .0001) between the relative amount of high–molecular weight multimers and levels of intact VWF was observed. Reduced levels of intact VWF were further found in plasmas from patients with severe aortic stenosis and patients receiving mechanical circulatory support. KB-VWF-D3.1 is thus a nanobody that detects changes in the exposure of its epitope within the collagen-binding site of the A3 domain. In view of its unique characteristics, it has the potential to be used as a diagnostic tool to investigate whether a loss of larger multimers is due to ADAMTS13-mediated proteolysis.
Introduction
von Willebrand factor (VWF) is a multimeric protein, the extent of which regulates its interaction with platelets. Multimerization of VWF occurs during its synthesis in megakaryocytes or endothelial cells.1,2 In this process, 2 VWF subunits (with the following domain structure: D1-D2-D'-D3-A1-A2-A3-D4-C1-C2-C3-C4-C5-C6-CK) are first covalently linked via disulfide bridging between 2 C-terminal CK domains. These pro-dimers are then processed into multimers via N-terminal coupling of the D'-D3 regions, with the D1-D2 portion (also known as VWF propeptide) being eliminated during this event. The multimer size in endothelial cells is highly variable and varies from dimers to ultra-large multimers with >40 subunits. Upon secretion, VWF multimers are susceptible to regulated proteolysis by ADAMTS13, a metalloprotease that cleaves VWF in its A2 domain at the Tyr1605-Met1606 peptide bond.3 Important to note is that proteolysis occurs only upon decryption of the cleavage site, which normally lies buried within the A2 domain.4
Several types of occurrence allow for the exposure of the ADAMTS13-cleavage site. First, multiple VWF multimers assemble at the endothelial surface upon stimulated secretion, forming elongated fibers that are proteolyzed by ADAMTS13.5-7 Second, VWF unfolds during circulation under conditions of increased shear stress or disturbed blood flow.8 When disturbed blood flow is more intense, as it is in patients experiencing severe aortic stenosis and those requiring mechanical circulatory support, excessive VWF degradation may occur, which is referred to as acquired von Willebrand syndrome (AVWS).9-11 Third, mutations within VWF may provoke exposure of the ADAMTS13 proteolytic site, and such mutations are most frequently found in von Willebrand disease (VWD)–type 2A–group 2 and VWD type 2B.12,13
Excessive proteolysis of VWF is associated with an increased loss of high–molecular weight (HMW) multimers, which results in reduced platelet binding and reduced collagen binding, thereby increasing the risk of bleeding.9 The classic approach used to visualize the extent of VWF degradation is to analyze the multimeric pattern using sodium dodecyl sulfate (SDS)–agarose electrophoresis.14 This approach is laborious, nonstandardized, and requires 24 to 72 hours, depending on the method used. Alternatively, collagen- or platelet-binding assays are used,15 which are less specific in that they do not distinguish between impaired multimerization and excessive degradation. Finally, Kato and colleagues described a monoclonal antibody that binds to the newly formed C-terminal end in the A2 domain (residues Asp1596-Tyr1605). This antibody was successfully used to measure ADAMTS13 activity in patients with thrombotic thrombocytopenic purpura.16 In addition, we and others used this antibody to monitor VWF proteolysis in patients.17,18 However, while using an antibody targeting residues Asp1596-Tyr1605, we noticed that this antibody was unable to detect degradation, as for instance, in samples from patients with nonsevere aortic stenosis or congenital VWD type 1 or 2M.
For the present study, we developed a nanobody (designated KB-VWF-D3.1) that distinguishes between intact and ADAMTS13-cleaved VWF. Interesting to note is that this nanobody binds to the A3 domain, showing that proteolysis within the A2 domain modifies the exposure of the nanobody’s epitope in the adjacent A3 domain. This nanobody proved that it has sufficient sensitivity to detect degradation in plasma from patients with AVWS and congenital VWD, including types 1 and 2M.
Materials and methods
An extensive description of the experimental procedures can be found in the supplemental Methods (available on the Blood website).
All volunteers and patients provided informed written consent according to the Declaration of Helsinki. Patients with VWD were selected from the French cohort multicentric database of VWD (Centre Reference Maladie Willebrand).19 The database and biobank of this cohort are declared to and approved by the French data protection authority (CNIL-1245379/DEC-19252, CODECOH-DC-2008-642). Patients with severe aortic stenosis (Von Willebrand Factor as a Biological Sensor of Blood Flow in Percutaneous Cardiac Procedure [WITAVI] trial, NCT02628509) and patients receiving extracorporeal mechanical oxidation (ECMO; Frequency of Hemorrhages Associated With the Functional Anomalies of Willebrand Factor in Emergency Patients (WITECMO-H) trial; NCT03070912) were included in the study. All protocols were approved by the local review and ethics committees.
Isolation of anti-VWF nanobodies
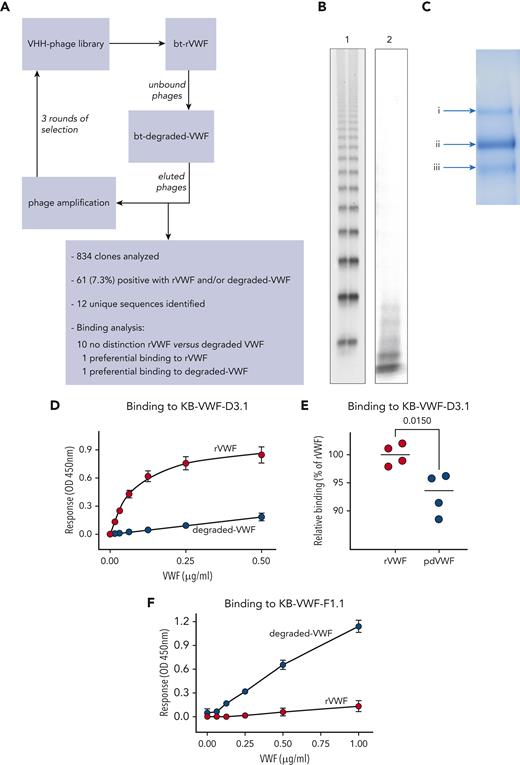
A synthetic nanobody-encoding phage library20 was used to isolate anti-VWF nanobodies. The library (3 × 109 clones) was incubated with streptavidin-coated beads loaded with biotinylated recombinant (r)VWF. Unbound phages were then incubated with beads loaded with biotinylated degraded VWF. Three rounds of phage display were performed, with the depletion step repeated every round. Twelve unique sequences were obtained via this procedure (Figure 1A).
Generation of anti-VWF nanobodies. (A) Flow diagram of screening approach using rVWF and degraded VWF for the isolation of anti-VWF nanobodies that distinguish between intact and degraded VWF. (B) SDS-agarose gel (2%) of rVWF (lane 1) and the degraded-VWF preparation (lane 2) used for screening. (C) Coomassie-staining of an SDS-polyacrylamide gel under reducing conditions containing the degraded-VWF preparation used for screening. Arrows indicate (Ci) intact VWF and (Cii, iii) degraded-VWF fragments. (D-F) Dose-response of rVWF (red circles) and degraded VWF (blue circles) to immobilized nanobody KB-VWF-D3.1 (5 μg/mL) (D) or KB-VWF-F1.1 (5 μg/mL) (F). (E) Comparison of rVWF to pdVWF, both added at a concentration of 5 μg/mL. Bound VWF was probed using peroxidase-labeled polyclonal anti-VWF antibodies and detected via hydrolysis of 3,3',5,5'-tetramethylbenzidine. Data represent mean ± SD of 4 to 8 experiments. bt, biotinylated; OD, optical density; VHH, variable domain of heavy chain-only antibodies.
Generation of anti-VWF nanobodies. (A) Flow diagram of screening approach using rVWF and degraded VWF for the isolation of anti-VWF nanobodies that distinguish between intact and degraded VWF. (B) SDS-agarose gel (2%) of rVWF (lane 1) and the degraded-VWF preparation (lane 2) used for screening. (C) Coomassie-staining of an SDS-polyacrylamide gel under reducing conditions containing the degraded-VWF preparation used for screening. Arrows indicate (Ci) intact VWF and (Cii, iii) degraded-VWF fragments. (D-F) Dose-response of rVWF (red circles) and degraded VWF (blue circles) to immobilized nanobody KB-VWF-D3.1 (5 μg/mL) (D) or KB-VWF-F1.1 (5 μg/mL) (F). (E) Comparison of rVWF to pdVWF, both added at a concentration of 5 μg/mL. Bound VWF was probed using peroxidase-labeled polyclonal anti-VWF antibodies and detected via hydrolysis of 3,3',5,5'-tetramethylbenzidine. Data represent mean ± SD of 4 to 8 experiments. bt, biotinylated; OD, optical density; VHH, variable domain of heavy chain-only antibodies.
Analysis of VWF binding to nanobodies
Wells coated with nanobody KB-VWF-D3.1 or KB-VWF-1.1 (both 5 μg/mL) were incubated with purified rVWF (0-0.5 μg/mL). Bound VWF was probed using polyclonal anti-VWF antibodies and detected via hydrolysis of 3,3',5,5'-tetramethylbenzidine.
Detecting intact VWF
Intact VWF is referred to as VWF that is recognized by KB-VWF-D3.1. Briefly, wells coated with KB-VWF-D3.1 (5 μg/mL) were incubated with samples containing nonproteolyzed VWF, ADAMTS13-proteolyzed VWF, or a mixture of both. Alternatively, plasma samples were used. Bound VWF was probed using polyclonal anti-VWF antibodies and detected via hydrolysis of 3,3',5,5'-tetramethylbenzidine.
Total VWF antigen
Total VWF antigen was measured in an enzyme-linked immunosorbent assay using polyclonal rabbit anti-VWF antibodies as described.21
Results
Selection of anti-VWF nanobodies
To isolate nanobodies that distinguish between intact vs proteolyzed VWF, a selection method using rVWF and degraded VWF was applied, generating 12 unique sequences (Figure 1A-C). Purified nanobodies were tested for interaction with rVWF and degradedVWF. Whereas 10 of 12 nanobodies displayed similar binding to both VWF preparations, 2 of them were characterized by differential binding. First, rVWF associated in a dose-dependent manner to immobilized nanobody KB-VWF-D3.1, whereas binding of degraded VWF to this nanobody was strongly reduced (Figure 1D). In these assays, plasma-derived (pd)VWF yielded responses that were consistently lower than those of rVWF (93% ± 4% compared to 100% ± 2%; P = .015; Figure 1E), probably due to degraded VWF being present in normal plasma. In complementary assays, KB-VWF-D3.1 bound 8-fold less efficiently to immobilized degraded VWF, compared to immobilized rVWF (supplemental Figure 1). As for KB-VWF-F1.1, efficient binding of degraded VWF was detected, whereas binding of rVWF approached background levels (Figure 1F). Therefore, these 2 nanobodies were selected for further analysis.
Determination of the binding epitope for KB-VWF-F1.1
Given that KB-VWF-F1.1 bound to degraded VWF but not intact VWF, we anticipated that this nanobody recognizes the region surrounding the Tyr1605-Met1606 cleavage site. This possibility was tested in an ADAMTS13-activity test utilizing its substrate FRETS-VWF73, which contains the VWF A2-domain sequence Asp1596-Arg1668 (supplemental Figure 2A). Whereas KB-VWF-D3.1 and control nanobody KB-VWF-004 (against the region VWF/D4-CK) left substrate proteolysis unaffected, KB-VWF-F1.1 efficiently interfered with substrate conversion by ADAMTS13. This finding suggests that the epitope of VWF-KB-F1.1 is located within the region Asp1596-Arg1668. Of note, KB-VWF-F1.1 and MAB27642, which targets residues Arg1596-Tyr1605, did not compete for binding to degraded VWF (supplemental Figure 2B), suggesting that they recognize different epitopes within this region.
Determination of the binding epitope for KB-VWF-D3.1
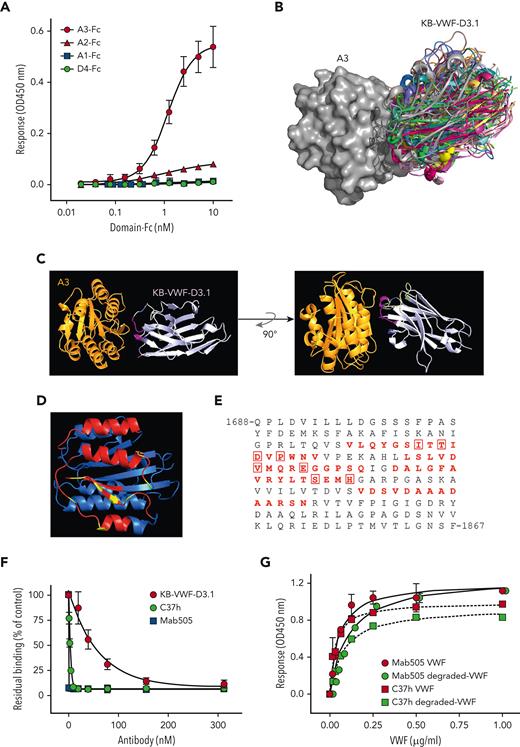
To determine the epitope of KB-VWF-D3.1, we first analyzed binding of this nanobody to a series of rVWF fragments, specifically, A1-Fc, A2-Fc, A3-Fc, and D4-Fc. Surprisingly, KB-VWF-D3.1 bound most efficiently to the A3-Fc fragment rather than to the A2-Fc fragment (Figure 2A). Binding was similar when binding of fragments to immobilized KB-VWF-D3.1 was assessed (supplemental Figure 3A). Moreover, no binding of rVWF lacking the A3 domain to KB-VWF-D3.1 could be detected, whereas deletion of other domains left binding unaffected (supplemental Figure 3B).
KB-VWF-D3.1 binds to the VWF A3 domain. (A) Binding of KB-VWF-D3.1 (1 μg/mL) to various concentrations of VWF domain-Fc fusion proteins (0-10 nM) that were captured onto anti-human Fc antibodies. Bound KB-VWF-D3.1 was probed using peroxidase-labeled polyclonal rabbit anti-cMyc antibodies and detected following hydrolysis of 3,3',5,5'-tetramethylbenzidine. Blue squares, A1-Fc; red triangles, A2-Fc; red circles, A3-Fc: green circles, D4-Fc. Data represent mean ± SD of 3 experiments. (B) In silico simulation of KB-VWF-D3.1 (colored structures) docking on the VWF A3 domain (grey structure). Shown are the top 30–ranked structures of KB-VWF-D3.1, which all bind in a similar fashion to the A3 domain. (C) Single-structure representation of KB-VWF-D3.1 binding to the A3 domain. (D) The VWF A3 domain, with residues in red representing amino acids predicted to be in the epitope of KB-VWF-D3.1. Residues known to affect collagen binding are indicated in yellow. (E) Amino acid sequence of the VWF A3 domain, with the residues predicted to harbor the epitope for KB-VWF-D3.1 in red. Residues previously reported to be involved in collagen binding22 are boxed. (F) Inhibition of pdVWF binding to collagen-type III by KB-VWF-D3.1 (red circles), monoclonal antibody Mab505 (blue squares) and nanobody C37h (green circles). Presented is residual pdVWF binding vs nanobody/antibody concentration. Data represent mean ± SD of 3 experiments. (G) Binding of pdVWF (red symbols) or degraded-VWF (green symbols) to immobilized Mab505 (5 μg/mL; circles) or C37h (5 μg/mL; squares). Bound pdVWF was probed using peroxidase-labeled polyclonal anti-VWF antibodies and detected via hydrolysis of 3,3',5,5'-tetramethylbenzidine. Data represent mean ± SD of 3 experiments. OD, optical density.
KB-VWF-D3.1 binds to the VWF A3 domain. (A) Binding of KB-VWF-D3.1 (1 μg/mL) to various concentrations of VWF domain-Fc fusion proteins (0-10 nM) that were captured onto anti-human Fc antibodies. Bound KB-VWF-D3.1 was probed using peroxidase-labeled polyclonal rabbit anti-cMyc antibodies and detected following hydrolysis of 3,3',5,5'-tetramethylbenzidine. Blue squares, A1-Fc; red triangles, A2-Fc; red circles, A3-Fc: green circles, D4-Fc. Data represent mean ± SD of 3 experiments. (B) In silico simulation of KB-VWF-D3.1 (colored structures) docking on the VWF A3 domain (grey structure). Shown are the top 30–ranked structures of KB-VWF-D3.1, which all bind in a similar fashion to the A3 domain. (C) Single-structure representation of KB-VWF-D3.1 binding to the A3 domain. (D) The VWF A3 domain, with residues in red representing amino acids predicted to be in the epitope of KB-VWF-D3.1. Residues known to affect collagen binding are indicated in yellow. (E) Amino acid sequence of the VWF A3 domain, with the residues predicted to harbor the epitope for KB-VWF-D3.1 in red. Residues previously reported to be involved in collagen binding22 are boxed. (F) Inhibition of pdVWF binding to collagen-type III by KB-VWF-D3.1 (red circles), monoclonal antibody Mab505 (blue squares) and nanobody C37h (green circles). Presented is residual pdVWF binding vs nanobody/antibody concentration. Data represent mean ± SD of 3 experiments. (G) Binding of pdVWF (red symbols) or degraded-VWF (green symbols) to immobilized Mab505 (5 μg/mL; circles) or C37h (5 μg/mL; squares). Bound pdVWF was probed using peroxidase-labeled polyclonal anti-VWF antibodies and detected via hydrolysis of 3,3',5,5'-tetramethylbenzidine. Data represent mean ± SD of 3 experiments. OD, optical density.
To refine its binding site within the A3 domain, molecular modeling was performed (see supplemental Materials). This procedure revealed that the top 30–ranked conformations of the complex all clustered similarly, with the nanobody docking onto 4 separate amino acid stretches within the VWF A3-domain region Val1731-Asn1818 (Figure 2B-D). Interestingly, 8 of the amino acids included in the epitope of KB-VWF-D3.1 have been recognized previously as being relevant for collagen binding (Figure 2E),22 indicating that the epitope of KB-VWF-D3.1 overlaps the collagen-binding site. Consequently, we compared the effect of KB-VWF-D3.1 to that of 2 known A3 domain–binding antibodies (the C37h nanobody23 and the Mab505 monoclonal antibody24) on binding of VWF to collagen type III. The positive controls C37h and Mab505 efficiently blocked pdVWF–collagen interactions (Figure 2F). KB-VWF-D3.1 also dose-dependently reduced binding of pdVWF to collagen, but less efficiently than antibodies C37h and Mab505 (Figure 2F). In addition, KB-VWF-D3.1 delayed VWF-dependent platelet adhesion to collagen under flow conditions (supplemental Figure 3C). Having their epitope overlapping the collagen-binding site in common raises the question of whether C37h and Mab505 can distinguish between intact and degraded VWF akin to KB-VWF-D3.1. However, C37h and Mab505 displayed similar binding to both intact rVWF and degraded VWF (Figure 2G). Thus, nanobody KB-VWF-D3.1 is unique in binding to an epitope within the A3 domain, the exposure of which is modulated upon proteolysis within the A2 domain.
Effect of multimer size on VWF binding to KB-VWF-D3.1
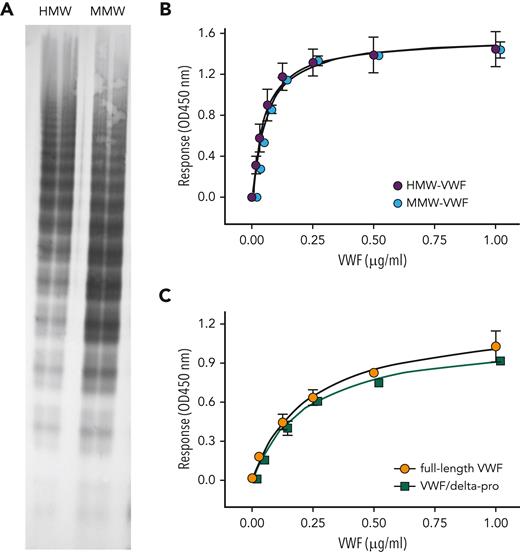
Proteolysis of VWF by ADAMTS13 results in loss of the Tyr1605-Met1606 peptide bond, thereby reducing multimer size. We therefore tested how multimer size affects binding of VWF to immobilized KB-VWF-D3.1. First, we analyzed 2 distinct pdVWF preparations that were obtained from pdVWF concentrates via gel-filtration chromatography. One had HMW multimers, and one was enriched in medium–molecular-weight multimers (Figure 3A). The 2 fractions displayed similar binding to KB-VWF-D3.1 (Figure 3B). Next, we compared binding of dimeric rVWF/delta-pro to that of full-length rVWF (Figure 3C). Both dimeric rVWF/delta-pro and rVWF bound to KB-VWF-D3.1 with similar half-maximal binding (0.2 ± 1 μg/mL vs 0.2 ± 0.1 μg/mL; P = .62). Apparently, binding of VWF to immobilized KB-VWF-D3.1 is independent of its multimer size. Reduced binding of degraded VWF to KB-VWF-D3.1 conceivably originates from proteolysis of the Tyr1605-Met1606 peptide bond rather than from a reduction in multimer size.
Binding of VWF with varying multimer size to KB-VWF-D3.1. (A) pdVWF concentrates were applied to gel-filtration chromatography using Bio-Gel-A-15m. Fractions enriched in HMW-multimers (HMW-VWF) and medium–molecular weight multimers (MMW-VWF) were analyzed for their multimeric pattern using SDS-agarose (2%) electrophoresis. (B) Binding of HMW-VWF (purple circles) and MMW-VWF (blue circles) to immobilized KB-VWF-D3.1 (5 μg/mL). (C) Binding of multimeric rVWF (orange circles) and the dimeric VWF/delta-pro variant (green squares) to immobilized KB-VWF-D3.1 (5 μg/mL). In both panels, bound VWF was probed using peroxidase-labeled polyclonal anti-VWF antibodies and detected via hydrolysis of 3,3',5,5'-tetramethylbenzidine. Data represent mean ± SD of 3 to 4 independent measurements. OD, optical density.
Binding of VWF with varying multimer size to KB-VWF-D3.1. (A) pdVWF concentrates were applied to gel-filtration chromatography using Bio-Gel-A-15m. Fractions enriched in HMW-multimers (HMW-VWF) and medium–molecular weight multimers (MMW-VWF) were analyzed for their multimeric pattern using SDS-agarose (2%) electrophoresis. (B) Binding of HMW-VWF (purple circles) and MMW-VWF (blue circles) to immobilized KB-VWF-D3.1 (5 μg/mL). (C) Binding of multimeric rVWF (orange circles) and the dimeric VWF/delta-pro variant (green squares) to immobilized KB-VWF-D3.1 (5 μg/mL). In both panels, bound VWF was probed using peroxidase-labeled polyclonal anti-VWF antibodies and detected via hydrolysis of 3,3',5,5'-tetramethylbenzidine. Data represent mean ± SD of 3 to 4 independent measurements. OD, optical density.
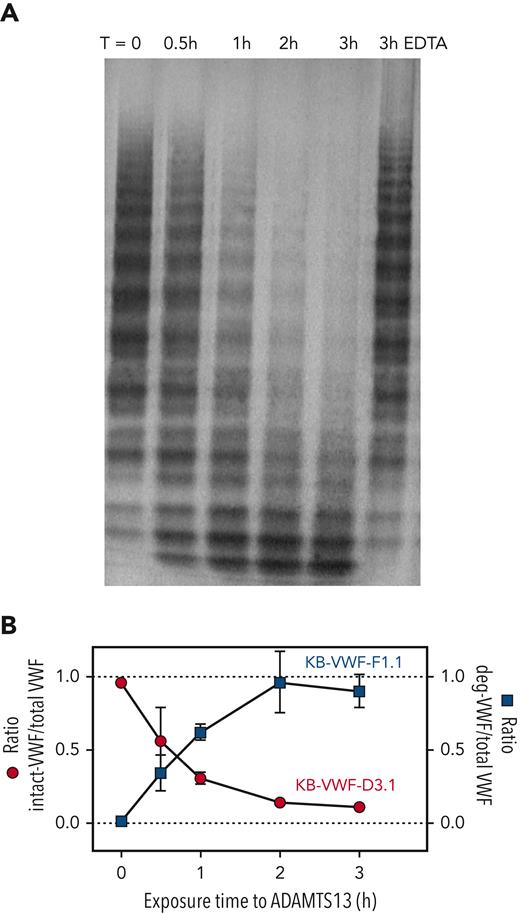
Proteolysis of VWF over time
We next investigated the effect of ADAMTS13 proteolysis on VWF binding to KB-VWF-D3.1 and KB-VWF-F1.1 in a time-dependent manner. Briefly, pdVWF was exposed to shear in the presence of recombinant ADAMTS13, and samples were taken at indicated time-points (0-3 hours). Multimeric pattern and binding to both nanobodies were analyzed. Exposure to ADAMTS13 resulted in a time-dependent decrease in pdVWF multimer size (Figure 4A). As expected, proteolysis was inhibited in the presence of EDTA, a metal-ion chelator that renders ADAMTS13 inactive. Concurrent with increased pdVWF proteolysis, increased binding to KB-VWF-F1.1 was observed (Figure 4B). In contrast, binding of pdVWF to KB-VWF-D3.1 disappeared in a complementary fashion (Figure 4B). These data validate that the binding of both nanobodies to VWF is dependent on the extent of proteolysis by ADAMTS13.
ADAMTS13-mediated proteolysis modulates binding of VWF to KB-VWF-D3.1 and KB-VWF-F1.1. (A) Purified pdVWF was incubated with recombinant ADAMTS13 and exposed to vortex-induced shear. Samples taken at indicated time (T)-points (0-3 hours) were analyzed via SDS-agarose electrophoresis. As a control, pdVWF was exposed to ADAMTS13 and shear for 3 hours in the presence of EDTA, a chelating agent that blocks ADAMTS13 activity. (B) Samples were analyzed for total VWF-antigen using polyclonal antibodies, for the presence of intact-VWF using KB-VWF-D3.1 and for the presence of degraded-VWF using KB-VWF-F1.1. Presented is the ratio of intact VWF to total VWF antigen (red circles; left y-axis) and the ratio of degraded VWF to total VWF antigen (blue squares; right y-axis) vs exposure time to ADAMTS13. Normal pooled plasma was used as calibrator for KB-VWF-D3.1, whereas a degraded-VWF preparation was used as calibrator for KB-VWF-F1.1. Data represent mean ± SD of 3 independent experiments.
ADAMTS13-mediated proteolysis modulates binding of VWF to KB-VWF-D3.1 and KB-VWF-F1.1. (A) Purified pdVWF was incubated with recombinant ADAMTS13 and exposed to vortex-induced shear. Samples taken at indicated time (T)-points (0-3 hours) were analyzed via SDS-agarose electrophoresis. As a control, pdVWF was exposed to ADAMTS13 and shear for 3 hours in the presence of EDTA, a chelating agent that blocks ADAMTS13 activity. (B) Samples were analyzed for total VWF-antigen using polyclonal antibodies, for the presence of intact-VWF using KB-VWF-D3.1 and for the presence of degraded-VWF using KB-VWF-F1.1. Presented is the ratio of intact VWF to total VWF antigen (red circles; left y-axis) and the ratio of degraded VWF to total VWF antigen (blue squares; right y-axis) vs exposure time to ADAMTS13. Normal pooled plasma was used as calibrator for KB-VWF-D3.1, whereas a degraded-VWF preparation was used as calibrator for KB-VWF-F1.1. Data represent mean ± SD of 3 independent experiments.
Measuring degraded VWF in mixtures of intact and degraded VWF
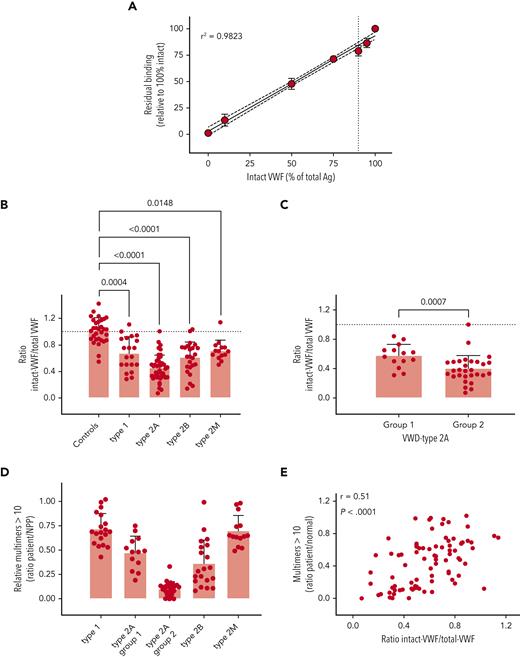
Given the specificity of both nanobodies for intact and degraded VWF, respectively, we anticipated that they could be useful in determining the extent of VWF proteolysis in patient samples. In preliminary experiments, KB-VWF-F1.1 lacked sufficient sensitivity to detect minor proteolysis of VWF in plasma, and we therefore focused for the remainder of the study on KB-VWF-D3.1. We first analyzed the extent to which increased proteolysis would be detectable. Different mixtures of purified rVWF and degraded VWF were prepared, and the ratio of intact VWF to total VWF antigen was determined. A dose-dependent decrease of this ratio was observed when the percentage of degraded VWF in the samples increased (Figure 5A). These experiments suggest that an increase of approximately 10% degraded VWF can be detected (P = .0009, compared to 100% intact VWF).
Detection of intact VWF in congenital VWD. (A) VWF-deficient plasma was spiked with different amounts of purified rVWF and degraded VWF, and incubated in microtiter plates coated with KB-VWF-D3.1. Bound VWF was probed using peroxidase-labeled polyclonal anti-VWF antibodies and detected via hydrolysis of 3,3',5,5'-tetramethylbenzidine. Data represent mean ± SD of 3 to 4 independent measurements. The solid line illustrates the best linear fit, with 95% confidence intervals indicated with the dotted lines. The vertical line indicates 90% intact rVWF supplemented with 10% degradedVWF. (B, C) Patient plasma samples were analyzed for total antigen using polyclonal antibodies, and for intact VWF using KB-VWF-D3.1. Normal pooled plasma (NPP) was used as a calibrator. Presented is the ratio of intact VWF to total VWF antigen. Each individual sample is represented by a closed symbol. Statistical analysis was performed via a one-way analysis of variance with Dunnett’s correction for multiple comparisons (B) or Mann-Whitney (C). (D) Multimers were analyzed via SDS-agarose (Ag) electrophoresis. The relative amount of multimers exceeding 10 bands was determined via comparison to NPP. (E) Plotted is the ratio of intact VWF to total VWF antigen vs the relative amount of large multimers. Correlation was determined using GraphPad Prism Software (GraphPad, La Jolla, CA).
Detection of intact VWF in congenital VWD. (A) VWF-deficient plasma was spiked with different amounts of purified rVWF and degraded VWF, and incubated in microtiter plates coated with KB-VWF-D3.1. Bound VWF was probed using peroxidase-labeled polyclonal anti-VWF antibodies and detected via hydrolysis of 3,3',5,5'-tetramethylbenzidine. Data represent mean ± SD of 3 to 4 independent measurements. The solid line illustrates the best linear fit, with 95% confidence intervals indicated with the dotted lines. The vertical line indicates 90% intact rVWF supplemented with 10% degradedVWF. (B, C) Patient plasma samples were analyzed for total antigen using polyclonal antibodies, and for intact VWF using KB-VWF-D3.1. Normal pooled plasma (NPP) was used as a calibrator. Presented is the ratio of intact VWF to total VWF antigen. Each individual sample is represented by a closed symbol. Statistical analysis was performed via a one-way analysis of variance with Dunnett’s correction for multiple comparisons (B) or Mann-Whitney (C). (D) Multimers were analyzed via SDS-agarose (Ag) electrophoresis. The relative amount of multimers exceeding 10 bands was determined via comparison to NPP. (E) Plotted is the ratio of intact VWF to total VWF antigen vs the relative amount of large multimers. Correlation was determined using GraphPad Prism Software (GraphPad, La Jolla, CA).
Analysis of plasma of congenital VWD patients
We then analyzed plasma samples obtained from controls (n = 31) and VWD patients included in the French reference center for VWD (n = 101).19 The cohort consisted of patients with VWD type 1 (n = 20), type 2A (n = 43), type 2B (n = 24), and type 2M (n = 14).
To determine the amount of intact VWF, the amount of antigen obtained using KB-VWF-D3.1 (= intact VWF) was divided by the amount of total VWF antigen, using normal pooled plasma as a calibrator. In doing so, we found that the ratio of intact VWF to total antigen for controls was 1.0 ± 0.2 (Figure 5B). The ratio of intact VWF to total VWF was decreased for each of the VWD types analyzed. The ratio was as follows: 0.7 ± 0.3 (P = .0004) for type 1; 0.5 ± 0.2 (P < .0001) for type 2A; 0.6 ± 0.2 (P < .0001) for type 2B; and 0.7 ± 0.2 (P = .0148) for type 2M (Figure 5B). A separate graph showing the ratio for each mutation is presented in supplemental Figure 4. Given that VWD type 2A is divided into 2 subtypes—ie, VWD type 2A–group 1 and VWD type 2A–group 2, in which the loss of multimers is dominated by impaired multimerization and increased proteolysis, respectively, we separately analyzed samples from patients with VWD-type 2A–group 1 (n = 14) and VWD-type 2A–group 2 (n = 29). The ratio of intact VWF to total VWF antigen was significantly lower in VWD-type 2A–group 2 (0.4 ± 0.2), compared with that in VWD-type 2A–group 1 (0.6 ± 0.2; P = .0007; Figure 5C).
Given that the finding of a decreased ratio of intact VWF to total VWF antigen in all VWD types was unexpected, we also verified whether this decreased ratio would correspond to a potential loss of HMW multimers. Multimer analysis was available for a subset of samples, and we indeed observed that in all patient groups, including VWD type 1 and type 2M, on average, a relative decrease occurred in the HMW multimers (>10 multimer bands) compared to normal pooled plasma (Figure 5D). An example of a multimeric pattern with reduced HMW multimers for a VWD type 1 and a type 2M patient is provided in supplemental Figure 5. Interestingly, a significant correlation was found between the ratio of intact VWF to total VWF antigen vs multimer size (r = 0.51; P < .0001; Figure 5E). Thus, the majority of VWD patients appear to have increased proteolysis, compared to that in the normal population.
Analysis of plasma of AVWS patients
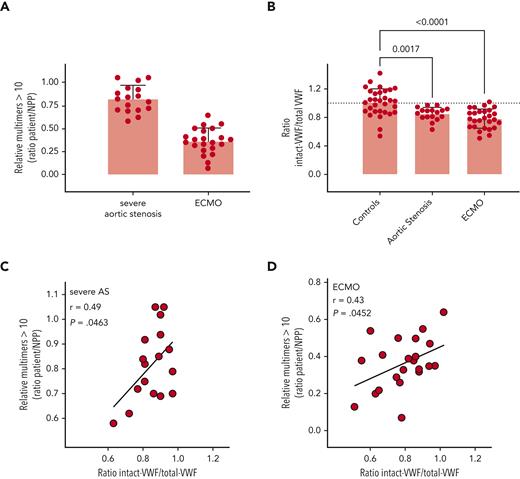
We next examined plasma from patients receiving extracorporeal membrane oxygenation (ECMO) support (n = 27) and patients with severe aortic stenosis (n = 17). Both patient groups are characterized by a loss of VWF HMW multimers (Figure 6A), potentially caused by increased ADAMTS13-mediated proteolysis. Compared to that in normal controls, the ratio of intact VWF (measured by binding to KB-VWF-D3.1) to total VWF antigen was significantly reduced for both patient groups, as follows (mean ± standard deviation [SD]): 0.85 ± 0.09 (P = .0017) and 0.78 ± 0.13 (P < .0001) for severe aortic stenosis and ECMO patients, respectively (Figure 6B). Of note, for both groups, a significant correlation was found between the ratio of intact to total antigen and the presence of HMW-multimers (>10), with P-values being P = .0463 for severe aortic stenosis-samples and P = .0452 for ECMO samples (Figures 6C-D). This finding suggests that the loss of larger multimers indeed is due predominantly to proteolysis rather than other mechanisms.
Detection of intact VWF in AVWS. (A) Multimers were analyzed via SDS-agarose electrophoresis. The relative amount of multimers exceeding 10 bands was determined via comparison to normal pooled plasma (NPP). (B) Patient plasma samples were analyzed for total antigen using polyclonal antibodies and for intact VWF using KB-VWF-D3.1. NPP was used as a calibrator. Presented is the ratio of intact VWF to total VWF antigen. Each individual sample is represented by a closed symbol. Statistical analysis was performed via a one-way analysis of variance with Dunnett’s correction for multiple comparisons. Control samples were identical to those presented in Figure 5. (C, D) Plotted is the ratio of intact VWF to total VWF antigen vs the relative amount of large multimers for samples from patients with severe aortic stenosis (AS) (C) and ECMO patients (D).
Detection of intact VWF in AVWS. (A) Multimers were analyzed via SDS-agarose electrophoresis. The relative amount of multimers exceeding 10 bands was determined via comparison to normal pooled plasma (NPP). (B) Patient plasma samples were analyzed for total antigen using polyclonal antibodies and for intact VWF using KB-VWF-D3.1. NPP was used as a calibrator. Presented is the ratio of intact VWF to total VWF antigen. Each individual sample is represented by a closed symbol. Statistical analysis was performed via a one-way analysis of variance with Dunnett’s correction for multiple comparisons. Control samples were identical to those presented in Figure 5. (C, D) Plotted is the ratio of intact VWF to total VWF antigen vs the relative amount of large multimers for samples from patients with severe aortic stenosis (AS) (C) and ECMO patients (D).
Discussion
In this study, we describe a novel nanobody, designated KB-VWF-D3.1, that loses its capacity to bind VWF upon proteolysis by ADAMTS13. We show that this nanobody displays reduced binding to VWF present in the plasma of congenital VWD patients, as well in those having an acquired deficiency of VWF. Unexpectedly, this nanobody revealed the presence of increased VWF degradation in some patients with VWD types 1 and 2M.
Structural conformational changes are key regulators of VWF function. Although several ways are available to monitor such structural changes, including functional assays and microscopic visualization, the use of specific antibodies is by far the most convenient and accessible approach. In 2005, a nanobody (AU-VWFa-11) was described that selectively binds to VWF in its active, glycoprotein Ibα-binding conformation, but not to globular inactive VWF.25 This nanobody indeed has been useful to identify the presence of active VWF under several pathological conditions.26-31 Following a similar strategy, we set out to identify nanobodies that distinguish between intact and ADAMTS13-proteolyzed VWF, leading to the successful identification of a nanobody that binds to intact—but not degraded—VWF (Figure 1).
Admittedly, others have reported antibodies that distinguish between intact and degraded VWF.16,32 These antibodies have in common that they recognize the VWF A2 domain and bind VWF only in its elongated or proteolyzed conformation. Our nanobody is different from these antibodies in 2 distinct ways. First, it binds to VWF in both globular and elongated conformations (allowing it to detect intact VWF in plasma samples). Second, it has its epitope in the VWF A3 domain. Of course, binding to the A3 domain is perhaps less relevant from a diagnostic point of view, but its unique epitope may provide new insights into the structure–function relationship of VWF. Indeed, the finding that the epitope of KB-VWF-D3.1 is in the A3 domain was unexpected in the sense that proteolysis of VWF by ADAMTS13 takes place in the adjacent A2 domain. Nevertheless, a series of experiments led to the clear conclusion that KB-VWF-D3.1 interacts with the A3 domain, and more specifically, that its epitope overlaps the collagen-binding site (Figure 2; supplemental Figure 3).
The notion that KB-VWF-D3.1 binds to the A3 domain clearly raises the question of how proteolysis in the A2 domain affects the exposure of the epitope of KB-VWF-D3.1 in the A3 domain. Previously, different domains within VWF have been shown to interact or communicate with each other. For instance, VWF has a 4-fold reduced affinity for FVIII when bound to collagen, suggesting that binding of the A3 domain to collagen affects the exposure of the FVIII binding site in the D'D3 region.33 Thus, one possibility is that proteolysis within the A2 domain results in conformational changes in the A3 domain. Second, the D'D3 region and the A2 domain can both bind to the A1 domain.34,35 This perspective reveals the possibility that proteolysis in the A2 domain allows a portion of this domain to move toward the A3 domain, binding to the adjacent epitope of KB-VWF-D3.1. Irrespective of the exact mechanism, given that this epitope overlaps the collagen-binding site, a tempting speculation is that an additional mechanism exists, by which ADAMTS13 reduces the hemostatic potential of VWF; not only would proteolysis reduce multimer size, but also, modulating the exposure of the collagen-binding site could reduce the capacity of VWF to bind collagen. In this scenario, the ratio of affected A3 domains to normal A3 domains would be determinant for the extent to which collagen binding is modulated. Additional experiments are warranted to explore this hypothesis in more detail.
Should the cleaved A2 domain start covering the epitope of KB-VWF-D3.1, then this may provide an explanation for why KB-VWF-D3.1, but not the concurrent antibodies C37h and Mab505, distinguishes between intact and degraded VWF. Both antibodies have a significant higher affinity for VWF. Given this, they would efficiently compete with the A2 domain for a common binding site within the A3 domain, and therefore be unable to detect proteolysis within the A2 domain.
Between the 2 nanobodies KB-VWF-D3.1 and KB-VWF-F1.1, the former proved to be more sensitive in detecting degradation of VWF. Both monovalent and bivalent variants of KB-VWF-F1.1 did bind degraded VWF only at relatively high concentrations. In this respect, KB-VWF-F1.1 seems similar to the antibody described by Kato and coworkers.16 A possibility is that the low affinity originates from the flexible nature of their epitope, the polypeptides at either end of the Tyr1605-Met1606 cleavage site.
Focusing on nanobody KB-VWF-D3.1, we investigated whether it could detect increased proteolysis in samples of patients. To do so, the nanobody should meet the following criteria: (i) binding should be independent of multimer size; (ii) binding should decrease dose-dependently upon an increased presence of proteolyzed VWF; and (iii) the nanobody should be sufficiently sensitive to detect low amounts of proteolyzed VWF. A number of control experiments indicate that KB-VWF-D3.1 does meet these 3 criteria (Figures 3-5). In particular, the notion that the presence of 10% degraded VWF could be detected is relevant.
Although this study was not designed to validate the KB-VWF-D3.1-based assay as a diagnostic tool, we did analyze a series of control and patient samples. We measured both the amount of antigen detected using KB-VWF-D3.1 (referred to as intact VWF) and the amount of antigen using polyclonal anti-VWF antibodies (referred to as total VWF antigen). Analysis of 31 control samples from volunteers showed that the ratio of intact VWF to total VWF antigen was 1.0 ± 0.2 (mean ± standard deviation), with a 95% confidence interval of 0.9 to 1.1 (Figure 5). A few of these samples were either particularly low, with a ratio of <0.82 (mean – 1 × SD; 3 of 31 samples) or particularly high (>1.20; mean + 1 × SD; 4 of 31 samples). Apparently, on average, the analyzed individual plasma samples do not contain less intact VWF, compared to the plasma calibrator. However, within this control group are a number of individuals in which proteolysis of VWF seems to be somewhat up- or down-regulated.
We also had access to a large group of congenital VWD patients. As expected, the ratio of intact VWF to total VWF antigen was lowest in samples from VWD type 2A–group 2 patients, who express mutated VWF that has increased sensitivity for ADAMTS13-mediated proteolysis.12 Also, for VWD type 2B, ADAMTS13 has been reported as a possible contributor to the loss of larger multimers in these patients, but this degradation is very much mutation-dependent.13 Indeed, ratios between 0.14 and 1.03 were obtained in these samples, suggesting a high variability among VWD type 2B patients. Unexpectedly, we also observed a decreased ratio of intact VWF to total VWF antigen in some of the patients with VWD type 1 and VWD type 2M, although to a lesser extent compared to the decrease in patients with VWD types 2A and 2B. This finding suggests that VWF degradation is increased in these samples, compared to that in normal controls. If so, one would anticipate that a concordant loss of HMW multimers occurs. We indeed observed that, compared to normal pooled plasma, all subtypes including VWD type 1 and VWD type 2M samples were characterized by reduced quantities of the larger multimers (Figure 5). Moreover, a highly significant correlation was seen between the ratio of intact VWF to total VWF antigen and the presence of these larger multimers. Therefore, conceivably, at least part of the loss of HMW multimers can be explained by an increased degradation of VWF by ADAMTS13.
A point of interest is that we noticed quite some variation in the extent of VWF degradation in patients with a similar mutation (supplemental Figure 4). This finding suggests that besides the mutation itself, other factors also may contribute to the extent to which VWF is proteolyzed by VWF. One potential hypothesis could be that mutations allow VWF to unfold more easily during circulation, for instance at sites of bifurcations, making them more susceptible to proteolysis. This effect would be amplified if secondary conditions, such as a developing atherosclerosis, further contribute to the presence of disturbed flow conditions. Whether this effect is associated with a more pronounced bleeding phenotype remains to be investigated.
Excessive VWF degradation associated with a loss of HMW multimers also is observed in AVWS, including in patients with severe aortic stenosis and ECMO patients (Figure 6). Therefore, the finding is perhaps unsurprising that levels of intact VWF were significantly reduced in both patient groups. As for the congenital VWD samples, both AVWS groups displayed a significant correlation between the ratio of intact VWF to total VWF antigen and the presence of larger multimers. Important to note is that the average loss of intact VWF was less pronounced in severe aortic stenosis and ECMO patients, compared to that in congenital VWD patients.
We noted that degradation of VWF was more pronounced in VWD type 2A–group 2 patients and in the in vitro degradation assays, compared to those of AVWS patients, although all are exposed to conditions that favor VWF degradation. This difference can be explained by the fact that VWF is constitutively in an ADAMTS13-sensitive conformation in VWD type 2A–group 2 and in the in vitro degradation assays, allowing both large and short multimers to be proteolyzed. In contrast, in AVWS, VWF is elongated during a short period of time, when passing the stenosed valve or the mechanical pump. Such conditions particularly favor proteolysis of the larger multimers only, with the highest chance of cleavage occurring within the middle region of these multimers (supplemental Figure 6).
In summary, we have developed a novel nanobody that binds to the A3 domain of VWF, a subunit that is not proteolyzed by ADAMTS13. This nanobody not only provided new insight into how ADAMTS13 may regulate the exposure of the collagen-binding site within the A3 domain, but also identified the presence of low-grade VWF degradation in a large number of patient samples. By using its sensitivity, we revealed an increase in VWF degradation in VWD type 1 and type 2M samples, which we were unable to do with assays currently available for detecting degraded VWF.
Whether KB-VWF-D3.1 could be useful as a diagnostic tool regarding congenital VWD would need to be established in additional studies designed for this specific purpose. However, we anticipate that our assay might be useful in a number of other situations. First, we now know that, in the majority of aortic stenosis patients who undergo transcatheter valve replacement, the VWF degradation defect is corrected within 5 to 10 minutes after valve placement.11,36 However, the VWF degradation defect is not corrected in 10% to 20% of the patients due to incorrect valve placement that causes regurgitation. Currently, no point-of-care assays are available to verify the correct placement of the valves. A point-of-care assay using KB-VWF-D3.1 possibly could be applied for this purpose, as it will detect an increase in levels of intact VWF. Second, this rapid point-of-care approach can also be used to test the opposite, that is, whether a loss of VWF degradation occurs due to a lack of ADAMTS13 activity. This test could eventually facilitate clinical decision-making for patients who arrive into the hospital with suspected thrombotic thrombocytopenic purpura.
Acknowledgments
The authors thank Jenny Goudemand, Edith Fressinaud, Mouhamed Moussa, Christophe Zawadzki and Pierre Boisseau for the development, patient recruitment, and genetic analysis of the patient cohort in the framework of the French Reference Center for von Willebrand disease.
This work was supported by funding from the National Research Agency (Agence Nationale de la Recherche, grant numbers ANR-17-RHUS-17-0011-WIllAssistHeart and ANR-21-CE14-0076-02-Vista), and the Fondation Crédit Agricole Nord de France.
Authorship
Contribution: C.K.M., I.P., S.G., G.L.F., S.M.L., S.D., M.C., S.R., T.D., C.C., and P.J.L. performed experiments and analyzed data; V.R., A.R., F.V., E.J., A.D., C.T., and E.vB. provided patient samples; S.S., O.D.C., P.J.L., and C.V.D. designed and supervised the study; P.J.L. wrote the first draft of the manuscript; and all authors contributed to the interpretation of the data and the editing of the final manuscript.
Conflict-of-interest disclosure: G.L.F., S.L., S.D., and T.D. are employees of Diagnostica Stago. The remaining authors declare no competing financial interests.
Correspondence: Peter J. Lenting, INSERM U1176, 80 rue du Général Leclerc, 94270 Le Kremlin-Bicetre, France; e-mail: peter.lenting@inserm.fr.
References
Author notes
∗C.K.M. and I.P. contributed equally to this study.
Data are available upon reasonable request to the corresponding author.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal