Key Points
First description of cell type characteristic NfκB dynamics in a primary mammalian tissue (blood) stem cell differentiation system.
Time-lapse imaging, single-cell RNA sequencing, plus signaling manipulation identify NfκB dynamics to influence cell behavior.
Abstract
Cells can use signaling pathway activity over time (ie, dynamics) to control cell fates. However, little is known about the potential existence and function of signaling dynamics in primary hematopoietic stem and progenitor cells (HSPCs). Here, we use time-lapse imaging and tracking of single murine HSPCs from green fluorescent protein-p65/H2BmCherry reporter mice to quantify their nuclear factor κB (NfκB) activity dynamics in response to tumor necrosis factor α and interleukin 1β. We find response dynamics to be heterogeneous between individual cells, with cell type–specific dynamics distributions. Transcriptome sequencing of single cells physically isolated after live dynamics quantification shows activation of different target gene programs in cells with different dynamics. Finally, artificial induction of oscillatory NfκB activity causes changes in granulocyte/monocyte progenitor behavior. Thus, HSPC behavior can be influenced by signaling dynamics, which are tightly regulated during hematopoietic differentiation and enable cell type–specific responses to the same signaling inputs.
Introduction
Signaling inputs control cell fates, including those of hematopoietic stem and progenitor cells (HSPCs).1-6 Nuclear factor κB (NfκB) is expressed in all mammalian cells, is activated by inflammatory cytokines like tumor necrosis factor α (TNFα) and interleukin 1β (IL1β), and controls numerous target genes7 and fates of most cell types, including HSPCs.2,8,9 NfκB signaling can affect cells in a context-dependent and cell type–specific way. For example, while increased TNFα exposure induces apoptosis in myeloid progenitors, it promotes survival and differentiation of HSCs.10
Cells integrate signaling inputs via different complex and interconnected biochemical signaling cascades. This information transmission was initially thought to be binary (only on/off), but time-lapse imaging of biosensors reporting signaling activity over time (ie, signaling dynamics)11-16 revealed heterogeneous dynamics in individual cells of cell lines.13,17-22 Sustained (SUS) vs transient (TRA) extracellular signal-regulated kinase (ERK) signaling correlated with future differentiation vs proliferation in the PC12 cell line, suggesting the functional relevance of signaling dynamics for controlling these fates.23 Pharmacological manipulation of ERK, NfκB, and p53 signaling dynamics by small molecules correlated with changed gene expression and cell fates in PC12 and MCF7 cell lines.17,23,24 In HeLa cells, SUS and persistent oscillatory (OSC) NfκB signaling correlate with the expression of early and late target genes, but TRA dynamics correlate only with early target genes.25,26
Thus, NfκB dynamics likely are important for cell fate control. However, due to a lack of suitable technologies,27 they have mostly been studied in easy-to-handle cell lines. Data for more challenging primary stem and progenitor cells and cell type-dependent dynamics during their differentiation are largely missing.14
We, therefore, develop a quantitative long-term imaging and data analysis pipeline to quantify TNFα-induced NfκB activity dynamics in different primary cell types of the hematopoietic differentiation hierarchy. We find different HSPC types to exhibit different NfκB dynamics distributions after the same stimulation, individual cells with different dynamics to activate different transcriptional target programs, and artificial induction of specific signaling dynamics to alter cell behavior.
Methods
Ethical statement
All experiments were done according to Swiss federal law and institutional guidelines of ETH Zurich, approved by the local animal ethics committee Basel-Stadt (approval number 2655).
Mice
For details about mice11 and mouse handling, see supplemental Materials.
Genotyping
Green fluorescent protein (GFP)-p65 and H2B-mCherry transgenes were verified by flow cytometry and kept homo/heterozygous, respectively.
Hematopoietic cell isolation
Primary cells were isolated and sorted as previously described.28-30 For details and fluorescence-activated cell sorting (FACS) antibodies, see supplemental Materials.
Primary cell culture
Cells were isolated from transgenic mice as described above and cultured before (∼30 to 90 minutes to let them settle) and during time-lapse movies in 4-well microinserts (ibidi) within a glass-bottom 24-well plate (Greiner). Before cultures, plates were coated with 10 ug/mL anti-CD43-biotin antibody for 2 hours at room temperature to reduce cell movement.31,32 After washing with phosphate-buffered saline (PBS), 1 mL of IB20/SI media (IB20 = custom Iscove’s modified Dulbecco’s medium without riboflavin [Thermo Fisher]) supplemented with 20% BIT 9500 Serum Substitute (Stem Cell Technologies), 50 U/mL penicillin, 50 µg/mL streptomycin (Gibco), GlutaMAX (Gibco), 2-mercaptoethanol (50 µM; Gibco); SI = 100 ng/mL murine stem cell factor (SCF) + 10 ng/mL murine IL-3 (both Peprotech) was added per well, and cells were cultured at 37°C and 5% CO2.
Macrophage differentiation
Lineage-biased granulocyte/monocyte progenitors (GMPs) (LinnegcKitposSca1negCD16/32posCD34posLy6CposCD115pos) were isolated as described above. Cells were cultured for 3 to 4 days in IB20 medium with 100 ng/mL murine macrophage colony-stimulating factor (M-CSF) (Peprotech) at 37°C and 5% CO2. Before imaging, M-CSF was washed out with PBS (3 times), and media was replaced by IB20/SI media. Macrophage identification by morphology.
Confocal time-lapse imaging
Time-lapse experiments were conducted at 37°C, 5% O2, and 5% CO2 with media conditions described above using Nikon NIS acquisition software on a Nikon A1 confocal microscope with a Hamamatsu Flash 4.0 camera. Blue (GFP, 3.5% intensity) and green (mCherry, 1%) lasers, custom GFP and mCherry emission filter settings, and a 20×/0.75 CFI Plan Apochromat λ objective were used. Images were acquired every 7 minutes for 12 hours. Movies were stopped after 63 minutes (9 time points) for ∼1 minute, and 1 mL of IB20/SI media supplemented with 80 ng/mL TNFα (final concentration in well = 40 ng/mL; no TNFα for controls) was added slowly to each well.
Liquid colony assays
Cells were cultured as described above. The medium was supplemented with 20 ng/mL in-culture live stain antibodies CD115-BV421 and CD24-APC (supplemental Data 1). Cells were first imaged for 12 hours as described above using 488 nm (GFP, 3.5% intensity) and 561 nm (mCherry, 1%) lasers with imaging frequencies of 9 minutes, and then every 30 minutes for another 48 hours using 405 nm (CD115-BV421, 1.5%), 488 nm, 561 nm, and 640 nm (CD24-APC, 2.5%) lasers. For stimulation after 1 hour, 1 mL of IB20/SI media supplemented with 80 ng/mL TNFα (final concentration, 40 ng/mL), 20 ng/mL IL1β (final concentration, 10 ng/mL), or a blank control was added slowly to each well. For repeated stimulation, a preset pipetting sequence was performed by the Pipetting Helper Imaging Lid (PHIL) robot (see below). For analysis and feature extraction (see supplemental Table 2), cells were tracked until at least generation 4.
Automated TNFα stimulation with PHIL robot
The liquid handling robot PHIL33 operated as described.33-35 For forced oscillations, after 1 hour of imaging, the liquid was aspired and replaced by media containing 40 ng/mL TNFα. After 20 minutes, stimulation media was washed out 3 times and replaced by control media without TNFα for 70 minutes before a new round of stimulation. This cycle of 20 minutes (+TNFα) and 70 minutes (−TNFα) was repeated 5 times. For the 2 control conditions, media with (transient control, reflecting manual stimulation) or without (control without stimulation) 40 ng/mL TNFα was added by PHIL only once after 1 hour of imaging without washing.
Image quantification and analyses
Image quantification and analyses were as previously described.36,37 See supplemental Materials.
Blind manual time-series classification
For “manual” classification, time series were assigned to the predefined categories nonresponsive (NON), SUS, TRA, OSC, and unclear/outlier by the experimenter. Time series were chosen randomly and displayed without any metadata (eg, cell type or stimulation information) to the experimenter by an algorithm implemented in R. All time series of associated replicates were classified at once.
Feature-based time-series analysis
Up to 23 features related to the dynamics/shape of time series were defined, and scores for each feature were calculated per cell. The distribution of cells within the feature space was evaluated by using uniform manifold approximation and projection (UMAP) [umap() from package “umap”, version 0.2.7.0 in R 3.4.2]. For details about features, their calculation, and meaning, please refer to supplemental Table 1.
Single-cell picking
After imaging, cells were washed 3 times with 1 mL PBS per well and placed on ice to be transferred to the picker. Washing is crucial since the cultivation media impedes complementary DNA production. Cells were selected randomly to be picked with the CellCelector from Automated Lab Solutions GmbH Jena. Isolated cells were directly transferred into individual wells of cooled 96-well PCR plates (Eppendorf, twin.tec) containing 2.3 μL lysis buffer (per well: 0.115 μL SUPERase-In RNase Inhibitor 20 U/µL [Promega], 0.046 μL 10% Triton X-100 [Sigma-Aldrich/Merck Kgaa], 2.139 μL DEPC-treated or RNase-free H2O [Ambion]). After picking, plates were kept at −80°C until processing for transcriptional profiling by single-cell RNA sequencing (scRNASeq). Images were taken before and after each picking and matched by eye to the images taken at the last time point during time-lapse movies to conserve cell IDs between the 2 experimental parts.
scRNASeq
After single-cell isolation by picking, we performed scRNASeq in an adaption from previously described protocols.38 For details, see supplemental materials.
Primary analysis of scRNASeq data
Analysis was done using established R packages.39-46 For details, see supplemental materials.
Quantification and statistical analysis
All quantification and statistical analyses were done in the programming environment R (R 3.4.2, R-Project). For details, see supplemental materials.
Results
Quantification of HSPC NfκB dynamics
p65 is part of the NfκB dimer and translocates into the nucleus during NfκB pathway activation. To enable quantification of NfκB signaling activity dynamics, we used homozygous B6.Bruce 4-Relatm2.1Mpa mice,11 where eGFP is knocked into the endogenous p65 gene locus, tagging all p65 proteins. NfκB activity dynamics can thus be quantified by the nuclear localization of GFP-p65. To improve nuclear segmentation, we crossed these mice with C57Bl/6-Tg(Gt[ROSA]H2B-mCherry) mice,47 where the chromatin is labeled by mCherry in all cells. Time-lapse confocal microscopy and adapting a custom software pipeline for single-cell tracking and dynamics quantification1-3,14,36,37,48-52 enabled quantification of GFP-p65 fusion protein nuclear localization dynamics in individual freshly isolated primary HSPCs (Figure 1A). HSPC were cultured in the presence of SCF and IL3 directly after FACS isolation, without prior cell cycle stage synchronization or selection. After 1 hour of time-lapse imaging the dynamics baseline of unstimulated cells, we added TNFα as a canonical NfκB signaling trigger and quantified its nuclear localization dynamics for 11 hours.
NfκB signaling dynamics quantification in single primary hematopoietic cells. (A) Quantification of nuclear fluorescence dynamics in a representative cell plotted as trace or “heat stripe.” The fading magenta pattern reflects limited TNFα stability after a single addition. (B) Four main NfκB response types upon TNFα challenge. (C) Features extraction from signaling traces for detailed objective analysis (supplemental Table 1). (D) Two-dimensional UMAP representation of cellular heterogeneity across extracted features. One dot represents 1 trace. (E) UMAP as in (D) but with colored dots indicating the localization of cells with a specific response pattern (n = 2721 cells, N = 3 biological replicates). See also supplemental Table 1.
NfκB signaling dynamics quantification in single primary hematopoietic cells. (A) Quantification of nuclear fluorescence dynamics in a representative cell plotted as trace or “heat stripe.” The fading magenta pattern reflects limited TNFα stability after a single addition. (B) Four main NfκB response types upon TNFα challenge. (C) Features extraction from signaling traces for detailed objective analysis (supplemental Table 1). (D) Two-dimensional UMAP representation of cellular heterogeneity across extracted features. One dot represents 1 trace. (E) UMAP as in (D) but with colored dots indicating the localization of cells with a specific response pattern (n = 2721 cells, N = 3 biological replicates). See also supplemental Table 1.
For dividing cells, 1 random daughter cell was quantified. p65 activation response dynamics were heterogeneous between individual cells in most analyzed populations, with 4 main dynamics: NON, SUS, TRA, and OSC (Figure 1B). These dynamics were classified by using both algorithmic scoring (based on 23 extracted time-series features) (see supplemental Table 1) and blinded manual assignment, which crossconfirmed each other (Figure 1C). Two-dimensional uniform manifold approximation and projection (UMAP) of single-cell NfκB signaling profiles revealed good separation between TNFα-stimulated and -unstimulated cells (Figure 1D) and between the different response classes (Figure 1E). Thus, NfκB dynamics can be robustly quantified even in small primary HSPCs, and time-series classification and scoring work robustly in a supervised or automated manner. TNFα concentration affected p65 activity and dynamics in GMPs (supplemental Figure 1A,B), with a steep increase in responding cells from 0.1 ng/mL TNFα (supplemental Figure 1A). Signaling amplitude and area under the curve (AUC) reach a plateau from 1 ng/mL TNFα (supplemental Figure 1B). Interestingly, OSCs are induced only from 10 ng/mL and only in specific cell types.
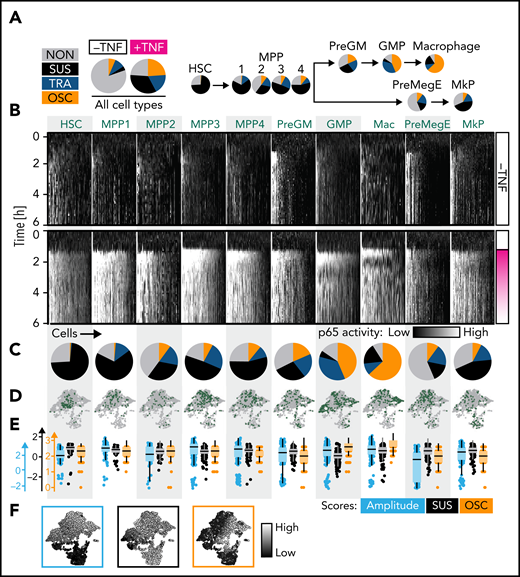
NfκB activity dynamics distributions are HSPC-type characteristic
We quantified NfκB response dynamics in 10 different myeloid cell types: HSCs, 4 multipotent progenitor populations (MPP1 to MPP4), premegakaryocytic/erythroid progenitors (PreMegEs), megakaryocyte progenitors (MkP), pregranulocyte/monocyte progenitors (PreGMs), GMPs, and macrophages (Ms) (Figure 2A-B; supplemental Figure 4A-B). After TNFα stimulation, 19.5% vs 76.1% of all un/stimulated cells showed a NfκB response (Figure 2A), with 76 ± 4% (mean ± standard deviation) HSCs, 83 ± 9 GMPs, and 90 ± 15% Ms responding. PreMegEs often do not respond (Figure 2C). These cells already have higher nuclear GFP-p65 levels before stimulation (supplemental Figure 2E), probably leading to fewer responders.
NfκB dynamics change throughout the hematopoietic hierarchy. (A) Main NfκB response dynamics type frequencies in different cell types (manual classification). (B-E) Cell type–specific signaling dynamics. (B) Single-cell heat stripes (vertical) of all measured cells and conditions. Brightness represents nuclear p65 signal intensity (n = 2721 cells: 166 HSC, 167 MPP1, 93 MPP2, 340 MPP3, 210 PreGM, 680 GMP, 329 Mas, 204 PreMegE, and 290 MkP; N = 3 biological replicates). The first 6 hours are shown (12 hours in supplemental Figure 2H). (C) Frequencies of main response types per cell type (manual classification). Mean ± standard deviation (3 independent replicates). P values in supplemental Figure 2G. Colors as in (A). (D) UMAP representation based on extracted signaling dynamics features. UMAP shape as in Figure 1D. Only TNFα-stimulated cells are shown. Green dots = cell type as indicated in (B). (E-F) Three cell type–specific signaling dynamics feature scores across cell types (E) and on the signaling dynamics UMAP representation (F). UMAP shape as in Figure 1E. See also supplemental Figure 2.
NfκB dynamics change throughout the hematopoietic hierarchy. (A) Main NfκB response dynamics type frequencies in different cell types (manual classification). (B-E) Cell type–specific signaling dynamics. (B) Single-cell heat stripes (vertical) of all measured cells and conditions. Brightness represents nuclear p65 signal intensity (n = 2721 cells: 166 HSC, 167 MPP1, 93 MPP2, 340 MPP3, 210 PreGM, 680 GMP, 329 Mas, 204 PreMegE, and 290 MkP; N = 3 biological replicates). The first 6 hours are shown (12 hours in supplemental Figure 2H). (C) Frequencies of main response types per cell type (manual classification). Mean ± standard deviation (3 independent replicates). P values in supplemental Figure 2G. Colors as in (A). (D) UMAP representation based on extracted signaling dynamics features. UMAP shape as in Figure 1D. Only TNFα-stimulated cells are shown. Green dots = cell type as indicated in (B). (E-F) Three cell type–specific signaling dynamics feature scores across cell types (E) and on the signaling dynamics UMAP representation (F). UMAP shape as in Figure 1E. See also supplemental Figure 2.
Importantly, these response dynamics were not equally distributed across cell types but changed from the most primitive cells (HSC) to the most differentiated cells (Ms). While HSCs predominantly exhibit sustained dynamics, more lineage-restricted cells are biased toward TRA (PreGMs/GMPs) or OSC (GMPs/Ms) responses (Figure 2B-C). Based on the blind manual classification of responders, 98%/1%/1% of HSCs, 5%/43%/52% of GMPs, and 20%/10%/70% of Ms showed SUS/TRA/OSC dynamics, respectively (Figure 2C). Mapping cell populations on our previously calculated UMAP (Figure 2D,F) confirmed decreasing probability of SUS dynamics along the hematopoietic hierarchy. Ms show the highest OSC score and a higher SUS score than GMPs (since OSC are also more sustained than TRA dynamics) (Figure 2E-F). Although individual cells show considerable heterogeneity, specific dynamics are enriched in specific populations (Figure 2D), as quantified by hierarchical clustering of feature scores (supplemental Figure 2A,B). This classification solely depends on the signaling dynamics and not on any other information like cell morphology or motility. Taken together, NfκB signaling dynamics change from SUS (primitive stem and progenitor cells) via TRA (more restricted progenitors) to OSC (most differentiated cells) in the HSC–monocyte/M axis.
NfκB dynamics can prospectively enrich GMP subtypes
Interestingly, GMPs (LinnegcKitposSca1negCD16/32posCD34pos) (supplemental Figure 5) are segregated into TRA vs OSC responders (Figure 3A-B), suggesting that dynamics are sharply regulated and may identify GMP subtypes and potentially influence cell fate choices. To address these points, we first used “trackSeq”53 to isolate individual HSPCs after time-lapse imaging and single-cell tracking with known identity for scRNAseq (Figure 3C). Full transcriptome data can thus be matched to the history of individual cells, including their signaling dynamics.
GMP subpopulations with different lineage potential show distinct NfκB dynamics. (A) Response dynamics frequencies of all GMPs. (B) Exemplary traces. (C) Experimental setup for (D-H). (D) Two-dimensional UMAP representation of the transcriptional space. OSC and TRA cells differ transcriptionally (n = 231 cells, N = 3 biological replicates, 2 independent sequencing runs). (E) Hierarchical clustering on UMAP coordinates is used to quantify distributions within the UMAP in (F-G). (F) Response type fractions in the different clusters. Colors as in (A). (G) Distribution of selected dynamics feature scores across clusters. Confirmation of manual scoring in (D,F). ACF, auto correlation function. (H) Identification of OSC vs TRA surface markers by scRNAseq. (I-N) NfκB dynamics in GMP subpopulations after TNFα stimulation (n = 1298 cells, N = 3 biological replicates). (I) Single-cell heat stripes (vertical) of all measured cells and conditions. Brightness represents nuclear p65 signal intensity. The first 6 hours are shown (12 hours in supplemental Figure 3H). (J) Dynamics frequencies (blind-manual classification). Colors as in (A). (K) The correlation of signaling dynamics and cell cycle stages does not explain HSPC characteristic dynamics. Cell cycle phases are inferred from transcriptomes ∼7 hours after stimulation (see Figure 4). Left: color coding as in (A). (L-N) UMAP graph based on 17 extracted signaling trace features. (L) Right: colors as in (A). (N) Green dots show GMP subpopulation UMAP location. Only TNFα-stimulated cells are shown. (O) Scores for selected features for the 3 subpopulations. See also supplemental Figure 3.
GMP subpopulations with different lineage potential show distinct NfκB dynamics. (A) Response dynamics frequencies of all GMPs. (B) Exemplary traces. (C) Experimental setup for (D-H). (D) Two-dimensional UMAP representation of the transcriptional space. OSC and TRA cells differ transcriptionally (n = 231 cells, N = 3 biological replicates, 2 independent sequencing runs). (E) Hierarchical clustering on UMAP coordinates is used to quantify distributions within the UMAP in (F-G). (F) Response type fractions in the different clusters. Colors as in (A). (G) Distribution of selected dynamics feature scores across clusters. Confirmation of manual scoring in (D,F). ACF, auto correlation function. (H) Identification of OSC vs TRA surface markers by scRNAseq. (I-N) NfκB dynamics in GMP subpopulations after TNFα stimulation (n = 1298 cells, N = 3 biological replicates). (I) Single-cell heat stripes (vertical) of all measured cells and conditions. Brightness represents nuclear p65 signal intensity. The first 6 hours are shown (12 hours in supplemental Figure 3H). (J) Dynamics frequencies (blind-manual classification). Colors as in (A). (K) The correlation of signaling dynamics and cell cycle stages does not explain HSPC characteristic dynamics. Cell cycle phases are inferred from transcriptomes ∼7 hours after stimulation (see Figure 4). Left: color coding as in (A). (L-N) UMAP graph based on 17 extracted signaling trace features. (L) Right: colors as in (A). (N) Green dots show GMP subpopulation UMAP location. Only TNFα-stimulated cells are shown. (O) Scores for selected features for the 3 subpopulations. See also supplemental Figure 3.
We isolated individual GMPs 7 hours after TNFα stimulation and p65 dynamics quantification to perform scRNAseq. OSC and TRA cells were found in different transcriptional states (Figure 3D). Hierarchical clustering on the transcriptome UMAP coordinates confirmed that the different dynamics are unequally distributed across transcriptome clusters (Figure 3E-F). This class-based analysis was confirmed by algorithmic extraction and quantification from dynamics curve features. OSC-associated features score highest in cluster 1, with features of other dynamics in clusters 2 and 3 (Figure 3G). This demonstrates the existence of GMP subtypes with different transcriptomes and correlated NfκB response dynamics.
Several surface markers, including Csf1r (CD115), associated with monocyte/M differentiation, were enriched in OSC over TRA cells (Figure 3H), suggesting that NfκB response dynamics may differ between lineage-biased GMP subtypes or early lineage-committed GMPs. CD115 and Ly6C were previously described54 to identify 3 GMP subpopulations: without lineage bias (Ly6CnegCD115low [GMPGM]), with monocyte lineage bias (Ly6CposCD115high [GMPM]), and with granulocyte lineage bias (Ly6CposCD115low [GMPG]) (supplemental Figure 5C). Indeed, 58% of OSC and only 13% of TRA GMPs are GMPM. In return, TNFα-induced NfκB dynamics differed between these GMP subtypes, with 77%/11% TRA/OSC in GMPGM and 22%/64% TRA/OSC in GMPM, respectively. GMPG subpopulations were more ambivalent (Figure 3I-J). Minor dynamics differences between cell cycle phases are much smaller than between GMP subtypes. Possible HSPC-specific cell cycle differences thus do not cause their signaling dynamics characteristics (Figure 3K). GMP subtype differences were confirmed by feature-based dynamics analysis (Figure 3L-O; supplemental Figure 3A-C), where GMPGM and GMPM clustered separately (Figure 3N). Analysis of single features confirmed the highest OSC score for GMPM, while scores of non-OSC features were similar across GMPs (Figure 3M,O; supplemental Figure 3D). Thus, there is a sharp switch to OSC NfκB responses during the fast1 differentiation from GMPGM to GMPM.
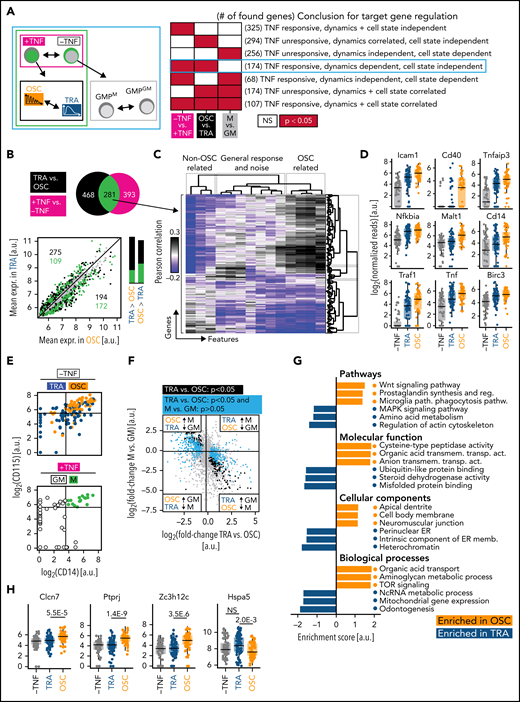
Identification of NfκB dynamics-specific target genes
The different transcriptome states between OSC and TRA GMPs could have 2 sources. (1) They could be differences between GMP subtypes. In this case, different NfκB signaling dynamics would only be a phenotype of a previously made fate choice and not influence future fate. (2) Alternatively, different NfκB dynamics could be functionally relevant to cause cell fate choices by activating different gene expression programs. However, this cause vs consequence distinction of simultaneously appearing phenotypes has been technically very difficult, in particular, because it was impossible to know dynamics patterns before they actually occurred after stimulation. Thus, differences between different initial cell states associated with distinct dynamics remained elusive.
However, trackSeq now allowed us to disentangle the 2 by combining 3 different differential gene expression analyses (DGEAs) (Figure 4A). (1) NfκB-dependent target genes can be identified by comparing cells with vs without TNFα stimulation. (2) Genes associated with distinct response dynamics can be identified by comparing cells that show OSC vs TRA behavior after TNFα stimulation. These differentially expressed genes might be independent of NfκB dynamics because of preexisting different molecular states in OSC and TRA GMPs. (3) To remove these initial cell type-dependent but NfκB dynamics-independent differences between OSC and TRA cells, we compared these GMP subpopulations without TNFα stimulation. This requires the identification of OSC vs TRA GMPs without actual TNFα stimulation, which has now become possible because of our finding that 67% of GMPM are OSC, while 76% of GMPGM show TRA behavior (Figures 3I-J and 4E). To that end, we determined CD14 and CD115 messenger RNA (mRNA) expression thresholds allowing GMPM vs GMPGM identification by their transcriptomes (Figure 4E). Combining these 3 DGEAs (results in supplemental Data 4-6) now removes NfκB-independent and cell type–specific TNFα target genes, and the remaining targets have no initial bias to OSC or TRA cells before stimulation.
mRNASeq of single cells with known signaling dynamics reveals dynamics-specific target genes. (A) DGEA comparisons and their meaning. The comparison identifying genes truly controlled by NfκB dynamics is circled in cyan (n = 231 cells, N = 3 biological replicates, 2 independent sequencing runs). (B) Top: DGEA between TNFα unstimulated vs stimulated (black) and between OSC vs TRA cells (both TNFα-stimulated) (magenta). Genes (n = 281) that are both upregulated by TNFα and differentially expressed between OSC and TRA dynamics (green) are the most promising candidates to be modulated by NfκB dynamics. Bottom: mean expression of all detected genes in OSC vs TRA cells. Most TNFα-responsive and dynamics-correlated genes are higher expressed in OSC cells, which activate a superset of TRA target genes. Bar graphs represent numbers above (left) and below (right) of the parity line. (C) Correlation of extracted features (supplemental Table 1) and the 281 gene candidates from (B). OSC-associated feature scores correlate with many of these genes. This confirms the visual scoring in (B) and suggests many NfκB target genes are more responsive to OSC than to TRA dynamics. (D) Dynamics-dependent expression of selected NfκB pathway genes. (E) Top: TRA vs OSC cells can be identified by CD14 and CD115 mRNA expression detected by scRNAseq (top right quadrant: 66% OSC; other quadrants: 77% TRA). Bottom: CD14 vs 115 gating of unstimulated cells to identify GMPM (67% OSC) vs GMPGM (76% TRA). (F) Gene expression changes between TRA and OSC cells (x-axis) and between unstimulated GMPM and GMPGM (y-axis) allow the identification of dynamics-specific target genes (cyan, significantly different between OSC and TRA but not between GMPM and GMPGM). Enrichment in the top left and bottom right quadrant confirms that GMPM/GMPGM are predominantly OSC/TRA, respectively. (G) GSEA of OSC and TRA cells, corrected for initial cell type differences (for details, see text). (H) Selected target genes that are activated by either TRA or OSC dynamics only (see also supplemental Figure 4C and supplemental Data 5).
mRNASeq of single cells with known signaling dynamics reveals dynamics-specific target genes. (A) DGEA comparisons and their meaning. The comparison identifying genes truly controlled by NfκB dynamics is circled in cyan (n = 231 cells, N = 3 biological replicates, 2 independent sequencing runs). (B) Top: DGEA between TNFα unstimulated vs stimulated (black) and between OSC vs TRA cells (both TNFα-stimulated) (magenta). Genes (n = 281) that are both upregulated by TNFα and differentially expressed between OSC and TRA dynamics (green) are the most promising candidates to be modulated by NfκB dynamics. Bottom: mean expression of all detected genes in OSC vs TRA cells. Most TNFα-responsive and dynamics-correlated genes are higher expressed in OSC cells, which activate a superset of TRA target genes. Bar graphs represent numbers above (left) and below (right) of the parity line. (C) Correlation of extracted features (supplemental Table 1) and the 281 gene candidates from (B). OSC-associated feature scores correlate with many of these genes. This confirms the visual scoring in (B) and suggests many NfκB target genes are more responsive to OSC than to TRA dynamics. (D) Dynamics-dependent expression of selected NfκB pathway genes. (E) Top: TRA vs OSC cells can be identified by CD14 and CD115 mRNA expression detected by scRNAseq (top right quadrant: 66% OSC; other quadrants: 77% TRA). Bottom: CD14 vs 115 gating of unstimulated cells to identify GMPM (67% OSC) vs GMPGM (76% TRA). (F) Gene expression changes between TRA and OSC cells (x-axis) and between unstimulated GMPM and GMPGM (y-axis) allow the identification of dynamics-specific target genes (cyan, significantly different between OSC and TRA but not between GMPM and GMPGM). Enrichment in the top left and bottom right quadrant confirms that GMPM/GMPGM are predominantly OSC/TRA, respectively. (G) GSEA of OSC and TRA cells, corrected for initial cell type differences (for details, see text). (H) Selected target genes that are activated by either TRA or OSC dynamics only (see also supplemental Figure 4C and supplemental Data 5).
Genes (n = 281) were induced by TNFα and differentially expressed between OSC and TRA cells, thus being candidates for NfκB dynamics-specific targets. Interestingly, most of these were expressed higher in OSC than TRA cells, suggesting that OSC signaling is required for their activation (Figure 4B). Indeed, most of these genes correlated with OSC-associated dynamics features (Figure 4C). Many OSC-activated genes are directly involved in NfκB signaling, including NfκB inhibitors Nfkbia and Tnfaip3 (Figure 4D), which complete negative feedback loops and reinforce oscillations.55
Genes (n = 468) were also differentially expressed between OSC and TRA GMPs after TNFα stimulation but not between GMPM and GMPGM before stimulation (of which 294 are not, and 174 are TNFα-responsive). Importantly, a large part of these genes was expressed at low or intermediate levels before stimulation, demonstrating the accessibility of their genomic loci (Figure 4F; supplemental Figure 4B). The 174 shared genes of these 281 and 486 gene-containing sets were, thus, TNFα responsive, dynamics (OSC vs TRA)-dependent, and cell state (GMPM vs GMPGM)-independent (Figure 4A, cyan box). Together with the accessibility of their genomic loci, these candidates thus are highly likely caused at least in part by differential signaling dynamics and do not merely reflect a molecular state of previously made fate choices. GSEA (see “Methods”) demonstrated increased expression of metabolic and inflammation genes by OSC NfκB activity (Figure 4G). Many of the highest-ranked OSC-activated genes showed barely any activation after TRA NfκB signaling (Figure 4H; supplemental Figure 4C), including Clcn7 (affects osteoclast differentiation),56,57 Ptprj (macrophage adhesion, myeloid leukemia),58,59 and Zc3h12c (macrophage regulation).60 Only very few genes like Hspa5 (PKB/AKT signaling)61 are activated by TRA but not by OSC signaling. This suggests that OSC and TRA NfκB dynamics have both shared and specific effects on gene expression in primary HSPCs.
Future cell fates correlate with NfκB activity dynamics
To quantify a possible correlation of NfκB dynamics with the future fates of individual GMPs, we imaged GMPs for 12 hours for signaling dynamics measurement (9-minute imaging frequency) and then for 48 hours (30-minute frequency) with live surface marker detection (anti-CD115-BV421 and anti-CD24-APC antibodies). In addition to TNFα, we used 10 ng/mL IL1β for NfκB stimulation, which triggered similar GMP p65 dynamics as TNFα but induced fewer OSC dynamics in GMPM (Figure 5A). Single cells were tracked (Figure 5B), and cell morphology, motility patterns, and surface marker expression were quantified. Indeed, signaling response dynamics correlated with some of the future fates of individual cell clonal progeny. As expected, OSC dynamics strongly correlated with CD115 expression (Figure 5C-D). Mean division time decreased with p65 activation. Motility and cell area strongly correlated with general fluctuations (volatility) of p65 signaling but were reduced in cells with OSC behavior (Figure 5C-D). Colony fates also correlated with stimulating cytokine and cell type: GMPGM-derived colonies show larger nuclei than GMPM-derived colonies and, as expected, lower CD115 expression. TNFα and IL1β induce similar but not identical colony development, leading to changed nuclear morphology, proliferation speed, and marker expression (Figure 5E). Importantly, cells with OSC vs TRA dynamics also show such differences, confirming that these dynamics and colony outcome are correlated. OSC-derived cells have smaller nuclear areas and perimeters than TRA-derived cells. Additionally, CD24, an OSC target (Figure 5E), is differentially expressed between OSC and TRA cells (P = .054) but not between GMPGM and GMPM, supporting our trackSeq data.
P65 dynamics predict GMP colony behavior. (A) Comparison of TNFα and IL1β as stimulating agents. (B) Pedigree and example images of a selected colony. Cells were tracked until generation 4 or higher. Imaging frequency 9 minutes (0-12 hours) and 30 minutes (12-60 hours). Calculation of fate readouts: supplemental Table 2. (C) Correlation of dynamics features with fate readouts. “Oscillation and responsiveness”: features associated with OSC and higher amplitude after stimulation; “trace volatility and noise”: features associated with general fluctuations in p65 signaling activity (n = 323 cells, N = 3 biological replicates). (D) Correlation of selected dynamics features with selected fate readouts. One dot represents 1 colony. Blue line = linear regression model, R = Pearson correlation. OSC correlates with increased future CD115 protein expression and a smaller nuclear area. (E) Selected fate readouts vs stimulating cytokine, cell type, and p65 response dynamics. CD115 and CD24 expressions are observed between 12 and 24 hours after the start of culture.
P65 dynamics predict GMP colony behavior. (A) Comparison of TNFα and IL1β as stimulating agents. (B) Pedigree and example images of a selected colony. Cells were tracked until generation 4 or higher. Imaging frequency 9 minutes (0-12 hours) and 30 minutes (12-60 hours). Calculation of fate readouts: supplemental Table 2. (C) Correlation of dynamics features with fate readouts. “Oscillation and responsiveness”: features associated with OSC and higher amplitude after stimulation; “trace volatility and noise”: features associated with general fluctuations in p65 signaling activity (n = 323 cells, N = 3 biological replicates). (D) Correlation of selected dynamics features with selected fate readouts. One dot represents 1 colony. Blue line = linear regression model, R = Pearson correlation. OSC correlates with increased future CD115 protein expression and a smaller nuclear area. (E) Selected fate readouts vs stimulating cytokine, cell type, and p65 response dynamics. CD115 and CD24 expressions are observed between 12 and 24 hours after the start of culture.
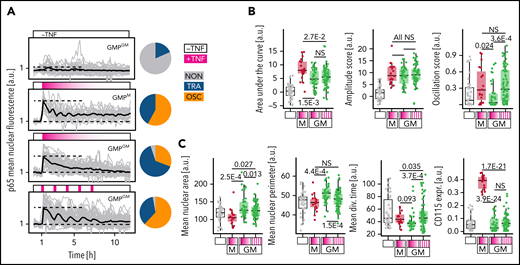
Manipulation of NfκB dynamics alters GMP behavior
The best proof that signaling dynamics do not just reflect, but cause changed cell states, is provided by demonstrating altered future single-cell fates following the manipulation of signaling dynamics. Unfortunately, no optogenetic tools for the manipulation of p65 signaling exist. We therefore aimed at manipulating NfκB dynamics through periodic culture media exchanges.20,23 Using a recently developed pipetting robot (PHIL),33 we were able to identify a temporal pattern of repeated TNFα stimulations (+TNFα for 20 minutes, then 70 minutes −TNFα, repeatedly for 7 hours) that induces OSC dynamics in GMPGM, which would show mostly TRA responses following a single stimulation (Figure 6A-B). This was confirmed by both manual (Figure 6A) and feature-based (Figure 6B) analyses. Importantly, only the type of dynamics, but not the AUC (ie, total signaling activity over time), was changed in forced oscillations (Figure 6B). Forced OSC dynamics indeed caused changed colony behavior, with decreased nuclear perimeter, longer mean division times, and a trend (P > .05) toward increased expression of CD115. These OSC-caused fates resemble those of naturally oscillating GMPM (Figures 6C and 5D). Therefore, NfκB dynamics do influence cell behavior, demonstrating that signaling dynamics can be used as an additional control layer to influence primary HSPCs fates.
Automated, repeated TNFα stimulations can artificially induce p65 oscillations and alter cell behavior. (A-B) Repeated addition and removal of TNFα (frequency: 20/70 minutes +/- TNFα) by a custom pipetting robot can force p65 to oscillate in GMPGM. Dynamics (for 12 hours) and fate quantification (for 48 hours, cells cultured −TNFα) as depicted in Figure 5A (n = 190 cells, N = 3 biological replicates). (A) Left: signaling dynamics for different stimulation conditions. Gray/black lines: individual cells/means. Right: response dynamics frequencies. (B) Selected dynamics features for different stimulation conditions. Pulsed TNFα stimulation selectively forces OSC dynamics without changing the AUC (total activity over time) of p65 responses. (C) Selected fate readouts for different stimulation conditions. Forced p65 oscillations lead to smaller cells with a trend (P > .05) toward increased CD115 expression, and longer average division time, same as naturally OSC (GMPM) cells (see also Figure 5D).
Automated, repeated TNFα stimulations can artificially induce p65 oscillations and alter cell behavior. (A-B) Repeated addition and removal of TNFα (frequency: 20/70 minutes +/- TNFα) by a custom pipetting robot can force p65 to oscillate in GMPGM. Dynamics (for 12 hours) and fate quantification (for 48 hours, cells cultured −TNFα) as depicted in Figure 5A (n = 190 cells, N = 3 biological replicates). (A) Left: signaling dynamics for different stimulation conditions. Gray/black lines: individual cells/means. Right: response dynamics frequencies. (B) Selected dynamics features for different stimulation conditions. Pulsed TNFα stimulation selectively forces OSC dynamics without changing the AUC (total activity over time) of p65 responses. (C) Selected fate readouts for different stimulation conditions. Forced p65 oscillations lead to smaller cells with a trend (P > .05) toward increased CD115 expression, and longer average division time, same as naturally OSC (GMPM) cells (see also Figure 5D).
Discussion
We demonstrate heterogeneous NfκB signaling activity dynamics in response to TNFα and IL1β stimulation in single primary HSPCs. The distributions of these response dynamics are HSPC type–specific, despite responding to the same stimulation. GMPs with OSC vs TRA dynamics activate different target gene expression programs and have different future cell behavior. NfκB signaling dynamics were previously studied mostly in cell lines and associated with different input stimuli: TNFα and PAM activate OSC, and lipopolysaccharide induces SUS or TRA signaling.13,62-64 Ligand concentration and temporal stimulation patterns caused different NfκB dynamics in fibroblasts.20 These stimuli lead to a distinct expression of key NfκB pathway molecules, including the negative feedback inhibitors nfkbia and tnfaip3, leading to different dynamics.13,55,64 Our data now show that differentiation states within the same tissue type can change signaling response dynamics even to identical input stimuli. To exclude that HSPC type–specific cell cycle differences cause their characteristic dynamics distributions, we inferred cell cycle states from their trackSeq transcriptomes (∼7 hours after stimulation). The observed minor dynamics shifts in different cell cycle phases were too small to explain the signaling dynamics differences between HSPC types. While cells will have progressed in cell cycle by 7 hours after stimulation, this still reflects their cell cycle phase proportions and dynamics correlation, and the lack of relevant effect sizes remains. The same was true when quantifying the time until division (as a proxy for cell cycle state) since the cell stimulation (data not shown).
NfκB target genes with different expression kinetics were previously identified.65 Single signaling bursts predominantly activated early and intermediate target genes, and persistent oscillations also triggered late gene expression.25 This agrees with our findings that OSC GMPs activate an overlapping but larger gene set than TRA GMPs. In the RAW264.7 cell line, NfκB dynamics correlated with gene expression,22 but it remained unclear if the correlation was based on causation, and the sum of total NfκB activity over time (as measured by AUC) was concluded to be the predominant predictor of gene expression. In contrast, since the AUC is very similar between different HSPC responder types, we find that the actual shape (eg, periodic oscillations, single peaks, etc) is relevant in primary HSPCs.
We used a novel method (trackSeq)53 for imaging, tracking, isolating, and sequencing single cells with known identity and history to match single-cell dynamics to transcriptomic data. DGEA combinations identified transcriptional target programs likely caused by NfκB dynamics and not only correlated with them, although preexisting (eg, epigenetic) differences could not be excluded as an alternative cause. This approach and the identified target genes can now be generalized to any cell system where closely related cells show heterogeneous signaling behavior.
While space restrictions prevent the discussion of all identified genes (supplemental Data 4-6), we highlight a few interesting candidates. Dynamics-regulated target genes include known HSPC fate regulators Gadd45b (involved in human granulocytic differentiation and murine stress myelopoiesis,66,67 chronic myelogenous leukemia)66-68 and Cebpa (mutations cause acute myeloid leukemia).69-71Cebpa primes GMP neutrophil differentiation, and, in line with its reduction after OSC dynamics, its knockdown promotes monopoiesis.72 In contrast, target genes of IRF8, a transcription factor executing monocytic lineage commitment,54 were enriched in OSC cells. While TNFα stimulation activates NfκB-dependent HSC survival, it induces apoptosis in myeloid progenitors.10 Apoptosis-related target genes Ddit373 and Lmna74 suggest the involvement of NfκB dynamics in influencing GMP death.
Differential gene expression and cell behavior may also be caused only by chromatin modifications or cofactor expression, which were established between GMP subtypes before NfκB signaling simulation and also lead to changed NfκB dynamics. Dynamics would then only correlate with future fates but not cause them. However, our induction of OSC dynamics in GMPGM cells by pulsed stimulation led to altered future colony behavior, demonstrating that signaling dynamics can indeed cause altered GMP development. It will be interesting to combine enforced dynamics with trackSeq in other HSPC types.75 If and how different signaling dynamics of other signaling pathways are induced and integrated with the more complex in vivo HSPC microenvironment will be interesting to analyze but requires further technological developments.76,77 These findings not only improve our understanding of HSPC fate control but will also guide the analysis of signaling dynamics and its relevance in other HSPCs, including HSCs, tissues, and signaling pathways.
Acknowledgments
The authors thank G. Camenisch, M.D. Hussherr, V. Jäggin, T. Lopes, M. di Tacchio, T. Lummen, and E. Montani for technical support. The authors thank Manolis Pasparakis and Shinichi Aizawa for providing the GFP-p65 knock-in and the R26-H2B-mCherry transgenic mouse line, respectively.
This work was supported by the Swiss National Science Foundation grant 179490 and a “Personalized Medicine Basel” grant to T.S.
Authorship
Contribution: T.S. and T.K. conceptualized the study; T.K., A.W., M.E., and P.D. created the methodology; T.K. and A.W. conducted the investigation; T.K., A.W., and M.A. curated the data; T.K. provided formal analysis; T.K., T.S., A.W., and P.D. provided software work; T.K. and T.S. provided the visualizations; T.S. supervised the study; T.S. and N.A. provided resources; T.K. and T.S. wrote the original draft; T.S., A.W., M.E., M.A., and P.D. reviewed and edited the manuscript; and T.S. conducted project administration and acquired funding.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.E. is Akasha International GmbH, Zug, Switzerland.
Correspondence: Timm Schroeder, Mattenstrasse 26, 4058 Basel, Switzerland; e-mail: timm.schroeder@bsse.ethz.ch.
Raw gene expression data will be uploaded to the Gene Expression Omnibus database (accession number GSE199404). Analysis scripts and complementary data used in this publication can be found at https://polybox.ethz.ch/index.php/s/ON7h7zmgsErR4hG (password: hspcdynamics).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal