Key Points
Human iPSC reprogramming and gene editing can be combined to generate an alternative source of rare RBCs.
Rh-engineered iPSC-derived RBCs provide a novel tool to aid in complex Rh antibody identification.
Abstract
Red blood cell (RBC) transfusion is one of the most common medical treatments, with more than 10 million units transfused per year in the United States alone. Alloimmunization to foreign Rh proteins (RhD and RhCE) on donor RBCs remains a challenge for transfusion effectiveness and safety. Alloantibody production disproportionately affects patients with sickle cell disease who frequently receive blood transfusions and exhibit high genetic diversity in the Rh blood group system. With hundreds of RH variants now known, precise identification of Rh antibody targets is hampered by the lack of appropriate reagent RBCs with uncommon Rh antigen phenotypes. Using a combination of human-induced pluripotent stem cell (iPSC) reprogramming and gene editing, we designed a renewable source of cells with unique Rh profiles to facilitate the identification of complex Rh antibodies. We engineered a very rare Rh null iPSC line lacking both RHD and RHCE. By targeting the AAVS1 safe harbor locus in this Rh null background, any combination of RHD or RHCE complementary DNAs could be reintroduced to generate RBCs that express specific Rh antigens such as RhD alone (designated D--), Goa+, or DAK+. The RBCs derived from these iPSCs (iRBCs) are compatible with standard laboratory assays used worldwide and can determine the precise specificity of Rh antibodies in patient plasma. Rh-engineered iRBCs can provide a readily accessible diagnostic tool and guide future efforts to produce an alternative source of rare RBCs for alloimmunized patients.
Introduction
Red blood cell (RBC) transfusion is a common therapy for individuals with sickle cell disease (SCD), thalassemia, bone marrow failure syndrome, and cancer. A major complication following transfusion is RBC alloimmunization, which is the production of antibodies in response to foreign proteins on donor RBCs. Alloantibodies to RBC antigens can lead to destruction or clearance of the transfused RBCs, compromising transfusion benefits or resulting in acute or delayed hemolytic transfusion reactions. Alloantibody production disproportionately affects patients with SCD because of the need for frequent RBC transfusion support, as well as the genetic diversity of blood group proteins in this population.1,2 Before each transfusion, the patient’s plasma is tested for the presence of antibodies to blood group antigens using commercial type O reagent donor-derived RBCs (dRBCs). If positive, additional testing is performed to identify the antibody target or specificity. Donor units lacking the target antigen must be selected for all future RBC transfusions.
The Rh blood group system is highly immunogenic and is often implicated in hemolytic disease of the newborn and hemolytic transfusion reactions. Routine RBC transfusion protocols consider the RhD-positive or RhD-negative type of the donor and the recipient. However, other Rh antigens (C, c, E, and e) are also immunogenic and along with the K antigen in the Kell system, are common targets of antibody complications. For patients with SCD, additional “extended” RBC antigen matching for Rh (C, E or C/c, E/e) and K between donor and patient is recommended.3 Extended matching has dramatically decreased the alloimmunization rates,3 but patients still form alloantibodies, and up to two-thirds are directed against the Rh system.4-7RH genotyping of patients and donors has demonstrated a high frequency of RH variants among Black individuals, which contributes to alloimmunization despite Rh antigen matching strategies.5,7,8
The RHD gene encodes the D antigen, and the RHCE gene encodes CE antigens in 4 combinations (ce, cE, Ce, or CE). RH variants result from the large number of single nucleotide polymorphisms and gene rearrangements that are facilitated by the inverted orientation and proximity of the duplicated RHD and RHCE genes (summarized in previous studies9,10). Patients with RH variants can become immunized when exposed to the common form of the Rh protein found on most donor RBCs (ie, D+ patient with variant RhD can form anti-D). Some RH genetic variants result in the loss of “high-prevalence” Rh antigens (ie, hrS, hrB), defined as antigens expressed by >99% of the general population, whereas others result in the expression of “low-prevalence” Rh antigens (ie, V, VS, Goa, DAK), expressed primarily on RBCs of Black individuals and both are associated with alloimmunization.4,5,7
Antibody identification in the Rh blood group system is more complex because of RH genetic diversity among patients with SCD and Black donors who are needed to sustain Rh and K matching programs.5,6,8 Identification of the antibody target, which is required for selection of compatible donor units, is limited by the availability of reagent RBCs that can distinguish challenging Rh specificities.11-13 Donors with the desired Rh phenotype are rare, and each donation has a limited life span (∼1 month). Rare Rh phenotype RBCs are currently only available in very limited quantities in specialized immunohematology laboratories. Wider availability would aid both hospital and reference laboratories to quickly focus on investigation of Rh antibody specificity and reduce delays in providing transfusion support to patients.
To address these challenges, we generated specialized human-induced pluripotent stem cells (iPSCs) by reprogramming rare donor cells or by gene editing and produced a renewable source of RBCs with unique Rh phenotypes, referred to as iRBCs. These include currently inaccessible “Rh null” cells lacking expression of all Rh antigens,14 and “D--” (dash dash) cells that express RhD but lack all RhCE antigens,15 which would immediately focus an antibody investigation on the Rh system (Rh null) or on the RhCE protein (D--). Specialized iRBCs lacking expression of the high-prevalence antigens hrS or hrB, and those expressing low-prevalence Rh antigens V, VS, Goa, or DAK can further focus on the antibody identification, and for the latter, detect potentially clinically relevant Rh antibodies not detected by current routine antibody screening assays.11,16 We refined erythroid differentiation of iPSCs to produce iRBCs that are compatible with routine antibody identification assays and represent a renewable source of cells for transfusion medicine.
Methods
RH and non-RH blood group genotyping
RH genotyping was performed on donor peripheral blood mononuclear cells (PBMCs), iPSCs, or embryonic stem cells (ESCs) using RHD and RHCE arrays (Immucor, GA) and laboratory-developed tests as previously described.5 Extended blood group antigen genotyping was performed on the PreciseType HEA (Human Erythrocyte Antigen, Immucor).
iPSC reprogramming and culture
PBMCs were reprogrammed into iPSCs as previously described to generate the R2R2 (DcE/DcE), rr (ce) lines (Table 1).17 Donors with uncommon Rh phenotypes (hrS−, hrB−, V/VS+) were recruited by New York Blood Center (NYBC) under informed consent. iPSCs were cultured on 0.1% gelatin and irradiated mouse embryonic fibroblasts or hESC-qualified Matrigel (Corning). Copy number variation analysis confirmed normal karyotypes (supplemental Table 1; supplemental Figure 1; available on the Blood website).
Rh phenotypes of iPSCs used to generate iRBCs
| Lines . | PSC . | ABO . | RHD∗ RHCE∗ genotypes and presumed haplotypes . | Rh antigen phenotype . | Application as reagent RBC in antibody identification . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D . | C . | c . | E . | e . | hrS . | hrB . | V . | VS . | |||||
| DCe (R1R1) | ESC | O | ∗D ∗Ce homozygous | + | + | 0 | 0 | + | + | + | 0 | 0 | Routinely used Rh phenotype lacking c and E |
| DcE (R2R2) | iPSC | O | ∗D ∗cE No RHD ∗cE | + | 0 | + | + | 0 | 0 | + | 0 | 0 | Routinely used Rh phenotype lacking C and e |
| ce (rr) | iPSC | O | No RHD ∗ce homozygous | 0 | 0 | + | 0 | + | + | + | 0 | 0 | Routinely used Rh phenotype lacking D, C, and E |
| hrS− | iPSC∗ | O | ∗DAU0 ∗ceMO ∗DOL ∗ceBI | + | 0 | + | 0 | + | 0 | + | 0 | 0 | Uncommon; lacks high-prevalence antigen hrS Anti-hrS would be (0) nonreactive |
| hrB− | iPSC∗ | O | ∗DIIIa-CE(4-7)-D ∗ceS homozygous | + | + | + | 0 | + | + | 0 | 0 | 0 | Uncommon; lacks high-prevalence antigen hrB Anti-hrB would be (0) nonreactive |
| V+VS+ | iPSC∗ | O | ∗D ∗ce733G ∗DFV ∗ce733G | + | 0 | + | 0 | + | + | + | + | + | Uncommon; expresses low-prevalence antigens V and VS Anti-V/VS would be (+) reactive |
| Rh null | iPSC | O | No RHD ∗ce (1kb del. KO) homozygous | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Exceedingly rare; lacks RhD and RhCE proteins (0) nonreactive suggests antibody in the Rh system |
| D-- | iPSC | O | RHD cDNA inserted at AAVS1 locus of Rh null iPSC | + | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Exceedingly rare; expresses RhD and no RhCE (0) nonreactive suggests antibody to RhCE antigens |
| Goa+ | iPSC | O | RHD∗DIVa cDNA inserted at AAVS1 locus of Rh null iPSC | + | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Expresses low-prevalence antigen Goa, RhD, and no RhCE Anti-Goa would be (+) reactive |
| DAK+ | iPSC | O | RHD∗DIIIa cDNA inserted at AAVS1 locus of Rh null iPSC | + | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Expresses low-prevalence antigen DAK, RhD, and no RhCE Anti-DAK would be (+) reactive |
| Lines . | PSC . | ABO . | RHD∗ RHCE∗ genotypes and presumed haplotypes . | Rh antigen phenotype . | Application as reagent RBC in antibody identification . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D . | C . | c . | E . | e . | hrS . | hrB . | V . | VS . | |||||
| DCe (R1R1) | ESC | O | ∗D ∗Ce homozygous | + | + | 0 | 0 | + | + | + | 0 | 0 | Routinely used Rh phenotype lacking c and E |
| DcE (R2R2) | iPSC | O | ∗D ∗cE No RHD ∗cE | + | 0 | + | + | 0 | 0 | + | 0 | 0 | Routinely used Rh phenotype lacking C and e |
| ce (rr) | iPSC | O | No RHD ∗ce homozygous | 0 | 0 | + | 0 | + | + | + | 0 | 0 | Routinely used Rh phenotype lacking D, C, and E |
| hrS− | iPSC∗ | O | ∗DAU0 ∗ceMO ∗DOL ∗ceBI | + | 0 | + | 0 | + | 0 | + | 0 | 0 | Uncommon; lacks high-prevalence antigen hrS Anti-hrS would be (0) nonreactive |
| hrB− | iPSC∗ | O | ∗DIIIa-CE(4-7)-D ∗ceS homozygous | + | + | + | 0 | + | + | 0 | 0 | 0 | Uncommon; lacks high-prevalence antigen hrB Anti-hrB would be (0) nonreactive |
| V+VS+ | iPSC∗ | O | ∗D ∗ce733G ∗DFV ∗ce733G | + | 0 | + | 0 | + | + | + | + | + | Uncommon; expresses low-prevalence antigens V and VS Anti-V/VS would be (+) reactive |
| Rh null | iPSC | O | No RHD ∗ce (1kb del. KO) homozygous | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Exceedingly rare; lacks RhD and RhCE proteins (0) nonreactive suggests antibody in the Rh system |
| D-- | iPSC | O | RHD cDNA inserted at AAVS1 locus of Rh null iPSC | + | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Exceedingly rare; expresses RhD and no RhCE (0) nonreactive suggests antibody to RhCE antigens |
| Goa+ | iPSC | O | RHD∗DIVa cDNA inserted at AAVS1 locus of Rh null iPSC | + | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Expresses low-prevalence antigen Goa, RhD, and no RhCE Anti-Goa would be (+) reactive |
| DAK+ | iPSC | O | RHD∗DIIIa cDNA inserted at AAVS1 locus of Rh null iPSC | + | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Expresses low-prevalence antigen DAK, RhD, and no RhCE Anti-DAK would be (+) reactive |
cDNA, complementary DNA; +, predicted positive for the antigen; 0, negative for the antigen.
Indicates iPSCs reprogrammed from rare donors.
Gene targeting
The CRISPR-Cas9 nuclease system was used to generate the Rh null iPSC line using a previously described protocol.18 Two gRNA vectors targeting RHCE intron 1 (TACACGTGCTTGATGCACATGG) and RHCE exon 2 (GTGTGATGACCACCTTCCCAGG) were used to knock out RHCE in an RhD-negative iPSC line. The Zinc Finger Nuclease (ZFN) system was used to generate the D--, DAK+, and Goa+ iPSC lines by inserting RHD∗RHD, ∗DIIIa, or ∗DIVa cDNA, respectively, into the AAVS1 locus as previously described,19 and in the supplemental Information. Copy number variation analysis confirmed normal karyotypes (supplemental Table 1; supplemental Figure 1). Polymerase chain reaction (PCR) primers used for screening are found in supplemental Table 2.
Differentiation of iPSCs to iRBCs
Hematopoietic differentiation of iPSCs to hematopoietic progenitor cells (HPCs) (CD41+235+) by embryoid body or adherent differentiation was performed as previously described.20,21 HPCs were collected from EB and adherent cultures on day 8 and subsequently cultured in Iscove’s Modified Dulbecco’s Media (IMDM, Corning), 2% human AB plasma (Seracare), 3% human AB serum (Millipore Sigma), 1% Penicillin-Streptomycin (Corning), 1 mg/mL holo-Transferrin (Sigma), 10 μg/mL Insulin (Sigma), and 3 U/mL Heparin (Sigma), supplemented with 3 U/mL Erythropoietin (Epo, Amgen) and 10 ng/μL stem cell factor (SCF, R&D Systems) on day 0 to 555; 3U/mL Epo only on day 6 to 12.12 On day 0, cultures were initiated with a concentration of 100 000 cells/mL. Fresh media was added every 3 days to target a concentration of ∼1 × 106 cells/mL, and cytokines were supplemented for the total volume.
Donor-derived red blood cells
Control dRBCs were obtained from the NYBC reference lab reagent RBC inventory, or from blood segments of donor units. Herein, dRBCs refer to primary erythroid cells that were not cultured.
Serologic Rh typing by gel card agglutination
Rh antigen typing (for cell surface expression) was performed using commercial monoclonal IgM typing reagents anti-D (Ortho Bioclone or Alba Alpha), anti-C, anti-c, anti-E, and anti-e (Immucor Gammaclone) on buffered gel cards (Ortho Clinical Diagnostics). Either 50 μL of 0.8% suspension of control dRBCs (NYBC) or 1 to 1.5 × 106 iRBCs resuspended in Micro Typing Systems 2 diluent (MTS2, Ortho Clinical Diagnostics) were added to each column of a buffered gel card and incubated with 25 μL of each typing reagent antibody for 30 minutes at room temperature before centrifugation on the MTS Workstation (Ortho Clinical Diagnostics) at 1032 rpm for 10 minutes.
Patient antibody detection by gel agglutination
Plasma containing previously identified Rh antibodies were from NYBC collections, or from patients after informed consent under a protocol approved by the institutional review board at the Children’s Hospital of Philadelphia. iRBCs were treated with 0.1% ficin solution (Immucor) for 10 minutes at 37°C and washed with PBS before use. Either 50 μL of 0.8% suspended control dRBCs or 1 to 1.5 million ficin-treated iRBCs were added to anti-IgG cards (Ortho Clinical Diagnostics) and incubated with 25 μL plasma at 37°C for 30 minutes followed by centrifugation on the MTS Workstation (Ortho Clinical Diagnostics) at 1032 rpm for 10 minutes.
The supplemental Methods includes procedures for ZFN targeting, flow cytometry, morphological analysis, quantitative reverse transcription-PCR, hemoglobin analysis, ektacytometry, oxygen dissociation assays, and serologic antigen typing for non-Rh blood group antigens.
Results
Customized pluripotent stem cell (PSC) panel for Rh antibody identification
We designed a panel of 10 PSC lines with distinct RH genotypes to produce ESC-derived RBCs (esRBCs) or iPSC-derived RBCs (iRBCs) that include common, uncommon, and rare Rh antigen phenotypes (Table 1). First, we performed ABO and RH genotyping to identify existing blood type O lines in our institutional PSC core laboratory with common Rh phenotypes consisting of (1) DCe/DCe (D-positive, Ce homozygous, ie, R1R1); (2) DcE/cE (D-positive, cE homozygous, ie, R2R2); and (3) ce/ce (D-negative, ce homozygous, ie, rr). If none were found for a specific Rh phenotype, PBMCs with these specific Rh phenotypes were reprogrammed into iPSCs. Second, we reprogrammed PBMCs from uncommon donors identified by RH genotyping to generate iPSCs that lack expression of the high-prevalence Rh antigens hrS or hrB or, conversely, express the low-prevalence Rh antigens V and VS. Third, we used gene editing to produce cells with rare Rh phenotypes: (1) Rh null, (2) D--, or (3) express low-prevalence Rh antigens Goa or DAK (Table 1).
Generation of Rh null, D--, Goa, and DAK iPSCs
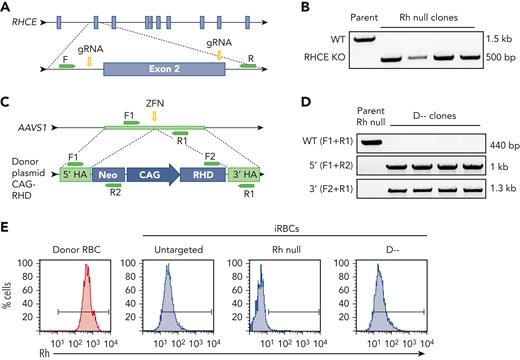
To generate the Rh null iPSCs, we used the iPSC line determined by genotyping to be Group O, RHCE∗ce and lacking the RHD gene (RhD negative). CRISPR-Cas9 was used to delete a 1-kb region surrounding exon 2 of RHCE that resulted in a premature stop codon (Figure 1A). iPSC clones with homozygous RHCE disruption were identified by a 500- bp PCR product compared with a 1500-bp product in untargeted alleles (Figure 1B). Designing the D-- iPSC line by targeting RHCE in RhD-positive iPSCs was precluded by the high homology of RHD and RHCE and lack of gRNAs that would inactivate RHCE but avoid RHD. As an alternative strategy, a construct containing the constitutive chicken actin (CAG) promoter driving RHD cDNA expression was inserted into the AAVS1 safe harbor locus (intron 1 of PPP1R12C) of the Rh null iPSC line by homologous recombination using a ZFN (Figure 1C).19 A combination of PCRs at the 5′ and 3′ sides of insertion confirmed appropriate integration and the absence of a 440-bp PCR product of the PPP1R12C gene identified clones with homozygous insertion (Figure 1C-D).
Generation of Rh null and D-- iPSCs. (A) Schematic for CRISPR-Cas9–mediated disruption of RHCE in RhD-negative iPSC to generate Rh null iPSCs. (B) PCR products amplified with primers targeting intron 1 and intron 2 identifies iPSC clones with Cas9-mediated large deletion of ∼1 kb. (C) Schematic for ZFN-mediated insertion of RHD cDNA into the safe harbor AAVS1 locus of Rh null iPSCs to generate D-- iPSCs. (D) PCR products amplified using primers indicated in panel C to identify clones with successful integration of the RHD cDNA cassette. (E) Cell surface Rh protein visualized by flow cytometry using a pan-Rh antibody in control donor RBCs (red), and untargeted parent, Rh null, and D-- iRBCs (blue). CAG, CAG promoter; F, forward primer; gRNA, guide RNA; HA, homology arm; KO, knockout; Neo, neomycin resistance cassette; R, reverse primer.
Generation of Rh null and D-- iPSCs. (A) Schematic for CRISPR-Cas9–mediated disruption of RHCE in RhD-negative iPSC to generate Rh null iPSCs. (B) PCR products amplified with primers targeting intron 1 and intron 2 identifies iPSC clones with Cas9-mediated large deletion of ∼1 kb. (C) Schematic for ZFN-mediated insertion of RHD cDNA into the safe harbor AAVS1 locus of Rh null iPSCs to generate D-- iPSCs. (D) PCR products amplified using primers indicated in panel C to identify clones with successful integration of the RHD cDNA cassette. (E) Cell surface Rh protein visualized by flow cytometry using a pan-Rh antibody in control donor RBCs (red), and untargeted parent, Rh null, and D-- iRBCs (blue). CAG, CAG promoter; F, forward primer; gRNA, guide RNA; HA, homology arm; KO, knockout; Neo, neomycin resistance cassette; R, reverse primer.
Following hematopoietic differentiation, Rh null iRBCs showed no cell surface Rh expression by flow cytometry using a pan-Rh antibody, whereas D-- iRBCs demonstrated Rh expression comparable to untargeted parental RhD-negative iRBCs that only express endogenous Rhce protein (Figure 1E). To generate iRBCs expressing the low-prevalence Goa and DAK antigens, Rh null iPSCs were targeted in the same fashion by inserting cDNA for RHD∗DIVa and ∗DIIIa, respectively, into the AAVS1 locus (supplemental Figure 2).
iRBC generation
To produce iRBCs from PSCs, we first generated HPCs (CD41+235+) via embryoid body or adherent differentiation (supplemental Figure 3A).20-22 HPCs released into the supernatant on day 8 were collected and further differentiated into iRBCs using a 12-day culture (Figure 2A). This time point was determined by Rh antigen expression and the size of iRBCs, which impacts its use with the gel card assay. For the first 6 days of erythroid differentiation, cells showed increasing expression of CD235, CD71, and Band3, but decreasing CD49d expression (supplemental Figure 3B). By day 12, 99% of cells were CD235+71low and CD49d−Band3+, approaching expression levels of adult donor-derived RBCs (dRBCs) that are CD235low71−49d−Band3+ (Figure 2B). By day 12, iRBCs reached the orthochromatic erythroblast stage (Figure 2C) but remained nucleated and larger than dRBCs. Using an automated cell analyzer, iRBCs had an average diameter of 9.678 μm (n = 205) compared with primary dRBCs with an average diameter of 6.982 μm (n = 1435, Figure 2D). iRBCs differentiated from cryopreserved HPCs were phenotypically identical to iRBCs generated from fresh HPCs based on cell surface antigen profiles, cell size, and maturation (supplemental Figure 4A-B). On average, day 12 erythroid cultures generated >60 iRBCs per fresh HPCs (n = 22; range, 37- to 117-fold, Figure 2E), whereas cryopreserved HPCs from the same iPSCs generated >35 iRBCs (n = 25; range, 24- to 55-fold, supplemental Figure 4C-D). Although iRBCs from different genetic backgrounds showed similar ability for RBC differentiation and maturation, the efficiency of HPC generation during EB differentiation and iRBC yield per HPC differed (supplemental Figure 5).
Generation of iRBCs. (A) Schematic of 12-day erythroid liquid culture of iPSC-derived HPCs into mature iRBCs. (B) Representative flow cytometric analysis of common cell surface erythroid maturation markers on day 12 of ce (rr) iRBC culture compared with control dRBCs. (C) Morphology of control dRBCs and day 12 ce iRBCs (May-Grunwald Giemsa stain). (D) Size distribution of day 12 ce iRBCs compared with control dRBCs measured by Countess Automated Cell Counter (n = 205 and 1435, respectively, Welch’s t test). (E) Fold expansion of iRBC per fresh HPC in erythroid-specific culture on days 3, 6, 9, and 12 (n = 22 independent assays). Ery, erythroid; IMDM, Iscove’s modified Dulbecco medium; pen strep, penicillin-streptomycin; Epo, erythropoietin, SCF, stem cell factor.
Generation of iRBCs. (A) Schematic of 12-day erythroid liquid culture of iPSC-derived HPCs into mature iRBCs. (B) Representative flow cytometric analysis of common cell surface erythroid maturation markers on day 12 of ce (rr) iRBC culture compared with control dRBCs. (C) Morphology of control dRBCs and day 12 ce iRBCs (May-Grunwald Giemsa stain). (D) Size distribution of day 12 ce iRBCs compared with control dRBCs measured by Countess Automated Cell Counter (n = 205 and 1435, respectively, Welch’s t test). (E) Fold expansion of iRBC per fresh HPC in erythroid-specific culture on days 3, 6, 9, and 12 (n = 22 independent assays). Ery, erythroid; IMDM, Iscove’s modified Dulbecco medium; pen strep, penicillin-streptomycin; Epo, erythropoietin, SCF, stem cell factor.
Rh typing of iRBCs by gel column agglutination
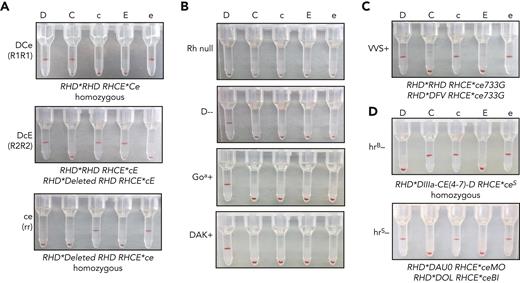
Gel column agglutination is a common method used to assess direct or indirect agglutination for RBC antigen phenotyping or for RBC antibody detection in plasma (supplemental Figure 6A). Agglutination is measured by the RBCs’ ability to pass through the gel matrix after centrifugation, from 4+ (strong agglutination that prevents RBCs from traveling to the bottom of the column after centrifugation) to 0 (no agglutination that allows cells to pellet to bottom of the column) (supplemental Figure 6B). To determine the feasibility of iRBCs as reagent RBCs, we immunophenotyped iRBCs with gel agglutination assays by incubating with monoclonal typing reagent antibodies. DCe/DCe (R1R1) esRBCs demonstrated strong agglutination (positive) with anti-D, anti-C, and anti-e, whereas no agglutination occurred with anti-c and anti-E as expected from the RH genotype (Table 1; Figure 3A), and similar to control dRBCs with the same Rh phenotype (supplemental Figure 6C). Rh typing of DcE/cE (R2R2) and ce/ce (rr) iRBCs also demonstrated agglutination patterns as expected by RH genotype. Rh null iRBCs showed no agglutination with antibodies against all 5 principal Rh antigens, whereas D--, Goa+, and DAK+ iRBCs were positive with anti-D and nonreactive with anti-C, anti-c, anti-E, and anti-e (Figure 3B). VVS+ iRBCs, derived from an iPSC line reprogrammed from an uncommon donor, were positive with anti-D, anti-c, and anti-e as expected (Figure 3C). Of note, hrB− and hrS− iRBCs, derived from iPSCs reprogrammed from rare donors whose RBCs are well-known to have decreased e antigen expression, showed minor or weak agglutination with anti-e (supplemental Figure 7A), but both were strongly positive after ficin treatment (Figure 3D, supplemental Figure 7B). Enzyme treatment of RBCs is used by clinical laboratories to enhance reactivity with Rh antibodies.
Detection of Rh antigen expression on iRBCs by gel column agglutination. (A) Rh antigen typing of DCe/DCe (R1R1), DcE/cE (R2R2), and ce/ce (rr) esRBCs or iRBCs using commercial monoclonal RBC typing reagents for the 5 common Rh antigens (anti-D, anti-C, anti-c, anti-E, and anti-e indicated above) on buffered gel cards. Agglutination prevents cells from traveling through the gel matrix to the bottom of each column upon centrifugation and indicates cell surface expression of the corresponding antigen. (B-D) (B) Rh typing for common Rh antigens of Rh null, D--, D+ Goa+, and D+ DAK+ iRBCs, and (C) D+c+e+, VVS+ iRBCs, and (D) C+c+e+, hrB− and D+c+e+, hrS− ficin-treated iRBCs.
Detection of Rh antigen expression on iRBCs by gel column agglutination. (A) Rh antigen typing of DCe/DCe (R1R1), DcE/cE (R2R2), and ce/ce (rr) esRBCs or iRBCs using commercial monoclonal RBC typing reagents for the 5 common Rh antigens (anti-D, anti-C, anti-c, anti-E, and anti-e indicated above) on buffered gel cards. Agglutination prevents cells from traveling through the gel matrix to the bottom of each column upon centrifugation and indicates cell surface expression of the corresponding antigen. (B-D) (B) Rh typing for common Rh antigens of Rh null, D--, D+ Goa+, and D+ DAK+ iRBCs, and (C) D+c+e+, VVS+ iRBCs, and (D) C+c+e+, hrB− and D+c+e+, hrS− ficin-treated iRBCs.
Detection of Rh antibodies in patient plasma
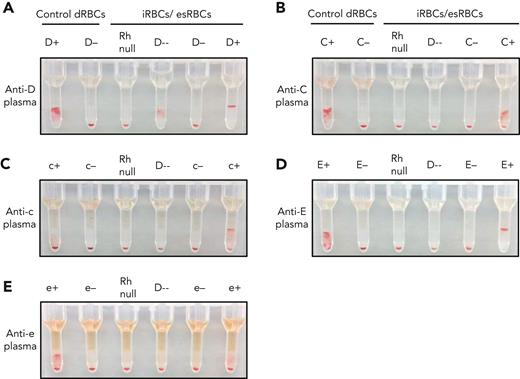
We tested the ability of iRBCs to detect Rh antibodies in patient plasma. The iRBCs were pretreated with ficin to enhance Rh reactivity (supplemental Figure 8). Untreated dRBCs and treated iRBCs were used as positive and negative controls. Using patient plasma with a previously detected anti-D, control D-positive and D-negative dRBCs showed 3+ agglutination and no agglutination, respectively (Figure 4A). The engineered Rh null iRBCs showed no agglutination and the D-- iRBCs showed 2+ agglutination, consistent with anti-D. D-negative and D-positive iRBCs showed the expected agglutination patterns. Patient plasma with antibodies against RhCE antigens, including anti-C, anti-c, anti-E, and anti-e, did not show reactivity with both the Rh null and D-- iRBCs, consistent with their known anti-CE specificity (Figure 4B-E). With each plasma sample, agglutination was observed only with iRBCs expressing the RhCE antigen to which the previously identified antibody was directed against. Plasma containing anti-S, anti-Fya, and anti-Jka did not react with ficin-treated iRBCs, and Rh reactivity was preserved (supplemental Figure 9C-G). Ficin-treated iRBCs did not agglutinate when incubated with plasma samples from patients with SCD who had no previously identified antibodies (supplemental Figure 10) or a plasma sample with paraproteins (supplemental Figure 11).
Detection of Rh antibodies in patient plasma using iRBCs and gel column agglutination. (A-E) Plasma containing (A) anti-D, (B) anti-C, (C) anti-c, (D) anti-E, and (E) anti-e were tested against a panel of control dRBCs and ficin-treated iRBCs or esRBCs. Each assay included control dRBCs expressing or lacking the corresponding Rh antigen (left) along with Rh null and D-- iRBCs, and iRBCs/esRBCs negative or positive for the corresponding Rh antigen (right). Agglutination prevents cells from traveling through the gel matrix to the bottom of each column upon centrifugation and indicates the presence of antibody in the plasma sample.
Detection of Rh antibodies in patient plasma using iRBCs and gel column agglutination. (A-E) Plasma containing (A) anti-D, (B) anti-C, (C) anti-c, (D) anti-E, and (E) anti-e were tested against a panel of control dRBCs and ficin-treated iRBCs or esRBCs. Each assay included control dRBCs expressing or lacking the corresponding Rh antigen (left) along with Rh null and D-- iRBCs, and iRBCs/esRBCs negative or positive for the corresponding Rh antigen (right). Agglutination prevents cells from traveling through the gel matrix to the bottom of each column upon centrifugation and indicates the presence of antibody in the plasma sample.
Identification of antibodies against high-prevalence Rh antigens is often a challenge because of limited availability of RH-genotyped dRBCs that lack these antigens, such as hrS− or hrB−. Antibodies against hrS and -hrB can be difficult to distinguish from anti-e in hospital laboratories. Using patient plasma with a previously identified anti-hrS, control DCe (R1R1) and ce (rr) dRBCs that are hrS+ showed 3+ agglutination, whereas control DcE (R2R2) dRBCs showed no agglutination because these cells lack hrS (Figure 5A). This pattern may be suggestive of anti-e present in the plasma. Rh null, and D-- iRBCs showed no agglutination with plasma containing anti-hrS, narrowing the specificity to the Rh system and specifically to RhCE rather than to RhD. Further testing with the rare iRBCs engineered to lack high-prevalence antigens with the e+ hrS− hrB+ phenotype or e+ hrS+ hrB− phenotype identifies the hrS specificity determined by the absence of agglutination and strong agglutination, respectively, indicating that anti-e has broader specificity (Figure 5A).
iRBCs identify antibodies against high-prevalence or low-prevalence Rh antigens in patient plasma. (A-B) Patient plasma containing antibody against (A) the high-prevalence antigen hrS or (B) low-prevalence antigens V or Goa were tested against a panel of control dRBCs and ficin-treated iRBCs. Each assay included the 3 control dRBCs routinely used for antibody screening: DCe (R1R1), DcE (R2R2), and ce (rr) phenotypes (left). In panel A, iRBCs that were Rh null, D--, e+ hrS− hrB+, and e+ hrS+ hrB− were selected to show hrS specificity (right). In panel B, patient plasma was tested against iRBCs that were Rh null, D--, or expressing low-prevalence Goa, DAK, or VVS antigens. Agglutination of cells prevents them from traveling through the gel matrix to the bottom of each column upon centrifugation and indicates the presence of antibody in the plasma sample.
iRBCs identify antibodies against high-prevalence or low-prevalence Rh antigens in patient plasma. (A-B) Patient plasma containing antibody against (A) the high-prevalence antigen hrS or (B) low-prevalence antigens V or Goa were tested against a panel of control dRBCs and ficin-treated iRBCs. Each assay included the 3 control dRBCs routinely used for antibody screening: DCe (R1R1), DcE (R2R2), and ce (rr) phenotypes (left). In panel A, iRBCs that were Rh null, D--, e+ hrS− hrB+, and e+ hrS+ hrB− were selected to show hrS specificity (right). In panel B, patient plasma was tested against iRBCs that were Rh null, D--, or expressing low-prevalence Goa, DAK, or VVS antigens. Agglutination of cells prevents them from traveling through the gel matrix to the bottom of each column upon centrifugation and indicates the presence of antibody in the plasma sample.
Antibodies against low-prevalence antigens are often missed on the antibody detection test because most commercial reagent dRBCs do not include these antigens. When dRBCs were incubated with patient plasma containing anti-V or anti-Goa, no reactivity was detected with DCe (R1R1), DcE (R2R2), or ce (rr) dRBCs that express only the common Rh antigens (Figure 5B). However, the VVS+ iRBCs showed 2+ agglutination with patient plasma containing anti-V, whereas there was no reactivity with Rh null, D--, Goa+, and DAK+ iRBCs. Similarly, Goa+ iRBCs specifically showed strong (4+) agglutination when incubated with patient plasma containing a previously identified anti-Goa, whereas Rh null, D--, DAK+, and VVS+ iRBCs showed no agglutination.
Functional characteristics of iRBCs
Individuals with Rh null phenotypes caused by inactivating mutations in RHAG, which is involved in cation transport and facilitates cell surface Rh expression, have mild hemolytic anemia and RBCs with stomatocyte morphology.23,24 The transporter function of RhAG motivated the generation of Rh null iPSCs here by disrupting the RHCE alleles rather than RHAG as by others.25,26 Rh null RBCs are invaluable as reagents and as a universal cell for transfusion. iRBCs, including the Rh null phenotype, were assessed for their globin chain expression, ability to bind oxygen, and deformability.
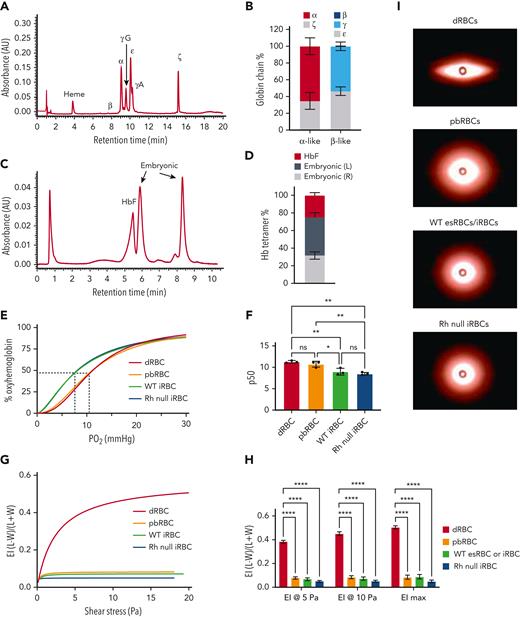
Globin chain analysis by high-performance liquid chromatography (HPLC) demonstrated that iRBCs most closely resemble RBCs that arise from the yolk sac, an early developmental stage. iRBCs expressed ∼35% ζ- and 49% ε-embryonic globin, 65% α- and 51% γ-fetal globin, and no adult-type β-globin (Figure 6A-B). Hemoglobin tetramer analysis showed 25% fetal (HbF) and 75% embryonic hemoglobin, correlating to 2 different tetramers, but presumed to be of embryonic origin (Hb Gower-1 [ζ2ε2] and Hb Goiwer-2 [α2ε2, Figure 6C-D]).27 Consistent with their increased embryonic and fetal globin chain content, untargeted (WT) iRBCs show stronger O2 binding affinity compared with adult noncultured dRBCs and adult CD34+ PBMC-derived cultured RBCs (pbRBCs) (Figure 6E-F, n = 3 per cell type). Rh null iRBCs showed comparable O2 binding affinity with WT iRBCs (Figure 6E-F).
Rh null iRBCs have comparable O2 binding capacity and deformability compared with WT iRBCs. (A) Representative and (B) quantification of alpha-like and beta-like globin chain analysis of day 12 iRBCs by HPLC (n = 5 independent assays). (C) Representative and (D) quantification of hemoglobin tetramer analysis of day 12 iRBCs by HPLC. (n = 5). (E) O2 dissociation curve of adult dRBCs, PB-RBCs, WT and Rh null iRBCs. (F) The partial pressure of O2 at 50% oxyhemoglobin (p50) interpolated from Sigmoidal 4PL curves of each type of RBC (n = 3 each, one-way ANOVA), ∗P < .05; ∗∗P < .01. (G) Average elongation indices of dRBCs, PB-RBCs, and WT or Rh null iRBCs determined by Lineweaver-Burk fitting for ektacytometry (n = 6 dRBCs, n = 3 for all others). (H) Elongation indices at 5 Pa, 10 Pa, and maximum elongation (n = 6 dRBCs, n = 3 for all others, two-way ANOVA), ∗∗∗∗P < .0001. (I) Representative images of fully “stretched” dRBCs, pbRBCs, and iRBCs obtained on the RheoScan AnD system. ANOVA, analysis of variance; HbF, fetal hemoglobin; O2, oxygen; Pa, Pascal; PB-RBCs, peripheral blood CD34+ cultured RBCs; WT, wild type.
Rh null iRBCs have comparable O2 binding capacity and deformability compared with WT iRBCs. (A) Representative and (B) quantification of alpha-like and beta-like globin chain analysis of day 12 iRBCs by HPLC (n = 5 independent assays). (C) Representative and (D) quantification of hemoglobin tetramer analysis of day 12 iRBCs by HPLC. (n = 5). (E) O2 dissociation curve of adult dRBCs, PB-RBCs, WT and Rh null iRBCs. (F) The partial pressure of O2 at 50% oxyhemoglobin (p50) interpolated from Sigmoidal 4PL curves of each type of RBC (n = 3 each, one-way ANOVA), ∗P < .05; ∗∗P < .01. (G) Average elongation indices of dRBCs, PB-RBCs, and WT or Rh null iRBCs determined by Lineweaver-Burk fitting for ektacytometry (n = 6 dRBCs, n = 3 for all others). (H) Elongation indices at 5 Pa, 10 Pa, and maximum elongation (n = 6 dRBCs, n = 3 for all others, two-way ANOVA), ∗∗∗∗P < .0001. (I) Representative images of fully “stretched” dRBCs, pbRBCs, and iRBCs obtained on the RheoScan AnD system. ANOVA, analysis of variance; HbF, fetal hemoglobin; O2, oxygen; Pa, Pascal; PB-RBCs, peripheral blood CD34+ cultured RBCs; WT, wild type.
Deformability of Rh null iRBCs was tested via ektacytometry. Compared with fully enucleated primary dRBCs, the average elongation indices of all in vitro–derived cells including adult cultured CD34+ pbRBCs, esRBCs, and iRBCs were significantly lower (Figure 6G-H), Lineweaver-Burk model, n = 3 each, one-way ANOVA). Rigidity of in vitro–derived RBCs can also be seen in the representative images at maximum shear stress (Figure 6I). Although Rh null iRBCs trended toward greater rigidity than WT iRBCs, the difference in the elongation indices was not significant, suggesting that their deformability is not due to loss of Rh antigens by RHCE disruption.
Discussion
Safe transfusion requires the patient’s plasma be tested for antibodies that could cause destruction of transfused cells and a hemolytic transfusion reaction. In vitro–generated RBCs represent an alternative renewable cell source for transfusion medicine antibody investigations.25,26,28 In this study, we generated a panel of customized iPSC lines with rare and uncommon RBC antigen phenotypes by combining CRISPR-Cas9 and ZFN gene editing technologies with reprogramming of uncommon donor cells. These specialized iRBCs can detect Rh specificities in patient plasma and have the potential to improve transfusion safety by facilitating complex antibody evaluations that are currently limited by the lack of appropriate reagent RBCs.29
To produce Rh null RBCs, prior work has focused on targeting RHAG,25,26 the gene encoding the Rh-associated glycoprotein, essential for stable surface expression of Rh and in which mutations result in Rh null phenotypes (regulator type).23 Our alternative approach to disrupt the RH locus circumvents the abnormal RBC morphology, notably stomatocytosis, and the hemolytic anemia associated with Rhag deficiency.24 Moreover, by targeting the AAVS1 safe harbor locus in the Rh null background, we were able to generate iRBCs that can express any Rh antigen.19 The D--, Goa+, and DAK+ iRBCs not only express conventional or variant RhD, but they also lack RhCE expression, which simplifies antibody evaluation. These RBC phenotypes are exceedingly rare, and generation of iPSCs with these Rh phenotypes via recruitment of rare donors would be near impossible. Moreover, preservation of RhAG in our Rh null iPSCs allows for reintroduction of any variant RH cDNA or any RhD-RhCE combination via heterozygous targeting of the AAVS1 loci. This approach is much more practical for generating iPSCs with RH variants than homology dependent recombination, which requires introduction of multiple single nucleotide polymorphisms across the genes.30
iRBCs detected antibodies against conventional Rh antigens (D, C, c, E, e), as well as those against high- and low-prevalence Rh antigens (hrS, Goa, V/VS), which are difficult but important to identify because they may lead to delayed hemolytic transfusion reactions.8,16 These rare donor-derived cells are limited to reference laboratories and not readily available in hospitals. In a study of Ghanian patients with SCD transfused with African donor units, up to one-third of antibodies were missed by the standard RBC antibody detection method, including against DAK, VVS, and Goa.29 In addition to detecting antibodies to antigens not represented on standard reagent RBCs, we propose that testing with Rh null and D-- iRBCs could expedite antibody identification for patients with SCD with a new positive antibody test or whose plasma reacts with all common cells tested and narrow the evaluation immediately. Use of iRBCs with rare or uncommon Rh phenotypes such as the e+hr +hrS− and e+hrB−hrS+ iRBCs could then aid identification of precise antibody specificity. In cases of a negative antibody test but a positive crossmatch, customized iRBCs could be used to test for antibodies against low-prevalence antigens such as V/VS, Goa, or DAK. The RH genotype of reagent iRBCs would be known, whereas the RH genotype of donor-derived reagents cells are not typically known in hospital laboratories.31,32 The ability to determine the fine specificity of antibodies would improve transfusion support for patients with SCD, especially when combined with donor matching based on the RH genotype.5,8,33
Because iRBCs recapitulate primitive erythroblasts, one limitation is that they do not express some RBC antigens in other blood group systems, including Duffy, Kidd, and Lutheran. We performed ficin treatment of iRBCs to enhance Rh reactivity, which also removed MNS antigen expression, allowing the focus of iRBC applications to be on the Rh and Kell blood group systems (supplemental Figure 9).34 Once the iPSC field develops differentiation protocols to efficiently produce developmentally definitive iRBCs (fetal liver or adult-like stages), their use can potentially be expanded to other blood group systems.
Immortalized human erythroblast lines are another potential source of renewable reagent RBCs, and combinatorial CRISPR editing has been used to generate cells lacking multiple blood group proteins.25,28,35 Immortalized erythroid lines have considerable potential for manufacture of novel RBC products, but impaired maturation capacity of these cells requires leukofiltration before their use with gel column agglutination technology,36 which substantially dampens the final cell yield. The use of viral oncoproteins to create these erythroblast lines is not a concern for reagent cells, and ongoing work is focused on producing an enucleated product for transfusion either with improved filter systems or with methods to enhance terminal erythroid differentiation.36
Banking of iPSCs has been proposed for generation of rare iRBCs for transfusion,37,38 or universal in vitro–generated RBCs with knockout of multiple blood group genes.25 The type O, Rh null, K− iRBCs we created would be an ideal phenotype for in vitro–derived universal donor RBCs. iPSCs provide a potentially unlimited progenitor source, but because 1 unit of blood contains ∼2 × 1012 RBCs,39 substantial hurdles must be overcome to reach clinical scale. The use of bioreactors,40-42 immortalized erythroblasts generated from rare iPSCs,28,35,36,43 or iRBCs mimicking definitive waves of hematopoiesis may allow improved proliferative capacity and/or enucleation efficiency to reach that goal.44,45 Additional protocols that support iRBC maturation and enucleation by coculture with murine stromal cells or longer culture duration may improve cell product quality and yield, but require prolonged cultures (42-120 days).46-51
Identifying the optimal storage conditions for readily accessible iRBCs is a remaining challenge to reach clinical application. Because the Rh null and D-- iRBCs would be useful for any patient with a new antibody detected on pretransfusion screening, storage at 4°C would be ideal for immediate use.52-54 Standard storage solutions including Alsever’s, MTS diluent, and additive solution 1 did not support storage of iRBCs. For customized iRBCs expressing variant Rh that would not be used frequently, possibilities include cryopreservation using glycerol and thawed as needed,53,55-57 or desiccation and rehydration as needed.58 Paraformaldehyde or glutaraldehyde fixation with or without desiccation has also been proposed as a way to preserve cells while maintaining adequate antigen expression.59,60
Our study harnessed iPSC technology with gene editing to create a specialized panel of iRBCs with rare antigen phenotypes to facilitate complex Rh antibody identification. These renewable RBC reagents provide a valuable diagnostic tool for use as an adjunct to current antibody identification panels, particularly for patients with SCD. For those who require immediate transfusion, the ability to expeditiously determine antibody specificity could significantly improve transfusion safety. In the future, these cells could offer universal transfusion products in place of rare blood, off-the-shelf RBCs to serve as drug carriers that increase targeted delivery61 or immunomodulatory vehicles expressing specific antigens to induce tolerance in autoimmune diseases.62
Acknowledgments
The authors thank Xiuli An of the New York Blood Center for providing the Band 3 antibody for flow cytometry, and members of the Human Pluripotent Stem Cell Core Laboratory at the Children’s Hospital of Philadelphia and the Immunogenomics Laboratory at the New York Blood Center for their expertise.
This work was supported by the National Institutes of Health, National Heart, Lung, and Blood Institute U01 HL134696 (S.T.C., P.G., D.L.F., C.M.W.), R01 HL147879 to 01 (S.T.C., C.M.W.), and a National Blood Foundation grant (S.T.C.).
Authorship
Contribution: S.T.C., C.M.W., P.G., and D.L.F. conceived the study, designed experiments, and analyzed results; H.H.A., A.L.G., J.A.M., G.P., and O.A. designed experiments, performed experiments, and analyzed results; C.M.W. provided critical material; H.H.A. and S.T.C. wrote the manuscript; and A.L.G., G.P., O.A., P.G., D.L.F. and C.M.W. read and edited the manuscript.
Conflict-of-interest disclosure: US Patent Application No. 16/757,815, entitled “Engineered Red Blood Cells Having Rare Antigen Phenotypes” by S.T.C. and C.M.W. in the name of The Children’s Hospital of Philadelphia and New York Blood Center, Inc (national stage entry of International Application No. PCT/US2018/057932); manuscript describes engineered red blood cells covered in patent application. The remaining authors declare no competing financial interests.
Correspondence: Stella T. Chou, Children’s Hospital of Philadelphia, 3615 Civic Center Blvd, Abramson Research Center Room 316D, Philadelphia, PA 19104; e-mail: chous@chop.edu.
References
Author notes
Requests for materials and methods should be sent to the corresponding author (chous@chop.edu).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal